Abstract
Cell-cell communication is a widespread phenomenon in nature, ranging from bacterial quorum sensing and fungal pheromone communication to cellular crosstalk in multicellular eukaryotes. These communication modes offer the possibility to control the behavior of an entire community by modifying the performance of individual cells in specific ways. Synthetic biology, i.e., the implementation of artificial functions within biological systems, is a promising approach towards the engineering of sophisticated, autonomous devices based on specifically functionalized cells. With the growing complexity of the functions performed by such systems, both the risk of circuit crosstalk and the metabolic burden resulting from the expression of numerous foreign genes are increasing. Therefore, systems based on a single type of cells are no longer feasible. Synthetic biology approaches with multiple subpopulations of specifically functionalized cells, wired by artificial cell-cell communication systems, provide an attractive and powerful alternative. Here we review recent applications of synthetic cell-cell communication systems with a specific focus on recent advances with fungal hosts.
Keywords: Cell-cell communication, Synthetic biology, Cellular consortia, Synthetic circuit engineering, Signal molecule pathways
Introduction
Cellular communication is a widespread phenomenon in nature, ranging from bacterial quorum sensing [1–4], communication of fungi by pheromones [5, 6] or quorum sensing molecules [7], interactions of microbes with their hosts [8, 9] or with each other [10] to cellular communication in multicellular eukaryotes [11]. These communication systems are either contact-based or rely on diffusible factors. Recently, a further communication mode acting via density pulses in lipid monolayers has been hypothesized [12].
Well characterized communication systems can be adopted into synthetic biology approaches to allow artificial cell-cell communication [13–16]. The design, the engineering and the implementation of artificial circuits and functions within biological systems are exciting fields of research that offer a broad range of applications [17–21]. Synthetic biological circuits have been implemented primarily in prokaryotic cells with Escherichia coli (E. coli) being the preferred host, but increasingly also in eukaryotic cells and in cellular consortia. A recent review covering a number of excellent studies in the emerging fields of biomedicine and tissue engineering explored the application of synthetic circuits in mammalian host cells [22]. In recent years, several synthetic biology tools – including synthetic riboswitches and ribozymes [23–25] as well as genome engineering tools [26] and a completely synthesized artificial designer chromosome [27] – have also been invented. However, within the fungal kingdom, these were almost exclusively restricted to yeast as a model organism. Despite the enormous importance of fungi in bioprocess engineering and the report of promising approaches for increased yields of biofuels and pharmaceuticals, synthetic biology tools for fungal cells are still in their infancy [28–30].
Bacterial communication systems based on quorum sensing utilize small diffusible molecules (referred to as autoinducers) that are released by bacterial cells and recognized via respective receptors. The extracellular concentration of autoinducers increases with increasing population density. Above a specific threshold, the autoinducer triggers a coordinated density-dependent response within the entire population. Quorum sensing systems have been reported to influence bacterial swarming, the secretion of exoenzymes, biofilm formation and genetic competence [1–4]. In several marine bacteria, bioluminescence is induced at high cell densities via a quorum sensing system, and numerous pathogenic bacteria utilize quorum sensing systems to control the expression of virulence genes [31–33]. Synthetic cellular communication in prokaryotic systems is mainly based on the engineering of bacterial quorum sensing systems [34–36].
Synthetic cellular communication in yeasts and fungi mostly focused on the mating pheromone systems of these organisms [5, 6]. Haploid cells of the commonly used baker’s yeast Saccharomyces cerevisiae (S. cerevisiae) exist in either of two opposite mating types, referred to as a and α. Both cell types secrete specific small diffusible peptides, acting as pheromones (termed a-factor and α-factor, respectively) and expose receptors for the respective pheromone of the opposite cell type at their surface. Pheromone communication between the haploid cells ultimately leads to zygote formation as part of the yeast’s sexual life cycle. Artificial communication systems in mammalian cells utilize amino acids [37, 38], second messengers [39], growth factors [40], contact-based signaling [41] or volatile compounds [42].
Here, we review recent approaches and applications as well as future challenges of synthetic genetic circuits utilizing artificial communication systems, especially highlighting the advances achieved with fungal host cells. We will focus on different fields of synthetic biology in which the implementation of cellular communication has proven to be beneficial or even mandatory.
Synthetic quorum sensing systems
In numerous studies, autoinducers have been applied to generate artificial cellular communication in order to synchronize the behavior of an entire population. Rewiring synthetic quorum sensing systems allows the control of target gene expression in a density-dependent manner (Fig. 1), which might be beneficial under certain conditions, e.g., when toxic proteins are produced or excessive protein production leads to a heavy metabolic burden. Such approaches can also circumvent the need for expensive inducer molecules to stimulate the expression of target genes in large-scale fermentations [43]. Rewiring of existing quorum sensing circuits provides the possibility to modify the cellular response to autoinducers (with the final response being graded, threshold-like or bistable [44]), hence synthetic quorum sensing systems may be used to fine-tune the response and to optimize the performance.
Fig. 1.
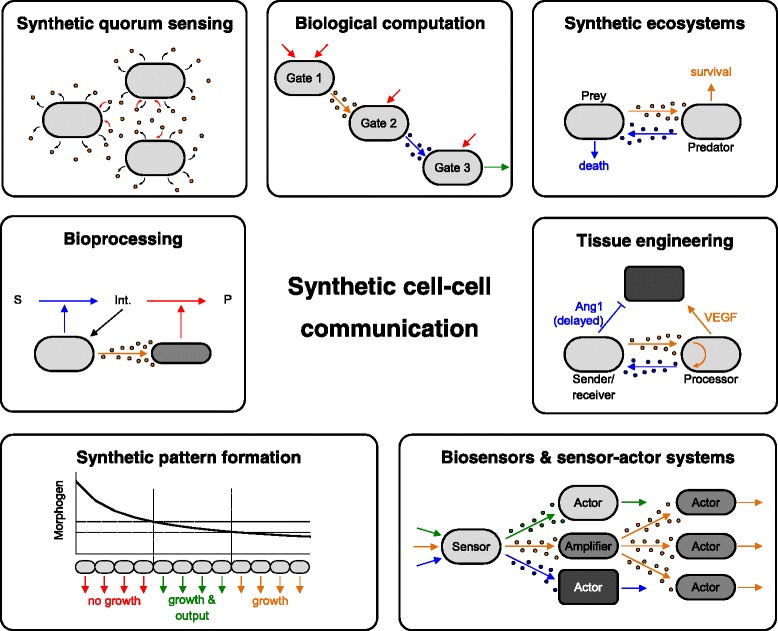
Applications of artificial cell-cell communication. Engineered cellular communication proved to be beneficial or even mandatory in numerous fields of application. These include the implementation of synthetic quorum sensing circuits, biological computation, the design of synthetic ecosystems, bioprocess engineering, biomedicine and tissue engineering as well as the formation of artificial patterns, biosensors and sensor-actor systems. Modified from [38, 78, 87, 133, 148]
Synthetic quorum sensing systems have been implemented in prokaryotic as well as in eukaryotic hosts. By triggering a synthetic quorum sensing circuit with acetate, a common intermediate in E. coli metabolism, Bulter et al. achieved the population-density dependent expression of a target gene [45]. Likewise, Chen and Weiss utilized artificial signaling elements to implement cellular communication and quorum sensing behavior in yeast [46]. Sender cells were engineered to secrete isopentenyladenine (IP), a signal molecule implicated in growth and development of Arabidopsis thaliana, whereas reporter cells expressed the IP receptor AtCRE1 and its downstream signaling elements. Activation of AtCRE1 in yeast led to the expression of GFP as an output gene from a synthetic promoter (a synthetic response element for SKN7, a nuclear aspartate response regulator of yeast activated by AtCRE1) solely in response to IP secreted by the sender cells. When cells were engineered to secrete IP and respond to it by increased IP synthesis (thus engineering a positive feedback loop), the output gene was expressed in a density-dependent manner. This circuit therefore closely resembled quorum sensing behavior.
Synthetic quorum sensing behavior can also be achieved by rewiring intrinsic intercellular communication systems. Recently, a tunable quorum sensing circuit was reported in yeast by utilizing the α-factor pheromone [43]. First, the pheromone response was coupled to increased pheromone synthesis and GFP expression leading to density-dependent pheromone secretion and fluorescence. Fine-tuning of the circuit’s performance was achieved by engineering of the promoter elements and the pheromone secretion rate. In a second approach, pheromone secretion - and in turn, quorum sensing behavior - was engineered to be inducible upon addition of aromatic amino acids [43]. The resulting circuit responds to the presence of these inducers and the population density, thus implementing a further control element for the desired design. As the yeast ARO9 promoter employed in this study shows specific response profiles to different aromatic amino acids, the overall circuit’s performance could be fine-tuned by choosing the type and concentration of the aromatic amino acid.
Synthetic quorum sensing behavior has also been implemented in mammalian cells using nitric oxide (NO) as an artificial quorum sensing molecule [39]. Human cells were engineered to synthesize NO, and a positive feedback loop triggered enhanced synthesis of NO upon detection of NO, thus creating a quorum sensing circuit. Fine-tuning was enabled by modifying the NO synthesis rate.
Biological computation
Biological computation, i.e., the ability of living matter to execute logic functions, has become an emerging issue in synthetic biology [47, 48]. Implementing logic gates within living cells enables them to respond to one or multiple trigger signals or environmental cues in a predefined and predictable manner (Fig. 1). Potential applications range from disease diagnosis, tissue engineering and cellular programming to bioprocessing and biosensing. Theoretically, cell populations or consortia performing logic functions may form autonomous systems that do not require human control, even under varying environmental conditions.
Computation of Boolean functions was achieved in clonal populations of specifically engineered prokaryotic and eukaryotic host cells. Synthetic Boolean gates have been successfully established in E. coli, ranging from synthetic AND gates [49–52] to more complex gates with multiple inputs [53] or analog functions [54]. Furthermore, synthetic circuits for counting events [55], push-on push-off switches [56] or oscillators [57], or the detection of the edges of illuminated patterns [58] have been designed. Similarly, logic operations can be performed by clonal populations of yeast [59, 60] or mammalian cells [61–65]. Recent approaches combined biological computation with synthetic memory, based on in vivo DNA recombination, thus achieving heritable computation as an exciting step towards the guidance of cellular differentiation in tissue engineering approaches [66, 67].
Performing logic operations in a single population often requires complex and multiple genetic elements to be engineered, which have to be transformed and tested extensively in the desired host. Besides being laborious, this approach may place a heavy metabolic burden on the cells. Distributed computation (logic performed by cellular consortia) might outcompete single-cell logic, especially if highly complex tasks have to be solved [68, 69]. Performing logic operations in microbial consortia allows the individual design and optimization of logic gates distributed throughout multiple subpopulations of the consortium. Physical separation of these gates ensures gate reusability without interference or crosstalk. Due to the reduction in the number of artificial genetic elements to be introduced into a single cell, the metabolic burden decreases, thus leading to enhanced genetic stability, reliability and long-term functionality. Furthermore, cellular consortia utilizing communication systems are able to calculate logic operations in a more robust manner, significantly suppressing misinterpretation of the output resulting from cellular noise [70, 71]. When combining cellular logic with quorum sensing, cells are enabled to calculate the desired function in a density-dependent manner [72–74].
Distributed computation requires the efficient communication of subpopulations in one or more directions. Typically, this is achieved by small diffusible factors (wiring molecules) that are secreted by one type and sensed by a second type of cells, thus allowing one-way communication. Metabolites [75] and bacterial autoinducers [58, 74, 76–78] as well as fungal pheromones [79] have been applied to generate cellular consortia calculating Boolean functions. In alternative approaches, signal transmission from one layer of cells to the next downstream layer was achieved by horizontal DNA transfer using conjugation [80] or bacteriophages [81]. The use of DNA stretches for artificial communication represents a promising strategy as it serves to increase the information capacity that can be forwarded to the receiver cells. Instead of just communicating via signal molecule concentrations and gradients, more complex messages can be passed on to the receivers [80, 81].
In E. coli [76, 77] and yeast cells [79] logic gates were implemented in individual populations, and two or more of these populations were wired by diffusible signals. The prokaryotic approaches were based on quorum sensing molecules, while in the eukaryotic system the α-factor pheromones of S. cerevisiae and Candida albicans were utilized. Employing two different wiring molecules and multiple specifically engineered subpopulations of cells allowed for the implementation of complex logic functions like multiplexers or 1-bit adders with carry in the yeast system. In-depth analysis and quantitative modelling of this approach revealed further potential for fine-tuning and optimization, based on alterations in cell density or by utilizing modified pheromone receptors with reduced affinity [82].
Given the increasing complexity of genetic programs and the need for more complex tasks to be solved, cellular consortia and the implemented communication systems between subpopulations become readily complex, typically associated with increasing noise affecting their reliability. Such systems require a high number of different subpopulations and wiring molecules that are independently secreted and sensed, without interfering with each other. Microfluidic platforms or spatial separation may provide options to use a single wiring molecule for more than one communication channel and thus to limit the required number of different communication systems [68]. Alternatively, distributed output production (i.e., multiple subpopulations are designed to synthesize a similar output in response to individual trigger signals) harbors great potential to efficiently reduce the number of wiring molecules and cellular populations required [69, 83].
Synthetic ecosystems
Natural ecosystems are intrinsically highly complex, and the vast majority of the interactions between different strains and species have not yet been explored in detail. Analyzing the interactions within such authentic ecosystems is very challenging as neither manipulation of the ecosystem nor in-depth analysis of multiple parameters can readily be achieved within the natural habitats. Synthetic ecosystems, preferably microbial communities, provide a valuable approach to overcome these limitations [84]. Several synthetic interactions between strains and species have been implemented, and some of them closely resemble naturally occurring ecosystems in specific parameters. Given the ease to manipulate synthetic ecosystems (either experimentally or via simulation) it is possible to dissect the major factors leading to a specific ecosystem behavior and to allow for conclusions to be drawn on naturally occurring communities. Although valuable insights were obtained by these synthetic ecologies, they still represent largely simplified communities, and there is still a massive gap to the enormous complexity of naturally occurring microbial ecosystems.
Synthetic circuits for tailored population control have been reported previously. Exploiting a quorum sensing system in E. coli by linking the response to the quorum sensing molecule with the expression of a killer gene allowed for a synthetic population control, i.e., predefined steady-state population densities were achieved [85, 86]. By implementing synthetic communication between two populations (prey and predator, Fig. 1), Balagaddé and coworkers established an E. coli consortium that exhibited population dynamics similar to authentic ecosystems [87]. Simulations have shown that this system might further be tuned by engineering autoinducer degradation or spatial separation [88, 89]. Rewired quorum sensing systems have also been utilized to control assembly and dispersal of artificial prokaryotic biofilms [90] or to implement obligate symbiosis within an E. coli consortium [91].
Simulations performed by Biliouris and coworkers indicated that synthetic obligate mutualism between E. coli and yeast might be engineered by utilizing quorum sensing systems [92]. In the respective design, E. coli cells secrete a first quorum sensing molecule, which is sensed by the yeast cells and triggers the expression of a resistance gene to detoxify an antibiotic compound. Similarly, yeast cells secrete a second quorum sensing molecule which induces the expression of a resistance gene in E. coli, thus leading to the degradation of a second antibiotic. Overall growth of the consortium is only promoted when both species are present in a sufficient density.
Obligate mutualism was also implemented in a synthetic yeast consortium [93]. Two auxotrophic strains, requiring adenine and lysine, respectively, were engineered to secrete high amounts of the supplement needed for growth of the respective other strain. The nutritional cross-feeding established in this study provided insights into mechanisms and population dynamics observed in naturally occurring cooperating communities. In a more recent study, this approach was extended to include a cheater strain requiring lysine secreted by the cooperator strain, but without paying the costs of secreting any common good [94]. Interestingly, some of the co-cultures were dominated by the cooperators, although the cheater strain initially had a higher relative fitness. In these cultures, the cooperators gained a fitness increase due to adaptation to the nutrient-limited environment (adaptive race), which consequently enabled them to dominate the cheaters. This approach provided valuable insights into the mechanisms driving the evolution and maintenance of cooperation in naturally occurring consortia.
Further insight into the population dynamics of cooperators and cheaters within an artificial ecosystem was gained with a yeast consortium in which histidine-requiring cooperator cells secreted invertase (an enzyme that hydrolyses sucrose extracellularly) to grow on sucrose as a single carbon source [95]. The cheater strains did not secrete invertase on their own, but benefitted from the sugars generated by the cooperators’ secreted invertase. Modifying the amount of supplemented histidine allowed for tuning of the costs of the cooperation. Thus, a tunable cooperation within an artificial ecosystem was created, which can contribute to our understanding of the mechanisms facilitating cooperation in natural environments.
Recently, the ability to tune the social behavior of yeast cells was impressively demonstrated [96]: in the experimental study, yeast cells were equipped to secrete and respond to the α-factor pheromone, and by additionally using “sense-only” strains, the authors were able to distinguish between self-communication (asocial behavior) and neighbor communication (social behavior). By tuning parameters such as the cell density, the pheromone secretion rate, expression of BAR1 (encoding a protease degrading the α-factor) and by implementing positive feedback circuits, the authors were able to achieve versatile ratios of both communication modes, some of which resembled phenomena observed in nature.
Synthetic ecosystems involving communication among different species, which rely on the utilization of volatile acetaldehyde as a common signaling molecule, have also been described [42]. Sender cells from various kingdoms, including prokaryotes, fungi and plants, were engineered to synthesize acetaldehyde that passively diffuses to mammalian receiver cells through the gas phase, thus facilitating artificial one-way communication. Due to the implementation of further interactions, the authors were able to generate synthetic inter-kingdom ecosystems with various behaviors, including commensalism, amensalism, mutualism, parasitism and predation.
Bioprocess engineering
The conversion of waste products into valuable materials, e.g., for the production of biofuels or biohydrogen, is an emerging field, aiming to increase the independence on fossil fuels. Of special interest is the processing of lignocellulose biomass, a major waste product from agriculture, to generate biofuels. To this end, it is desired to degrade lignocellulose-containing material into soluble sugars and to ferment these sugars to yield biofuels with minimal costs and process times. Exploiting the potential of microorganisms is one of the most promising approaches towards this goal. So far, neither naturally occurring nor genetically engineered organisms proved to perform this task sufficiently. Not only are the costs associated with single culture fermentations still too high to meet industrial demands, it also remains questionable whether such an ideal strain can be engineered at all given the high number of biochemical pathways that need to be tightly controlled for this purpose [30, 97, 98]. Metabolic engineering and the optimization of heterologous expression of numerous enzymes simultaneously might be very time-consuming, and the metabolic burden resulting from the expression of foreign genes may limit further strain optimization. Mixed culture fermentations, including specialists for cellulose degradation and specialists for soluble sugar fermentation, could be the better alternative [99]. Distributing the diverse tasks within a consortium significantly reduces the metabolic burden placed on each single cell type. Individual strains could be optimized in advance and finally integrated into the consortium. Consortia-based approaches allow biochemical reactions with multiple steps to be catalyzed. Compartmentalization of these reactions may reduce the risk of unwanted side-reactions [100]. In comparison to clonal cultures, microbial consortia show enhanced stability against environmental fluctuations or invasion by contaminating species, thus increasing the overall reliability of the process [101]. The use of microbial consortia is expected to enhance product yields by increasing the efficiency of substrate utilization and by removing or converting deleterious byproducts. This strategy may allow obtaining multiple valuable products from a single substrate source [102].
Naturally existing as well as synthetic consortia have been intensively studied in order to convert cellulose biomass into biofuels [99]. Recently, a yeast consortium was engineered for the conversion of cellulose into bioethanol [103, 104]. One strain displayed a scaffold protein at its cell surface, whereas three other strains were engineered to secrete one cellulolytic enzyme each. These enzymes included a specific protein domain which allowed their assembly at the scaffold protein, thus leading to an artificial mini-cellulosome displayed at the surface of yeast cells. By optimizing the ratio of the four strains involved, the authors achieved high ethanol titers (approx. 87 % of the theoretical value). The consortium approach was chosen to reduce the metabolic burden that prevented the engineering of a single strain for scaffold display and enzyme secretion. In a later study, the same authors generated even more complex cellulosomes including several anchor and adapter scaffolds to assemble more cellulolytic enzymes into the protein complex [105]. However, they utilized purified proteins for ex vivo assembly as a consortium approach would include numerous strains that have to be co-cultured in a fixed ratio over long time scales.
Synthetic consortia have also been applied to other fields of bioprocessing, including bioremediation, detoxification of byproducts of the chemical industry, food industry and the synthesis of chemicals [106, 107]. In contrast to the intrinsic robustness of naturally occurring microbial communities, synthetic consortia often suffer from a lack of long-term stability that may prevent some of these approaches from entering industrial processes. Several strategies have been proposed to generate stable microbial communities, including the engineering of symbiotic interactions among the consortium members, e.g., by cross-feeding of essential nutrients [93, 94, 108] or by removal of deleterious metabolic waste products [109]. Alternatively, external factors have to be applied, e.g., antibiotics [91, 92] or predefined oxygen tension [110], to maintain a synthetic consortium. Furthermore, spatial [111] or temporal [112, 113] separation as well as embedding of different species into biofilms [114] may provide tools to reach consortium stability. Alternative approaches to control consortia have been reported, including flipping of DNA elements [115], fitness engineering of individual strains [116], or the use of synthetic toggle switches [117–119] that enable cells to switch between two alternative states in response to a trigger signal. Recently, we showed that pheromone-based communication between two yeast species triggers a cell cycle arrest of the pheromone-responding strain, which might provide an option to control the composition of a synthetic consortium over long time scales (see below) [120].
Besides achieving a stable consortium composition, the behavior of individual strains within the microbial community needs to be tightly controlled to ensure maximum yield and reduced process time [99]. With an increasing number of different strains and species included in a synthetic consortium, this task is increasingly challenging. Engineering artificial cellular communication might provide a way to control individual subpopulations within the community to ensure that the sequential action of enzymes (each being expressed by a different strain or species) can be properly timed (Fig. 1). Adapted control of enzyme expression may serve to reduce the metabolic burden and to increase the overall process yield of consortium-based bioprocessing approaches.
In order to achieve an integrated community control by cellular communication, the response of the respective receiver strains to the trigger signals might require further tuning. Using yeast cells as a proof-of-principle, Bashor et al. demonstrated that the response to a pheromone can be modified [121]. Modification of the Mitogen Activated Protein Kinase (MAPK) signaling pathway via scaffold protein engineering allowed for direct coupling of positive or negative regulators to the cascade, thus leading to a modified sensitivity or timing of the cellular response (including accelerated, delayed or pulse-like pheromone response). This opens the way for further options to fine-tune concerted or subsequent events in microbial consortia in a sophisticated manner.
Biomedicine and tissue engineering
Synthetic biology devices and engineered cells have also been shown to be valuable tools in biomedicine. Engineered prokaryotic cells have been reported which specifically invade cancer cells [122] or detect NO released by inflamed gut tissue [123]. E. coli cells were designed to respond to quorum sensing molecules synthesized by Pseudomonas aeruginosa (P. aeruginosa), a harmful human pathogen [124–126]. In one of these approaches, sensing of the autoinducer induced the synthesis of pyocin S5 and subsequent self-lysis of E. coli. The released pyocin S5 acts as a killing agent for P. aeruginosa [124]. In another study, engineered E. coli cells secreted a novel pathogen-specific toxin upon detecting the autoinducer molecules of P. aeruginosa [125]. Furthermore, rewiring the chemotaxis response of the engineered E. coli cells enabled them to migrate along the autoinducer gradient towards the pathogen, thus exerting the antimicrobial and biofilm-degrading activities in the close vicinity of the pathogen [126]. These approaches proved to be highly efficient in inhibiting growth and biofilm formation of P. aeruginosa, making them attractive tools for future biomedicine applications.
Numerous pathogens utilize quorum sensing systems to initiate the expression of virulence genes or the formation of biofilms [1–4, 31–33]. Interference with the quorum sensing systems of pathogens, e.g., via quorum sensing inhibitors or local excess of quorum sensing molecules, could be a promising strategy to confuse pathogens [2, 3, 31–33, 127, 128]. This approach is particularly interesting because compounds affecting quorum sensing systems typically do not inhibit bacterial growth and thus do not create a high selective pressure. Therefore, the probability to develop resistance against such compounds is rather low. E. coli cells secreting quorum sensing molecules of the human pathogen Vibrio cholera were shown to significantly reduce the harm of the pathogen’s infection by interfering with the expression of its virulence genes [129].
Most of the potential applications in biomedicine so far rely on engineered E. coli cells which are part of the human gut microbiome. Future approaches may also utilize eukaryotic or artificial cells [130, 131] as sensing or actuating entity. Engineering artificial communication between the (bio)medical agent and eukaryotic host tissues has the potential to further improve the success of several therapies, especially for multi-factorial diseases [132].
Tissue development depends on precisely adjusted cellular responses, e.g., the division or differentiation of selected cells. This orchestration requires - in combination with appropriate scaffolds - numerous distinct signal molecules and physicochemical cues to be present at defined time points in a balanced concentration and position. Synthetic biology approaches might provide tools to achieve in vitro tissue engineering with the necessary precision and reliability. Recently, a synthetic two-way communication system with two output genes that are expressed with a different timing was applied to guide cellular differentiation in vitro [38]. Sender/receiver cells were engineered to synthesize and release tryptophan which was sensed by processor cells, leading to the expression of the Vascular Endothelial Growth Factor (VEGF) as a first output. The response of the processor cells to tryptophan was further coupled to the synthesis and release of acetaldehyde, which was sensed by the sender/receiver cell population and triggered the expression of angiopoietin-1 (Ang1) as a second output in a time-delayed manner. This approach allowed the timing of VEGF and Ang1 synthesis, respectively, eventually inducing transient permeability in vascular endothelial cells, a process that closely resembles blood vessel formation in vivo (Fig. 1).
Synthetic pattern formation
The ability of cells to form patterns is of crucial importance for the development of multicellular eukaryotes. However, the underlying mechanisms that drive these processes are not yet fully understood. The high number of signaling systems (both contact-based and diffusible signals, including formation and maintenance of their gradients), which are active in a developing tissue, is a major hurdle to gain further insights. A valuable approach to simulate these processes in a simplified manner is by engineering synthetic communities that form patterns on their own. The formation of predefined patterns by engineered cells or consortia can help to dissect the mechanisms leading to the development of tissues and to facilitate tissue engineering.
Synthetic patterns have been generated by clonal populations of E. coli in response to predefined gradients of antibiotics and inducers. Growth was only possible in a narrow range of concentrations, thus generating a bacterial band-pass filter [133]. Synthetic intercellular communication has also been applied to form predefined patterns of cell growth and reporter gene expression (Fig. 1). By modifying the cellular response to quorum sensing molecules, engineered E. coli receiver cells were able to produce a transient output depending on their distance from the autoinducer-releasing sender cells [134] or to build up a “bull’s eye” pattern around the sender cells [135]. Moreover, engineering of E. coli cells by the use of synthetic cellular communication resulted in artificial stripe- [136] and ring patterns [137].
Synthetic pattern formation has recently also been shown in a fungal system [138]. Yeast sender cells secreting the α-factor pheromone and responding receiver cells were immobilized in separate compartments within a hydrogel matrix. Diffusion of the low molecular weight peptide pheromone within the matrix enabled cell-cell communication. Expression of the red fluorescent protein (RFP) was used to visualize pheromone response of the receiver cells. RFP expression was dependent on the distance of receiver cells to the compartment boundary and the diffusion time. Tuning was achieved by modifying the density of sender cells and allowed to precisely adjust the area of pheromone responding cells. This approach might be extended to create more complex artificial patterns in hydrogel matrices, e.g., by controlled pheromone degradation or by employing different receiver cells with modified pheromone response profiles [121].
In mammalian cells, synthetic contact-based signaling has been used to propagate a signal along a closed layer of cells. This approach may be utilized for tissue engineering and/or synthetic pattern formation [41]. A mammalian system exploiting a diffusible factor (hepatocyte growth factor, HGF) has been described to yield a synthetic “bull’s eye” pattern in a 3D system [40]. In this study, single cysts of Madin-Darby Canine Kidney cells were locally transfected to secrete HGF (and concomitantly synthesize RFP) in the microenvironment. The response of the receiver cells was coupled to GFP expression, thus yielding a pattern of GFP expressing cells arranged around the HGF sender cells.
Biosensors and sensor-actor systems
Biological sensor devices are of great importance in numerous fields, including environmental monitoring and human healthcare. Whole-cell sensors, which utilize living cells as a biological recognition unit, represent an emerging branch within the field of biosensors. The sensor entity detects the presence of bioavailable analytes and translates it into a measurable output signal by utilizing analyte-induced or -repressed promoter elements [139, 140]. Besides bacterial whole-cell sensors, yeast-based systems were generated to detect copper ions [141], glucose [142], mycotoxins [143], estrogens [144], organic pollutants in sediments [145] or DNA damaging agents [146]. Devices based on a single sensor cell population might suffer from low signals and/or high background signals due to cellular noise. However, the use of cellular communication may overcome this limitation [70, 71].
In a microfluidic device, Prindle et al. synchronized 2.5 million prokaryotic cells in 500 entities (referred to as biopixels) in a sophisticated manner with a highly constant oscillatory period [147]. Oscillations within a biopixel were synchronized by an engineered quorum sensing system, whereas long-range synchronization of all biopixels was achieved by volatile hydrogen peroxide. By introducing arsenite-dependent regulatory elements in their design, the authors were able to generate a sensor device that tuned its oscillatory amplitude in response to trace amounts of arsenic, thus yielding a biosensor for arsenic contaminations.
Recently, a pheromone-based system for signal amplification in yeast relying on engineered cell-cell communication was reported [148]. Detection of the trigger signal by a population of sender cells was coupled to the secretion of the α-factor pheromone, which in turn induced GFP expression in a population of engineered receiver cells. This consortium approach allowed for signal amplification as compared to the respective device consisting of a single cell type. Further amplification could be achieved by inclusion of a third population of amplifier cells which responded to the presence of the α-factor by increased pheromone secretion, thus leading to higher pheromone concentrations (Fig. 1).
This approach has the potential to engineer autonomous sensor-actor systems that combine sensing of analytes with the expression of target proteins, which in turn result in the processing of the respective analytes. Such systems might be especially interesting for environmental monitoring by coupling the detection of a pollutant with its degradation via the formation of respective enzymes. Utilizing cellular communication would not only amplify the signal, but also reduce the metabolic burden for each cell type and prevent premature expression of the target protein due to cellular noise. Furthermore, by employing different communication molecules, this approach can pave the way to control multiple actor populations by a single sensor cell type, thus generating sensor-actor systems for numerous analytes (Fig. 1). Control of individual subpopulations of actor cells in such a design requires individual targeting, which could be achieved by the use of distinct signal molecules. This might also be a promising approach to include different species in these cellular consortia and to control their behavior individually by the use of proper signals.
Yeast cells have been successfully engineered to communicate via heterologous signaling components, e.g., by utilizing the IP-signaling of Arabidopsis thaliana (see above) [46]. Gonçalves-Sá and Murray used the pheromone-coding genes and receptors from Sordaria macrospora and Schizophyllum commune to engineer cellular communication in S. cerevisiae [149]. Upon replacement of the native pheromone and receptor genes by the heterologous genes, the yeast cells were able to mate when matching pairs of pheromones and receptors were expressed. Likewise, pheromones and pheromone receptors of different fungal species were previously expressed in yeast to provide insight into fungal pheromone systems [150–153]. Recently, we reported on an inter-species communication system based on the heterologous expression of pheromones [120]. By expressing native or chimeric pheromone-coding genes, S. cerevisiae cells were enabled to secrete a pheromone of the fission yeast Schizosaccharomyces pombe (S. pombe). Likewise, S. pombe cells were engineered to secrete the α-factor pheromone of S. cerevisiae, thus allowing a two-way communication between both yeast species using their respective pheromones (Fig. 2). Generating further inter-species communication modules, e.g., involving the pheromones of other fungal species, will provide a platform to individually control the behavior of multiple subpopulations in sensor-actor approaches or mixed-species consortia.
Fig. 2.
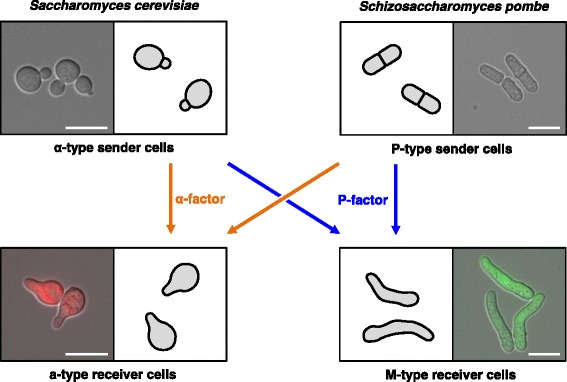
Synthetic inter-species communication in yeast. The yeast species S. cerevisiae and S. pombe have been designed to communicate artificially via the functional expression and secretion of different pheromones [120, 148]. S. cerevisiae cells were engineered to secrete the α-factor pheromone of S. cerevisiae or the P-factor pheromone of S. pombe, thus providing a possibility for artificial inter-species communication. Likewise, S. pombe cells were engineered to secrete either the P-factor or the α-factor pheromone. The pheromone response of the receiver cells of both species is linked to a cell cycle arrest in G1 phase, a characteristic change in morphology (shmoo effect) and to the expression of reporter genes controlled by pheromone-responsive promoters (e.g., the S. cerevisiae FIG1 promoter controlling the RFP reporter gene and S. pombe sxa2 promoter controlling the GFP reporter gene). Microscopic images were captured utilizing an Axio Observer Z1 (Carl Zeiss, Germany). Scale bars represent 10 μm
Conclusions
Synthetic biology approaches allow the implementation of numerous devices and systems, within the fields reviewed here and far beyond. With the increasing complexity of the designs and functions to be realized, engineering of the circuits in a single host cell is limited. The rapidly emerging consortium approaches may provide an attractive alternative. They could serve to overcome a number of limitations of synthetic biology tools that act on a single cell level, including intrinsic cellular noise, circuit crosstalk, metabolic burden and genetic instability. It is to be expected that tailored consortia become more and more important for industrial and medical purposes. However, maximum system robustness, product safety and yield can only be assured by a tight control of the systems’ performance.
Transferring designs from the laboratory scale to the conditions required within industrial or medical setups is challenging. The utilization of different strains and growth conditions may lead to deterioration or even failure of synthetic circuits at industrial scales [154], necessitating laborious optimization of the circuits’ components and cultivation conditions. Likewise, the functionality and specificity of synthetic devices for human healthcare need to be assured in order avoid side effects. Communication between the host tissue and the biomedical tool may be used to activate or shut down the implemented synthetic circuits.
Engineering of cellular consortia provides a promising and powerful tool to solve increasingly complex tasks requiring more and more distinct subpopulations to act in a predictable and reliable manner. To engineer cellular communication between the strains and/or species involved in such consortia, a palette of well characterized and standardized modules is indispensable. Although there is a wide spectrum of applications, the number of communication molecules utilized for their realization is still small. Thus, there is a need to integrate novel communication systems into host cells, with minimal crosstalk to their metabolism or to already existing natural and artificial communication systems [100, 155]. We expect new classes of molecules, derived from naturally occurring cell-cell communication systems, to be adopted into artificial cellular communication systems. Quorum sensing inhibitors [9], fungal quorum sensing molecules [7] and fungal pheromones [5] are attractive candidates to open new communication channels within microbial consortia. Evidently, there is a great need to identify the genes required for signal molecule synthesis, signal perception and transduction. Eventually, artificial communication systems may also function across the border of kingdoms, thus allowing for more versatile applications.
Acknowledgements
This work was supported by the Research Training Group “Nano- and Biotechnologies for the Packaging of Electronic Systems” (German Research Foundation, DFG 1401/2). We acknowledge support by the German Research Foundation and the Open Access Publication Funds of the TU Dresden.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SH, GR and KO planned the structure of the manuscript, designed the figures and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Stefan Hennig, Email: Stefan.Hennig1@tu-dresden.de.
Gerhard Rödel, Email: Gerhard.Roedel@tu-dresden.de.
Kai Ostermann, Email: Kai.Ostermann@tu-dresden.de.
References
- 1.Garg N, Manchanda G, Kumar A. Bacterial quorum sensing: circuits and applications. Antonie Van Leeuwenhoek. 2014;105:289–305. doi: 10.1007/s10482-013-0082-3. [DOI] [PubMed] [Google Scholar]
- 2.LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77:73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popat R, Cornforth DM, McNally L, Brown SP. Collective sensing and collective responses in quorum-sensing bacteria. J R Soc Interface. 2015;12:20140882. doi: 10.1098/rsif.2014.0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153:3923–38. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 5.Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merlini L, Dudin O, Martin SG. Mate and fuse: how yeast cells do it. Open Biol. 2013;3:130008. doi: 10.1098/rsob.130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albuquerque P, Casadevall A. Quorum sensing in fungi - a review. Med Mycol. 2012;50:337–45. doi: 10.3109/13693786.2011.652201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–20. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harder T, Campbell AH, Egan S, Steinberg PD. Chemical mediation of ternary interactions between marine holobionts and their environment as exemplified by the red alga Delisea pulchra. J Chem Ecol. 2012;38:442–50. doi: 10.1007/s10886-012-0119-5. [DOI] [PubMed] [Google Scholar]
- 10.Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr Opin Microbiol. 2009;12:205–14. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blagovic K, Gong ES, Milano DF, Natividad RJ, Asthagiri AR. Engineering cell-cell signaling. Curr Opin Biotechnol. 2013;24:940–7. doi: 10.1016/j.copbio.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griesbauer J, Bössinger S, Wixforth A, Schneider MF. Propagation of 2D pressure pulses in lipid monolayers and its possible implications for biology. Phys Rev Lett. 2012;108:198103. doi: 10.1103/PhysRevLett.108.198103. [DOI] [PubMed] [Google Scholar]
- 13.Bacchus W, Fussenegger M. Engineering of synthetic intercellular communication systems. Metab Eng. 2013;16:33–41. doi: 10.1016/j.ymben.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Chuang JS. Engineering multicellular traits in synthetic microbial populations. Curr Opin Chem Biol. 2012;16:370–8. doi: 10.1016/j.cbpa.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Song H, Ding MZ, Jia XQ, Ma Q, Yuan YJ. Synthetic microbial consortia: from systematic analysis to construction and applications. Chem Soc Rev. 2014;43:6954–81. doi: 10.1039/c4cs00114a. [DOI] [PubMed] [Google Scholar]
- 16.Song H, Payne S, Tan C, You L. Programming microbial population dynamics by engineered cell-cell communication. Biotechnol J. 2011;6:837–49. doi: 10.1002/biot.201100132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–79. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat Biotechnol. 2009;27:1139–50. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherji S, van Oudenaarden A. Synthetic biology: understanding biological design from synthetic circuits. Nat Rev Genet. 2009;10:859–71. doi: 10.1038/nrg2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rollié S, Mangold M, Sundmacher K. Designing biological systems: Systems engineering meets synthetic biology. Chem Eng Sci. 2012;69:1–29. [Google Scholar]
- 21.Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014;157:151–61. doi: 10.1016/j.cell.2014.02.039. [DOI] [PubMed] [Google Scholar]
- 22.Bacchus W, Aubel D, Fussenegger M. Biomedically relevant circuit-design strategies in mammalian synthetic biology. Mol Syst Biol. 2013;9:691. doi: 10.1038/msb.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berens C, Suess B. Riboswitch engineering - making the all-important second and third steps. Curr Opin Biotechnol. 2015;31:10–5. doi: 10.1016/j.copbio.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Groher F, Suess B. Synthetic riboswitches - A tool comes of age. Biochim Biophys Acta. 1839;2014:964–73. doi: 10.1016/j.bbagrm.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Jensen MK, Keasling JD. Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res. 2014; in press. [DOI] [PubMed]
- 26.David F, Siewers V. Advances in yeast genome engineering. FEMS Yeast Res. 2014; in press. [DOI] [PubMed]
- 27.Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–8. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blount BA, Weenink T, Ellis T. Construction of synthetic regulatory networks in yeast. FEBS Lett. 2012;586:2112–21. doi: 10.1016/j.febslet.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 29.Martínez EJ, Raghavan V, González-Andrés F, Gómez X. New biofuel alternatives: integrating waste management and single cell oil production. Int J Mol Sci. 2015;16:9385–405. doi: 10.3390/ijms16059385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Q, Singh A, Himmel ME. Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr Opin Biotechnol. 2009;20:364–71. doi: 10.1016/j.copbio.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 31.Helman Y, Chernin L. Silencing the mob: disrupting quorum sensing as a means to fight plant disease. Mol Plant Pathol. 2015;16:316–29. doi: 10.1111/mpp.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm A, Vikström E. Quorum sensing communication between bacteria and human cells: signals, targets, and functions. Front Plant Sci. 2014;5:309. doi: 10.3389/fpls.2014.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 34.Choudhary S, Schmidt-Dannert C. Applications of quorum sensing in biotechnology. Appl Microbiol Biotechnol. 2010;86:1267–79. doi: 10.1007/s00253-010-2521-7. [DOI] [PubMed] [Google Scholar]
- 35.Hooshangi S, Bentley WE. From unicellular properties to multicellular behavior: bacteria quorum sensing circuitry and applications. Curr Opin Biotechnol. 2008;19:550–5. doi: 10.1016/j.copbio.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell RJ, Lee SK, Kim T, Ghim CM. Microbial linguistics: perspectives and applications of microbial cell-to-cell communication. BMB Rep. 2011;44:1–10. doi: 10.5483/BMBRep.2011.44.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Weber W, Schuetz M, Dénervaud N, Fussenegger M. A synthetic metabolite-based mammalian inter-cell signaling system. Mol Biosyst. 2009;5:757–63. doi: 10.1039/b902070p. [DOI] [PubMed] [Google Scholar]
- 38.Bacchus W, Lang M, El-Baba MD, Weber W, Stelling J, Fussenegger M. Synthetic two-way communication between mammalian cells. Nat Biotechnol. 2012;30:991–6. doi: 10.1038/nbt.2351. [DOI] [PubMed] [Google Scholar]
- 39.Wang WD, Chen ZT, Kang BG, Li R. Construction of an artificial intercellular communication network using the nitric oxide signaling elements in mammalian cells. Exp Cell Res. 2008;314:699–706. doi: 10.1016/j.yexcr.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 40.Carvalho A, Menendez DB, Senthivel VR, Zimmermann T, Diambra L, Isalan M. Genetically encoded sender-receiver system in 3D mammalian cell culture. ACS Synth Biol. 2014;3:264–72. doi: 10.1021/sb400053b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuda M, Koga M, Nishida E, Ebisuya M. Synthetic signal propagation through direct cell-cell interaction. Sci Signal. 2012;5:ra31. doi: 10.1126/scisignal.2002764. [DOI] [PubMed] [Google Scholar]
- 42.Weber W, Daoud-El Baba M, Fussenegger M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proc Natl Acad Sci U S A. 2007;104:10435–40. doi: 10.1073/pnas.0701382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams TC, Nielsen LK, Vickers CE. Engineered quorum sensing using pheromone-mediated cell-to-cell communication in Saccharomyces cerevisiae. ACS Synth Biol. 2013;2:136–49. doi: 10.1021/sb300110b. [DOI] [PubMed] [Google Scholar]
- 44.Haseltine EL, Arnold FH. Implications of rewiring bacterial quorum sensing. Appl Environ Microbiol. 2008;74:437–45. doi: 10.1128/AEM.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulter T, Lee SG, Wong WW, Fung E, Connor MR, Liao JC. Design of artificial cell-cell communication using gene and metabolic networks. Proc Natl Acad Sci U S A. 2004;101:2299–304. doi: 10.1073/pnas.0306484101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen MT, Weiss R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat Biotechnol. 2005;23:1551–5. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- 47.Benenson Y. Biomolecular computing systems: principles, progress and potential. Nat Rev Genet. 2012;13:455–68. doi: 10.1038/nrg3197. [DOI] [PubMed] [Google Scholar]
- 48.Purcell O, Lu TK. Synthetic analog and digital circuits for cellular computation and memory. Curr Opin Biotechnol. 2014;29:146–55. doi: 10.1016/j.copbio.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldberg SD, Derr P, DeGrado WF, Goulian M. Engineered single- and multi-cell chemotaxis pathways in E. coli. Mol Syst Biol. 2009;5:283. doi: 10.1038/msb.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramalingam KI, Tomshine JR, Maynard JA, Kaznessis YN. Forward engineering of synthetic bio-logical AND gates. Biochem Eng J. 2009;47:38–47. [Google Scholar]
- 52.Sayut DJ, Niu Y, Sun L. Construction and enhancement of a minimal genetic AND logic gate. Appl Environ Microbiol. 2009;75:637–42. doi: 10.1128/AEM.01684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–53. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497:619–23. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- 55.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lou C, Liu X, Ni M, Huang Y, Huang Q, Huang L, et al. Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Mol Syst Biol. 2010;6:350. doi: 10.1038/msb.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danino T, Mondragón-Palomino O, Tsimring L, Hasty J. A synchronized quorum of genetic clocks. Nature. 2010;463:326–30. doi: 10.1038/nature08753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, et al. A synthetic genetic edge detection program. Cell. 2009;137:1272–81. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchisio MA. In silico design and in vivo implementation of yeast gene Boolean gates. J Biol Eng. 2014;8:6. doi: 10.1186/1754-1611-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–60. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ausländer S, Ausländer D, Müller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–7. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 62.Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol Bioeng. 2004;87:478–84. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 63.Lienert F, Torella JP, Chen JH, Norsworthy M, Richardson RR, Silver PA. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 2013;41:9967–75. doi: 10.1093/nar/gkt758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40:5180–7. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 66.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 67.Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31:448–52. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 68.Li B, You L. Synthetic biology: Division of logic labour. Nature. 2011;469:171–2. doi: 10.1038/469171a. [DOI] [PubMed] [Google Scholar]
- 69.Macía J, Posas F, Solé RV. Distributed computation: the new wave of synthetic biology devices. Trends Biotechnol. 2012;30:342–9. doi: 10.1016/j.tibtech.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 70.Koseska A, Zaikin A, Kurths J, García-Ojalvo J. Timing cellular decision making under noise via cell-cell communication. PLoS One. 2009;4:e4872. doi: 10.1371/journal.pone.0004872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanouchi Y, Tu D, Kim J, You L. Noise reduction by diffusional dissipation in a minimal quorum sensing motif. PLoS Comput Biol. 2008;4:e1000167. doi: 10.1371/journal.pcbi.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci U S A. 2007;104:17300–4. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, et al. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–9. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shong J, Collins CH. Quorum sensing-modulated AND-gate promoters control gene expression in response to a combination of endogenous and exogenous signals. ACS Synth Biol. 2014;3:238–46. doi: 10.1021/sb4000965. [DOI] [PubMed] [Google Scholar]
- 75.Silva-Rocha R, de Lorenzo V. Engineering multicellular logic in bacteria with metabolic wires. ACS Synth Biol. 2014;3:204–9. doi: 10.1021/sb400064y. [DOI] [PubMed] [Google Scholar]
- 76.Ji W, Shi H, Zhang H, Sun R, Xi J, Wen D, et al. A formalized design process for bacterial consortia that perform logic computing. PLoS One. 2013;8:e57482. doi: 10.1371/journal.pone.0057482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature. 2011;469:212–5. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang B, Barahona M, Buck M. A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens Bioelectron. 2013;40:368–76. doi: 10.1016/j.bios.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Regot S, Macia J, Conde N, Furukawa K, Kjellén J, Peeters T, et al. Distributed biological computation with multicellular engineered networks. Nature. 2011;469:207–11. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 80.Goñi-Moreno A, Amos M, de la Cruz F. Multicellular computing using conjugation for wiring. PLoS One. 2013;8:e65986. doi: 10.1371/journal.pone.0065986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ortiz ME, Endy D. Engineered cell-cell communication via DNA messaging. J Biol Eng. 2012;6:16. doi: 10.1186/1754-1611-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoffman-Sommer M, Supady A, Klipp E. Cell-to-cell communication circuits: quantitative analysis of synthetic logic gates. Front Physiol. 2012;3:287. doi: 10.3389/fphys.2012.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Macia J, Sole R. How to make a synthetic multicellular computer. PLoS One. 2014;9:e81248. doi: 10.1371/journal.pone.0081248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mee MT, Wang HH. Engineering ecosystems and synthetic ecologies. Mol Biosyst. 2012;8:2470–83. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z, Wu X, Peng J, Hu Y, Fang B, Huang S. Artificially constructed quorum-sensing circuits are used for subtle control of bacterial population density. PLoS One. 2014;9:e104578. doi: 10.1371/journal.pone.0104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.You L, Cox RS, 3rd, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–71. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 87.Balagaddé FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, et al. A synthetic Escherichia coli predator–prey ecosystem. Mol Syst Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dalchau N, Smith MJ, Martin S, Brown JR, Emmott S, Phillips A. Towards the rational design of synthetic cells with prescribed population dynamics. J R Soc Interface. 2012;9:2883–98. doi: 10.1098/rsif.2012.0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song H, Payne S, Gray M, You L. Spatiotemporal modulation of biodiversity in a synthetic chemical-mediated ecosystem. Nat Chem Biol. 2009;5:929–35. doi: 10.1038/nchembio.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hong SH, Hegde M, Kim J, Wang X, Jayaraman A, Wood TK. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device. Nat Commun. 2012;3:613. doi: 10.1038/ncomms1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hu B, Du J, Zou RY, Yuan YJ. An environment-sensitive synthetic microbial ecosystem. PLoS One. 2010;5:e10619. doi: 10.1371/journal.pone.0010619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Biliouris K, Babson D, Schmidt-Dannert C, Kaznessis YN. Stochastic simulations of a synthetic bacteria-yeast ecosystem. BMC Syst Biol. 2012;6:58. doi: 10.1186/1752-0509-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shou W, Ram S, Vilar JM. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci U S A. 2007;104:1877–82. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Waite AJ, Shou W. Constructing synthetic microbial communities to explore the ecology and evolution of symbiosis. Methods Mol Biol. 2014;1151:27–38. doi: 10.1007/978-1-4939-0554-6_2. [DOI] [PubMed] [Google Scholar]
- 95.Gore J, Youk H, van Oudenaarden A. Snowdrift game dynamics and facultative cheating in yeast. Nature. 2009;459:253–6. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Youk H, Lim WA. Secreting and sensing the same molecule allows cells to achieve versatile social behaviors. Science. 2014;343:1242782. doi: 10.1126/science.1242782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alper H, Stephanopoulos G. Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol. 2009;7:715–23. doi: 10.1038/nrmicro2186. [DOI] [PubMed] [Google Scholar]
- 98.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23:396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 99.Zuroff TR, Curtis WR. Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol. 2012;93:1423–35. doi: 10.1007/s00253-011-3762-9. [DOI] [PubMed] [Google Scholar]
- 100.Shong J, Jimenez Diaz MR, Collins CH. Towards synthetic microbial consortia for bioprocessing. Curr Opin Biotechnol. 2012;23:798–802. doi: 10.1016/j.copbio.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 101.Brenner K, You L, Arnold FH. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–9. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 102.Sabra W, Dietz D, Tjahjasari D, Zeng AP. Biosystems analysis and engineering of microbial consortia for industrial biotechnology. Eng Life Sci. 2010;10:407–21. [Google Scholar]
- 103.Goyal G, Tsai SL, Madan B, DaSilva NA, Chen W. Simultaneous cell growth and ethanol production from cellulose by an engineered yeast consortium displaying a functional mini-cellulosome. Microb Cell Fact. 2011;10:89. doi: 10.1186/1475-2859-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsai SL, Goyal G, Chen W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2010;76:7514–20. doi: 10.1128/AEM.01777-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsai SL, DaSilva NA, Chen W. Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth Biol. 2013;2:14–21. doi: 10.1021/sb300047u. [DOI] [PubMed] [Google Scholar]
- 106.Bader J, Mast-Gerlach E, Popović MK, Bajpai R, Stahl U. Relevance of microbial coculture fermentations in biotechnology. J Appl Microbiol. 2010;109:371–87. doi: 10.1111/j.1365-2672.2009.04659.x. [DOI] [PubMed] [Google Scholar]
- 107.Brune KD, Bayer TS. Engineering microbial consortia to enhance biomining and bioremediation. Front Microbiol. 2012;3:203. doi: 10.3389/fmicb.2012.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kerner A, Park J, Williams A, Lin XN. A programmable Escherichia coli consortium via tunable symbiosis. PLoS One. 2012;7:e34032. doi: 10.1371/journal.pone.0034032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bayer TS, Widmaier DM, Temme K, Mirsky EA, Santi DV, Voigt CA. Synthesis of methyl halides from biomass using engineered microbes. J Am Chem Soc. 2009;131:6508–15. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 110.Zuroff TR, Xiques SB, Curtis WR. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture. Biotechnol Biofuels. 2013;6:59. doi: 10.1186/1754-6834-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A. 2008;105:18188–93. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Park EY, Naruse K, Kato T. One-pot bioethanol production from cellulose by co-culture of Acremonium cellulolyticus and Saccharomyces cerevisiae. Biotechnol Biofuels. 2012;5:64. doi: 10.1186/1754-6834-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shin HD, McClendon S, Vo T, Chen RR. Escherichia coli binary culture engineered for direct fermentation of hemicellulose to a biofuel. Appl Environ Microbiol. 2010;76:8150–9. doi: 10.1128/AEM.00908-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li L, Yang C, Lan W, Xie S, Qiao C, Liu J. Removal of methyl parathion from artificial off-gas using a bioreactor containing a constructed microbial consortium. Environ Sci Technol. 2008;42:2136–41. doi: 10.1021/es702631x. [DOI] [PubMed] [Google Scholar]
- 115.Ramadas R, Thattai M. Flipping DNA to generate and regulate microbial consortia. Genetics. 2010;184:285–93. doi: 10.1534/genetics.109.105999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bayer TS, Hoff KG, Beisel CL, Lee JJ, Smolke CD. Synthetic control of a fitness tradeoff in yeast nitrogen metabolism. J Biol Eng. 2009;3:1. doi: 10.1186/1754-1611-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen S, Zhang H, Shi H, Ji W, Feng J, Gong Y, et al. Automated design of genetic toggle switches with predetermined bistability. ACS Synth Biol. 2012;1:284–90. doi: 10.1021/sb300027y. [DOI] [PubMed] [Google Scholar]
- 118.Hayat S, Ostermann K, Brusch L, Pompe W, Rödel G. Towards in vivo computing: quantitative analysis of an artificial gene regulatory network behaving as a RS flip-flop and simulating the system in silico. Proceedings of the 1st international conference on Bio inspired models of network, information and computing systems. doi: 10.1145/1315843.1315850.
- 119.Moon TS, Clarke EJ, Groban ES, Tamsir A, Clark RM, Eames M, et al. Construction of a genetic multiplexer to toggle between chemosensory pathways in Escherichia coli. J Mol Biol. 2011;406:215–27. doi: 10.1016/j.jmb.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hennig S, Clemens A, Rödel G, Ostermann K. A yeast pheromone-based inter-species communication system. Appl Microbiol Biotechnol. 2015;99:1299–308. doi: 10.1007/s00253-014-6133-5. [DOI] [PubMed] [Google Scholar]
- 121.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–43. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 122.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–27. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 123.Archer EJ, Robinson AB, Süel GM. Engineered E. coli that detect and respond to gut inflammation through nitric oxide sensing. ACS Synth Biol. 2012;1:451–7. doi: 10.1021/sb3000595. [DOI] [PubMed] [Google Scholar]
- 124.Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SS, et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gupta S, Bram EE, Weiss R. Genetically programmable pathogen sense and destroy. ACS Synth Biol. 2013;2:715–23. doi: 10.1021/sb4000417. [DOI] [PubMed] [Google Scholar]
- 126.Hwang IY, Tan MH, Koh E, Ho CL, Poh CL, Chang MW. Reprogramming microbes to be pathogen-seeking killers. ACS Synth Biol. 2014;3:228–37. doi: 10.1021/sb400077j. [DOI] [PubMed] [Google Scholar]
- 127.Masák J, Čejková A, Schreiberová O, Rezanka T. Pseudomonas biofilms: possibilities of their control. FEMS Microbiol Ecol. 2014;89:1–14. doi: 10.1111/1574-6941.12344. [DOI] [PubMed] [Google Scholar]
- 128.Wu H, Moser C, Wang HZ, Høiby N, Song ZJ. Strategies for combating bacterial biofilm infections. Int J Oral Sci. 2015;7:1–7. doi: 10.1038/ijos.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Duan F, March JC. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc Natl Acad Sci U S A. 2010;107:11260–4. doi: 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ding Y, Wu F, Tan C. Synthetic Biology: A Bridge between Artificial and Natural Cells. Life (Basel) 2014;4:1092–116. doi: 10.3390/life4041092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lentini R, Santero SP, Chizzolini F, Cecchi D, Fontana J, Marchioretto M, et al. Integrating artificial with natural cells to translate chemical messages that direct E. coli behaviour. Nat Commun. 2014;5:4012. doi: 10.1038/ncomms5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ye H, Aubel D, Fussenegger M. Synthetic mammalian gene circuits for biomedical applications. Curr Opin Chem Biol. 2013;17:910–7. doi: 10.1016/j.cbpa.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 133.Sohka T, Heins RA, Phelan RM, Greisler JM, Townsend CA, Ostermeier M. An externally tunable bacterial band-pass filter. Proc Natl Acad Sci U S A. 2009;106:10135–40. doi: 10.1073/pnas.0901246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. Spatiotemporal control of gene expression with pulse-generating networks. Proc Natl Acad Sci U S A. 2004;101:6355–60. doi: 10.1073/pnas.0307571101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–4. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 136.Liu C, Fu X, Liu L, Ren X, Chau CK, Li S, et al. Sequential establishment of stripe patterns in an expanding cell population. Science. 2011;334:238–41. doi: 10.1126/science.1209042. [DOI] [PubMed] [Google Scholar]
- 137.Payne S, Li B, Cao Y, Schaeffer D, Ryser MD, You L. Temporal control of self-organized pattern formation without morphogen gradients in bacteria. Mol Syst Biol. 2013;9:697. doi: 10.1038/msb.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jahn M, Mölle A, Rödel G, Ostermann K. Temporal and spatial properties of a yeast multi-cellular amplification system based on signal molecule diffusion. Sensors (Basel) 2013;13:14511–22. doi: 10.3390/s131114511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Baronian KH. The use of yeast and moulds as sensing elements in biosensors. Biosens Bioelectron. 2004;19:953–62. doi: 10.1016/j.bios.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 140.D’Souza SF. Microbial biosensors. Biosens Bioelectron. 2001;16:337–53. doi: 10.1016/s0956-5663(01)00125-7. [DOI] [PubMed] [Google Scholar]
- 141.Shetty RS, Deo SK, Liu Y, Daunert S. Fluorescence-based sensing system for copper using genetically engineered living yeast cells. Biotechnol Bioeng. 2004;88:664–70. doi: 10.1002/bit.20331. [DOI] [PubMed] [Google Scholar]
- 142.Thierfelder S, Ostermann K, Göbel A, Rödel G. Vectors for glucose-dependent protein expression in Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2011;163:954–64. doi: 10.1007/s12010-010-9099-5. [DOI] [PubMed] [Google Scholar]
- 143.Välimaa AL, Kivistö AT, Leskinen PI, Karp MT. A novel biosensor for the detection of zearalenone family mycotoxins in milk. J Microbiol Methods. 2010;80:44–8. doi: 10.1016/j.mimet.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 144.Baronian KH, Gurazada S. Electrochemical detection of wild type Saccharomyces cerevisiae responses to estrogens. Biosens Bioelectron. 2007;22:2493–9. doi: 10.1016/j.bios.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 145.Leskinen P, Hilscherova K, Sidlova T, Kiviranta H, Pessala P, Salo S, et al. Detecting AhR ligands in sediments using bioluminescent reporter yeast. Biosens Bioelectron. 2008;23:1850–5. doi: 10.1016/j.bios.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 146.Benton MG, Glasser NR, Palecek SP. The utilization of a Saccharomyces cerevisiae HUG1P-GFP promoter-reporter construct for the selective detection of DNA damage. Mutat Res. 2007;633:21–34. doi: 10.1016/j.mrgentox.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 147.Prindle A, Samayoa P, Razinkov I, Danino T, Tsimring LS, Hasty J. A sensing array of radically coupled genetic ‘biopixels’. Nature. 2011;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gross A, Rödel G, Ostermann K. Application of the yeast pheromone system for controlled cell-cell communication and signal amplification. Lett Appl Microbiol. 2011;52:521–6. doi: 10.1111/j.1472-765X.2011.03035.x. [DOI] [PubMed] [Google Scholar]
- 149.Gonçalves-Sá J, Murray A. Asymmetry in sexual pheromones is not required for ascomycete mating. Curr Biol. 2011;21:1337–46. doi: 10.1016/j.cub.2011.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Böhm J, Hoff B, O’Gorman CM, Wolfers S, Klix V, Binger D, et al. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc Natl Acad Sci U S A. 2013;110:1476–81. doi: 10.1073/pnas.1217943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fowler TJ, DeSimone SM, Mitton MF, Kurjan J, Raper CA. Multiple sex pheromones and receptors of a mushroom-producing fungus elicit mating in yeast. Mol Biol Cell. 1999;10:2559–72. doi: 10.1091/mbc.10.8.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mayrhofer S, Pöggeler S. Functional characterization of an alpha-factor-like Sordaria macrospora peptide pheromone and analysis of its interaction with its cognate receptor in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:661–72. doi: 10.1128/EC.4.4.661-672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Olesnicky NS, Brown AJ, Dowell SJ, Casselton LA. A constitutively active G-protein-coupled receptor causes mating self-compatibility in the mushroom Coprinus. EMBO J. 1999;18:2756–63. doi: 10.1093/emboj/18.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moser F, Broers NJ, Hartmans S, Tamsir A, Kerkman R, Roubos JA, et al. Genetic circuit performance under conditions relevant for industrial bioreactors. ACS Synth Biol. 2012;1:555–64. doi: 10.1021/sb3000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pai A, Tanouchi Y, Collins CH, You L. Engineering multicellular systems by cell-cell communication. Curr Opin Biotechnol. 2009;20:461–70. doi: 10.1016/j.copbio.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


