Abstract
Valvular heart disease is associated with significant morbidity and mortality and often the result of congenital malformations. However, the prevalence is increasing in adults not only because of the growing aging population, but also because of improvements in the medical and surgical care of children with congenital heart valve defects. The success of the Human Genome Project and major advances in genetic technologies, in combination with our increased understanding of heart valve development, has led to the discovery of numerous genetic contributors to heart valve disease. These have been uncovered using a variety of approaches including the examination of familial valve disease and genome-wide association studies to investigate sporadic cases. This review will discuss these findings and their implications in the treatment of valvular heart disease.
Keywords: Heart valve, Heart valve development, Genetics, Bicuspid aortic valve, Aortic valve stenosis, Mitral valve prolapse, Myxomatous valve disease, Pulmonic valve stenosis, Ebstein anomaly, Aortic valve calcification, Valvular heart disease
Introduction
Valvular heart disease (VHD), which encompasses both congenital and acquired forms, results in significant morbidity and mortality [1]. Over the past several decades, there has been a significant change in the subtypes of VHD with rheumatic heart disease becoming less common and calcific (or degenerative) disease being more prevalent in light of the increased lifespans in industrialized populations. The incidence of congenital valve defects has remained relatively constant and accounts for 10 % of all congenital heart defects (CHD) and found in over 50 % of CHD cases [1, 2]. Regardless of the etiology, therapies for diseased heart valves are limited and severe dysfunction ultimately requires surgical replacement with mechanical or bioprosthetic valves. The downside to this is the lack of durability of bioprosthetic valves that often need replacing, and the requirement of anticoagulation therapy for patients receiving mechanical valves [3]. Increased understanding of the mechanisms underlying cardiac valve development along with recent advances in sequencing of the human genome, have resulted in the identification of numerous genetic etiologies for congenital valvular malformations in humans. In addition, there is increasing evidence to suggest that adult-onset VHD has origins during embryonic development.
The emphasis of this review will be on the genetic contributors of congenital valvular malformations, and accordingly includes a brief overview of heart valve development. We have focused on the most common congenital anomalies of each of the 4 heart valves: bicuspid aortic valve, mitral valve prolapse, pulmonary valve stenosis, and Ebstein anomaly of the tricuspid valve. For each these lesions, there have been recent discoveries into their genetic basis and these etiologic human genetic mutations will be discussed. In addition, this review will include a discussion on the recent mutant mouse models that mimic human disease phenotypes, as these provide additional genetic links to disease onset. Finally, we will review recent studies investigating the genetic contributors to adult-onset VHD.
Development of the Cardiac Valves
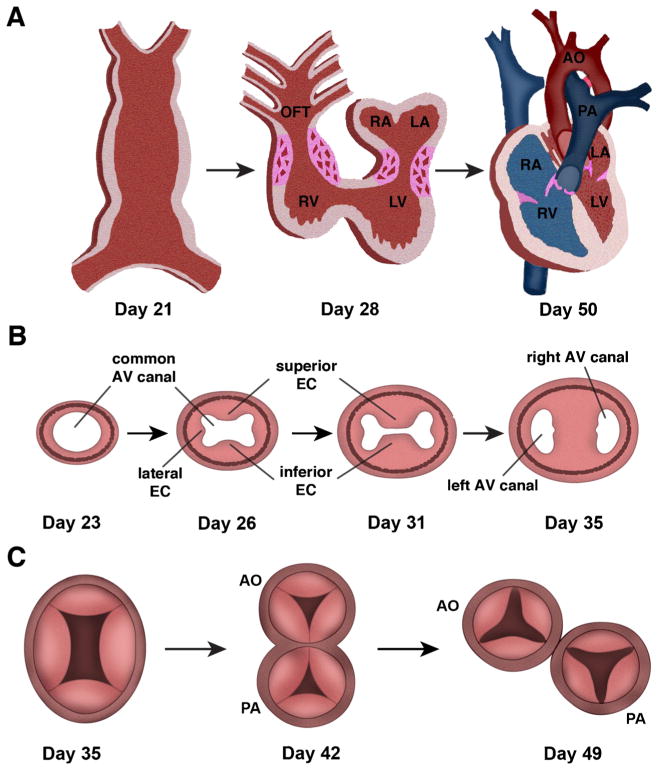
Heart valve development is a complex process that involves the interplay of several cell lineages and cellular process all tightly controlled by multiple genetic programs [4, 5]. Development of the atrioventricular (AV) valves precedes semilunar valve development but both begin with the formation of swellings, referred to as endocardial cushions, within the wall of the linear heart tube of the outflow tract (OFT) and AV regions. The AV cushions rise to the mature mitral and tricuspid valves, whereas the OFT cushions will go on to form the aortic and pulmonary valves (Fig. 1A).
Fig. 1.
Cardiac valve development. A, At day 21 of gestation, the linear heart tube is formed and the primitive heart completes rightward looping by day 28 of human gestation. Significant remodeling of the inner curvature and growth of the ventricular chambers then occurs to result in the maturely developed heart with partitioned systemic (red) and pulmonary circulations (blue) by day 50. Regions of atrioventricular and outflow tract (OFT) cushion formation and development are shown in pink. LA left atrium, LV left ventricle, RA right atrium, RV right ventricle. B, Growth of the superior, inferior, and lateral endocardial cushions in the common atrioventricular canal is shown. Days of human gestation are noted. By day 35, the superior and inferior cushions have fused to divide to result in 2 atrioventricular openings. AV atrioventricular, EC endocardial cushions. C, Transverse section through the common outflow tract (truncus arteriosus) with 4 truncal swellings at day 35 of gestation. At day 42 of gestation, the common outflow tract has started to divide into the aorta (AO) and pulmonary artery (PA) and leaflet formation has begun. By day 49, the aorta and pulmonary artery are septated and each has 3 leaflets. Adapted from: Garg V. “Growth of the Normal Human Heart” In: Preedy VR, editor. Handbook of Growth and Growth Monitoring in Health and Disease. New York: Springer; 2011; and Garg V. “Insights into the Genetic Basis of Congenital Heart Disease”. Cell Mol Life Sci. 2006;63:1141–8 [4, 6]
Endocardial cushion formation is first evident at human embryonic day (E)31–E35 and in the mouse around E9.5 [7, 8] and begins with endothelial to mesenchymal transformation (EMT). This process is initiated by signals emanating from the endocardial cells and adjacent myocardium that promote a subset of endocardial cells to lose cell-cell contacts and undergo transformation and migrate into the hyaluronan-rich matrix within the developing cushion. The resulting population of mesenchyme cells contributes to the progenitor cell pool that, in addition to other cell lineages, will give rise to the mature valve structures. Previous studies have shown that members of the Tgf-β family are required for EMT, and secretion of BMP2 from the myocardium acts synergistically with endothelial derived Tgf-β to enhance mesenchyme cell formation [9–11]. In addition to these growth factors, the Notch and Ras signaling pathways have also been implicated in endocardial cushion development of the AVC and OFT [12, 13].
Following active stages of EMT, newly transformed mesenchymal cells within the AV region continue to proliferate resulting in expansion of the endocardial cushions and elongation into valve primordia (Fig. 1B). The expanding endocardial cushions divide the common atrioventricular canal, first by growth of the superior (anterior) and inferior (posterior) endocardial cushions and then by growth of the 2 lateral cushions (Fig. 1B). Finally, the AV canal is divided into the left and right AV valve orifices when the superior and inferior cushions fuse [4]. In addition to endothelial-derived cells, there is evidence that cells from the epicardium populate the cushions and parietal leaflets of the valve primordia and cells from the dorsal mesenchymal protrusion, a derivative of the second heart field, contribute to atrioventricular septation [14–16]. The final morphogenetic steps that lead to formation of the mature atrioventricular septum and valve leaflets is less defined, but NFAT signaling is involved [17, 18].
Development of the OFT cushions involves not only EMT but also requires contribution from a population of migratory neural crest cells, referred to as the cardiac neural crest (CNC), which arise from the neural tube between the otic placode and third somite [4]. The CNC cells populate the aortic sac after migrating through the pharyngeal arches. In the aortic sac, the CNC cells are required for the proper septation of the common outflow tract (truncus arteriosus) into the aorta and pulmonary artery. The CNC cells also contribute to the development of the semilunar valve leaflets and the superior aspect of the ventricular septum. As semilunar valve development occurs in a synchronized manner with OFT development, the CNC cells contribute to formation of the truncal swellings. A small eminence on each swelling, along with a third precursor region, will grow to form the valve cusps in the aorta and pulmonary artery (Fig. 1C).
Individual valve leaflets are initially evident in human fetuses at around weeks 5 and 6 of development and in mice at around E13.5. The valve primordia will continue to develop by a process of thinning, reshaping, and elongation, as well as remodeling of the ECM. This occurs well into postnatal development, until the valve has become well organized with a stratified trilaminar structure made up of organized ECM layers, which contains elastin, which makes up the atrialis of the AV and the ventricularis of the OFT, fibrillar collagen that form the fibrosa, and proteoglycans, which make up the spongiosa [19–21].
Genetics of Bicuspid Aortic Valve
Bicuspid aortic valve (BAV) is the most common valvular malformation, and it affects between 1 % and 2 % of the population [22, 23]. BAV occurs when there are 2 cusps, rather than the 3 that are found in a normal aortic valve. Over 35 % of affected individuals will develop severe complications from BAV, such as aortic valve stenosis and regurgitation, infective endocarditis, ascending aortic aneurysms, and dissection [24, 25]. BAV has the largest health burden of all other CHD. BAVs are known to prematurely calcify leading to calcific aortic valve disease (CAVD), the second most common cause of aortic valve stenosis. In addition, BAV predisposes affected individuals to aortic aneurysms and infective endocarditis [26]. BAV has strong genetic components, as evidenced by reports of familial clustering and calculated heritability [27, 28]. In the past few years, mutations in several genes have been linked to BAV in humans and numerous mouse models with BAV have been identified.
Although the initial potential genetic link for human BAV was mutations in the gene KCNJ2 in the setting of Anderson syndrome, no mutations in this gene had been reported in nonsyndromic BAV. The first genetic etiology of nonsyndromic BAV was identified through the use of genome-wide linkage analysis by studying families with autosomal-dominant disease. NOTCH1 is a transmembrane receptor known to function in highly conserved signaling pathways that play important roles in cell fate and cardiovascular developmental processes [29, 30]. Interestingly, mutations in NOTCH1 were found to segregate with affected individuals, and direct sequencing of the gene in another unrelated family also showed segregation of the mutation with individuals affected with aortic valve disease [23]. Following this study, several mutations in NOTCH1 have been found to be associated with aortic valve disease [31–33]. These findings demonstrate evidence for NOTCH1 haploinsufficiency as a causative agent in a subset of familial aortic valve disease.
In addition to NOTCH1, rare sequence variants in 2 genes implicated from murine studies have also been implicated in human BAV. GATA5 belongs to the family of GATA transcription factors, and several of these factors have been implicated in human disease. It was recently shown that targeted deletion of GaTa5 in mice leads to a partially penetrant BAV phenotypes [34]. Examination of a cohort of 100 unrelated individuals with BAV identified 4 individuals with rare sequence variations that predicted nonsynonymous amino acid substitutions of highly conserved residues [35•]. Our group has performed similar studies in a well-phenotyped population of BAV and demonstrated similar results (Bonachea and Garg, unpublished data). Mutations in SMAD6, a member of the Bmp signaling pathway, display functional deficits in vitro and have been found in humans with BAV [36, 37•]. Consistent with this, cardiac cushion abnormalities have also been observed in Smad6-null mice [36].
Numerous additional genes have also been implicated as potential contributors to human BAV from animal models. Although one of the most studied animal models is the inbred Syrian hamster model, which exhibits BAVs with a fusion of the right and left coronary aortic leaflets, the genetic etiology for the malformation in hamsters remains unknown [38, 39]. One of the first mouse models of BAV was the observation that mice with targeted deletion of endothelial nitric oxide synthase (Nos3) displayed partially penetrant right-noncoronary fusion leaflet fusion BAV along with a broad spectrum of CHD [40]. Interestingly, the BAV phenotype displayed an almost 100 % penetrance when mice were null for Nos3 and heterozygous for NOTCH1 (Nos3−/−;NOTCH1+/−), demonstrating that the nitric oxide and NOTCH1 signaling pathways displayed a genetic interaction during aortic valve development [41•]. Recently, deletion of the proteoglycanase Adamts5 in mice has also been shown to lead to BAV. This model demonstrates the ability of uncleaved versican and insufficient Smad2 phosphorylation to cause bicuspid aortic and pulmonic valves [42]. Another member of the Bmp signaling pathway has also been implicated in BAV in mice as recent studies have also shown that the tissue specific deletion of Activin Receptor Type I (Alk2), in the endocardial cushion mesenchyme at post-EMT stages, resulted in BAV [43]. Additional investigations of nitric oxide and Bmp signaling pathways are required to determine if these findings will translate to the genetics of human BAV.
Genetics of Mitral Valve Prolapse
Mitral valve prolapse (MVP) is a common cardiac valvular disorder that affects about 2 %–3 % of the general population, and occurs when there is abnormal bulging and displacement of the mitral valve leaflets during ventricular systole. The mitral valve is a bi-leaflet atrioventricular valve on the left side of the heart. It allows blood to flow from the left atria to the left ventricle during diastole. MVP predisposes the affected individual to a greater risk for mitral valve regurgitation, congestive heart failure, arrhythmia, and infective endocarditis [44–47]. In MVP, fibromyxomatous degeneration occurs in the valve leaflets, causing them to become thickened and lengthened, therefore unable to properly function. These thickened leaflets, combined with weakened or ruptured chordae tendineae, are able to abnormally protrude superiorly into the left atrium during systole, leading to mitral regurgitation and cardiac insufficiency [48]. Normal mitral valves are composed of a trilaminar ECM structure: the fibrosa, which is tensile and composed of collagen fibers, the spongiosa, which is compressive and composed of collagen and proteoglycans, and the atrialis, which has the ability to stretch and is composed of elastic fibers. In contrast, diseased myxomatous valves show an abnormal expansion of the spongiosa caused by excess proteoglycan deposition that contributes to the ‘floppy’ functional phenotype. This is accompanied by diminished collagen fibers, elastin fragmentation, myofibroblast activation, and overexpression of proteolytic enzymes such as matrix metalloproteinase (MMP)-1, MMP-2, and MMP-13, that together leads to pathologic remodeling of the valve connective tissue [49, 50].
The genetic basis of MVP in humans has primarily occurred through investigation of MVP associated with Marfan syndrome, a well-known connective tissue disorder affecting multiple tissues including the heart, blood vessels, eyes, bones, and lungs [51]. Marfan syndrome, is caused by mutations in Fibrillin 1 (FBN1), a key component of ECM microfibrils [51–53]. Originally thought to be a structural disorder, subsequent investigation elegantly demonstrated that the primary pathophysiology is due to alterations in Tgf-β signaling, which plays key roles in the embryonic development of valves as well as in adult valve disease [54]. FBN1 limits the activation of Tgf-β through its interaction and stabilization with a large latent complex containing latent Tgf-β binding proteins (LTBPs), latency associated peptides (LAPs), and bound Tgf-β. The loss of function mutations, present in Marfan syndrome patients, prevent the interaction and the stabilization of Tgf-β with the large latent complex, thereby allowing excessive amounts of active Tgf-β [55]. Ng et al have described a mouse model carrying a C1039G mutation in Fbn1, which exhibit thickened valves soon after birth associated with increased Tgf-β activity indicated by Smad2 phosphorylation. The valve phenotype in these mice was rescued by pharmacologic Tgf-β antagonism further supporting a role for Tgf-β dysregulation in Marfan syndrome and MVP [55, 56]. A similar syndrome that has been associated with MVP is Loeys-Dietz (LDS). LDS has a similar phenotype to Marfan syndrome and interestingly is caused by mutations in TGF-β receptors 1 and 2 (TGFBR1 and TGFBR2), further implicating Tgf-β signaling in MVP [57].
Connective tissue dysplasias caused by mutations in a variety of collagen genes are also characterized by MVP and include Ehlers-Danlos (EDS), Stickler, and osteogenesis imperfecta [58–60] (Table 1). Collagen is one of the main components of valvular ECM [61, 62]. Deletion of collagen genes, ColVa1+/− and ColXa1−/−, in mice mutant mice recapitulate the EDS phenotype and also exhibit thickened valves with disorganized ECM [63]. Although the family of collagen genes represents exciting candidate genes for nonsyndromic MVP, human genetic studies do not support this, although studies have identified linkage to regions on chromosomes 11, 13, and 16 [64–66].
Table 1.
Genetic etiologies of valvular heart disease in humans
| Bicuspid aortic valve | |
| Syndromic | KCNJ2 |
| Nonsyndromic | NOTCH1, GATA5, SMAD6 |
| Myxomatous mitral valve | |
| Syndromic | FBN1, TGFBR1, TGFBR2, Collagen types I–III, V, XI |
| Nonsyndromic | Filamin A |
| Pulmonic valve stenosis | |
| Syndromic | PTPN11, RAF1, SOS1, KRAS |
| Nonsyndromic | GATA4 |
| Ebstein anomaly | |
| Nonsyndromic | MHY7 |
To date, the only gene linked to nonsyndromic myxomatous valve disease in humans is Filamin A (FLNA). By studying families with an X-linked form of valvular dystrophy, which predominantly affects the mitral and aortic valves in multiple families, FLNA was identified as having a role in nonsyndromic MVP [67, 68]. FLNA is a ubiquitous cytoplasmic phosphoprotein that has a structural role in the cytoskeleton and interacts with ECM bound cell-surface integrins [68]. FLNA has also been shown to affect TGF-β signaling through its association with Smads [68, 69•, 70]. FLNA-null mice exhibit a range of cardiovascular abnormalities including valve defects, OFT septation defects, and atrial and ventricular septal defects, as well as embryonic lethality [71, 72]. FLNA is expressed in the endocardium, epicardium, and interstitial cells of the valves throughout development, and mouse cell lineage studies have demonstrated that loss of endothelial FLNA expression leads to myxomatous mitral valves [69•]. Loss of FLNA leads to an impaired contractile phenotype with failed compaction and tissue remodeling of the valve leaflets. Further, a direct interaction has been shown between FLNA and serotonin during the fetal stage, which is considered to be an important time point for valvular remodeling and maturation required for proper valvular organization [69•].
In addition to these human studies, murine models for myxomatous mitral valve disease have been recently described. Mice haploinsufficient for Adamts9, which encodes a versican cleaving protease, exhibit abnormal myxomatous phenotypes with an abundance of proteoglycans along with other cardiovascular abnormalities involving the aortic valve and wall and ventricular myocardium. The thickened valves have an increase in uncleaved versican, as well as activated valve interstitial cells, which are normally quiescent in the absence of disease [73]. MVP phenotypes are also observed in mice overexpressing the ECM-degrading enzyme, matrix metalloproteainase-2 (MMP-2) in cardiac myocytes. Affected mice exhibit severely myxomatous mitral valves characterized by a disorganization of collagen bundles and a substantial accumulation of glycosoaminoglycans within the spongiosa [74]. It has been postulated that if the mechanism by which MMP-2 expression leads to MVP can be determined, then inhibition could be an alternative effective therapy for MVP patients [75]. Although yet to proven in vivo, the bHLH transcription factor Scleraxis is sufficient to promote proteoglycan secretion of valve interstitial cells in vitro and increased expression was observed in myxomatous valves from human patients and Fbn1 mutant mice [75]. By using these and other mouse models the molecular pathways that regulate the myxomatous changes will become increasingly defined.
Genetics of Pulmonary Valve and Tricuspid Valve Disease
The genetic contributors to disease involving the pulmonary and tricuspid valves are not as well defined. Pulmonary valve stenosis (PVS) occurs because of thickening of the valve leaflets causing stenosis, and is one of the more common types of CHD after cardiac septal defects [76]. The most well-studied genetic contributor to pulmonary valve stenosis in humans is in the setting of Noonan syndrome; a pleomorphic autosomal dominant disorder that is characterized by cardiac defects, typically pulmonary valve stenosis and hypertrophic cardiomyopathy, as well as cognitive disability, characteristic facies, and bleeding disorders [77]. Initially, mutations in PTPN11, which encodes protein tyrosine phosphatase SHP-2 involved in Ras signaling, were identified to be the cause of 50 % of Noonan Syndrome cases [78, 79]. Analysis of a knock-in mouse harboring a human associated mutation in PTPN11 recapitulates the human syndrome and exhibits abnormal endocardial cushion development and pulmonary stenosis [80]. Further analysis has determined that this mutation functions in the endocardium and the potential disease mechanism involves increased MAPK activation [81]. Subsequent studies have found that mutations of other genes involved in the Ras signaling pathway including RAF1, SOS1, and KRAS are also associated with Noonan syndrome in addition to LEOPARD and Costello syndromes, which exhibit similar phenotypes as a result of RAS mutations [82–84].
Although familial forms of nonsyndromic PVS have been described in the literature, the genetic contributors have not been identified. We previously reported that mutations in GATA4, are linked to cardiac septation defects [85]. Interestingly, in 1 family with a specific mutation (G296S), there were several members who were affected with PVS and subsequent families with the same mutation were reported to have a similar valve anomaly in conjunction with atrial septal defect [85–87]. We recently generated a mouse harboring this knock-in human mutation and found that affected mice also displayed pulmonary and aortic valve stenosis [88]. Future work will determine if mutations in GATA4 contribute to nonsyndromic isolated PVS in humans.
Anomalies of the tricuspid valve are quite uncommon but familial forms of Ebstein anomaly have been reported [89]. Ebstein anomaly of the tricuspid valve is characterized by downward displacement of the tricuspid valve into the right ventricle and is associated with varying degrees of valve dysfunction. Although the genetic basis of isolated Ebstein anomaly remains unknown, recent investigations have identified mutations in the MYH7, which encodes β-myosin heavy chain, in individuals with Ebstein anomaly that is associated with left ventricular noncompaction cardiomyopathy [90•, 91]. Future work is required to determine the role of MYH7 in isolated Ebstein anomaly.
Genetics of Adult-Onset Valve Disease
Although the focus has been on the genetics of congenital valve abnormalities, there is increasing evidence pointing toward genetic contributors in valvular diseases in adults. The first evidence of this was based upon familial clustering of death because of aortic and mitral valve diseases based upon a population based study [92]. The current evidence has focused on genetic contributors to calcific aortic valve disease (CAVD), in which valvular interstitial cells calcify preventing the valve from functioning properly, leading to valve stenosis and regurgitation [93]. Initial insights came from studying human families with NOTCH1 mutations and BAV, which were discussed above. In these families, there were individuals who had normal tri-leaflet aortic valves but developed calcification suggesting a role for NOTCH1. NOTCH1 was shown to interact with and activate hairy-like transcriptional repressors, which downregulate Runx2, a key transcriptional regulator of osteoblast fate [21, 94]. Subsequent studies have shown inhibition of Notch signaling prevents calcification in vitro and that NOTCH1 regulates Sox9, a transcription factor that when deleted leads to valve calcification in mice [95]. Inhibition of NOTCH1 causes a downregulation of Sox9, leading to an increase in calcification in vitro [96]. Along with these more molecular approaches, human genetic studies have attempted to identify potential genetic contributors to CAVD [97, 98]. Recently, a large genome-wide association study with 6942 participants with aortic valve calcification and 3795 participants with mitral valve calcification was performed [99••]. It led to the identification of 1 single nucleotide polymorphism (SNP) in the lipoprotein(a) (LPA) locus linked to CAVD. This finding was reproducible in multiple ethnicities and the SNP correlated with Lp(a) levels. Although potentially exciting, additional studies are required to confirm this association and determine if lowering of Lp(a) levels will serve as a new therapy.
Conclusions
The significant role that genetic factors play in human cardiac valve disease is becoming increasingly evident. Though only a handful of genetic mutations have been established as disease-causing, these findings have allowed a link to be created between congenital valvulopathies and human genetics. Mouse models have also been of great importance as the ability to recapitulate human disease will allow for the analysis of molecular pathways. In addition, the murine models allow for the elucidation of disease mechanisms and the identification of the roles that different cell types play in valve development and disease progression. The availability of new genetic technologies that allow for screening of whole exome/genomes and array comparative genome hybridization to identify subtle chromosomal deletions/duplications should allow for detection of additional genes implicated in human valvular disease.
Footnotes
Conflict of Interest Stephanie LaHaye, Joy Lincoln, and Vidu Garg declare that they have no conflict of interest.
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
This article is part of the Topical Collection on Cardiovascular Genomics
Contributor Information
Stephanie LaHaye, Center for Cardiovascular and Pulmonary Research and The Heart Center, Room WB4221, Nationwide Children’s Hospital, Columbus, OH 43205, USA. Department of Molecular Genetics, The Ohio State University, Columbus, OH, USA.
Joy Lincoln, Center for Cardiovascular and Pulmonary Research and The Heart Center, Room WB4239, Nationwide Children’s Hospital, Columbus, OH 43205, USA. Department of Pediatrics, The Ohio State University, Columbus, OH, USA.
Vidu Garg, Email: vidu.garg@nationwidechildrens.org, Center for Cardiovascular and Pulmonary Research and The Heart Center, Room WB4221, Nationwide Children’s Hospital, Columbus, OH 43205, USA. Department of Pediatrics, The Ohio State University, Columbus, OH, USA. Department of Molecular Genetics, The Ohio State University, Columbus, OH, USA.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics—2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffman JI, Kaplan S. The incidence of congenital heart disease. JACC. 2002;12:1890–900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 3.Maganti K, Rigolin VH, Sarano ME, Bonow RO. Valvular heart disease: diagnosis and management. Mayo Clin Proc. 2010;85:483–500. doi: 10.4065/mcp.2009.0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garg V. Growth of the normal human heart. In: Preedy VR, editor. Handbook of growth and growth monitoring in health and disease. New York: Springer; 2011. [Google Scholar]
- 5.Garg V. Molecular basis of cardiac development and congenital heart disease. In: Patterson C, Willis M, editors. Translational Cardiology. New York: Humana Press; 2012. [Google Scholar]
- 6.Garg V. Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci. 2006;63:1141–8. doi: 10.1007/s00018-005-5532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105:408–21. doi: 10.1161/CIRCRESAHA.109.201566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishman MC, Chien KR. Fashioning the vertebrate heart: earliest embryonic decisions. Development. 1997;124:2099–117. doi: 10.1242/dev.124.11.2099. [DOI] [PubMed] [Google Scholar]
- 9.Brown CB, Boyer AS, Runyan RB, Barnett JV. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science. 1999;283:2080–2. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima Y, Yamagishi T, Hokari S, Nakamura H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP) Anat Rec. 2000;258:119–27. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 11.Sugi Y, Yamamura H, Okagawa H, Markwald RR. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev Biol. 2004;269:505–18. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 12.Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yutzey KE, Colbert M, Robbins J. Ras-related signaling pathways in valve development: ebb and flow. Physiology. 2005;20:390–7. doi: 10.1152/physiol.00035.2005. [DOI] [PubMed] [Google Scholar]
- 14.Wessels A, van den Hoff MJ, Adamo RF, Phelps AL, Lockhart MM, Sauls K, et al. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol. 2012;366:111–24. doi: 10.1016/j.ydbio.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–52. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 16.Briggs LE, Phelps AL, Brown E, Kakarla J, Anderson RH, van den Hoff MJ, et al. Expression of the BMP receptor Alk3 in the second heart field is essential for development of the dorsal mesenchyme protrusion and atrioventricular septation. Circ Res. 2013;112:1420–32. doi: 10.1161/CIRCRESAHA.112.300821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, et al. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell. 2004;118:649–63. doi: 10.1016/j.cell.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Wu B, Wang Y, Lui W, Langworthy M, Tompkins KL, Hatzopoulos AK, et al. Nfatc1 coordinates valve endocardial cell lineage development required for heart valve formation. Circ Res. 2011;109:183–92. doi: 10.1161/CIRCRESAHA.111.245035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirrig EE, Yutzey KE. Transcriptional regulation of heart valve development and disease. Cardiovas Pathol. 2011;20:162–7. doi: 10.1016/j.carpath.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinton RB, Yutzey KE. Heart valve structure and function in development and disease. Annu Rev Physiol. 2011;73:29–46. doi: 10.1146/annurev-physiol-012110-142145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinton RB, Lincoln J, Deutsch GH, Osinska H, Manning PB, Benson DW, et al. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res. 2006;98:1431–8. doi: 10.1161/01.RES.0000224114.65109.4e. [DOI] [PubMed] [Google Scholar]
- 22.Ward C. Clinical significance of the bicuspid aortic valve. Heart. 2000;83:81–5. doi: 10.1136/heart.83.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–4. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 24.Vallely MP, Semsarian C, Bannon PG. Management of the ascending aorta in patients with bicuspid aortic valve disease. Heart Lung Circ. 2008;17:357–63. doi: 10.1016/j.hlc.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Siu SC, Silversides CK. Bicuspid aortic valve disease. J Am Coll Cardiol. 2010;55:2789–800. doi: 10.1016/j.jacc.2009.12.068. [DOI] [PubMed] [Google Scholar]
- 26.Fedak PW, Verma S, David TE, Leask RL, Weisel RD, Butany J. Clinical and pathophysiological implications of a bicuspid aortic valve. Circulation. 2002;106:900–4. doi: 10.1161/01.cir.0000027905.26586.e8. [DOI] [PubMed] [Google Scholar]
- 27.Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW. Bicuspid aortic valve is heritable. J Am Coll Cardiol. 2004;44:138–43. doi: 10.1016/j.jacc.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 28.McBride KL, Garg V. Heredity of bicuspid aortic valve: is family screening indicated? Heart. 2011;97:1193–5. doi: 10.1136/hrt.2011.222489. [DOI] [PubMed] [Google Scholar]
- 29.de la Pompa JL, Epstein JA. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–54. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- 31.Foffa I, Ait Ali L, Panesi P, Mariani M, Festa P, Botto N, et al. Sequencing of NOTCH1, GATA5, TGFBR1 and TGFBR2 genes in familial cases of bicuspid aortic valve. BMC Med Genet. 2013;14:44. doi: 10.1186/1471-2350-14-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TM. Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg. 2007;134:290–6. doi: 10.1016/j.jtcvs.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 33.Mohamed SA, Aherrahrou Z, Liptau H, Erasmi AW, Hagemann C, Wrobel S, et al. Novel missense mutations (p.T596M and p.P1797H) in NOTCH1 in patients with bicuspid aortic valve. Biochem Biophys Res Commun. 2006;345:1460–5. doi: 10.1016/j.bbrc.2006.05.046. [DOI] [PubMed] [Google Scholar]
- 34.Laforest B, Andelfinger G, Nemer M. Loss of Gata5 in mice leads to bicuspid aortic valve. J Clin Invest. 2011;121:2876–87. doi: 10.1172/JCI44555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Padang R, Bagnall RD, Richmond DR, Bannon PG, Semsarian C. Rare non-synonymous variations in the transcriptional activation domains of GATA5 in bicuspid aortic valve disease. J Mol Cell Cardiol. 2012;53:277–81. doi: 10.1016/j.yjmcc.2012.05.009. This study is the first to identify rare sequence variants in the gene, GATA5, in humans with BAV. [DOI] [PubMed] [Google Scholar]
- 36.Galvin KM, Donovan MJ, Lynch CA, Meyer RI, Paul RJ, Lorenz JN, et al. A role for smad6 in development and homeostasis of the cardiovascular system. Nat Genet. 2000;24:171–4. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 37•.Tan HL, Glen E, Topf A, Hall D, O’Sullivan JJ, Sneddon L, et al. Nonsynonymous variants in the SMAD6 gene predispose to congenital cardiovascular malformation. Hum Mutat. 2012;33:720–7. doi: 10.1002/humu.22030. This study is the first to report an association between rare sequence variants with functional deficits in the gene, SMAD6, in humans with bicuspid aortic valve. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sans-Coma V, Cardo M, Thiene G, Fernandez B, Arque JM, Duran AC. Bicuspid aortic and pulmonary valves in the Syrian hamster. Int J Cardiol. 1992;34:249–54. doi: 10.1016/0167-5273(92)90021-t. [DOI] [PubMed] [Google Scholar]
- 39.Sans-Coma V, Carmen Fernandez M, Fernandez B, Duran AC, Anderson RH, Arque JM. Genetically alike Syrian hamsters display both bifoliate and trifoliate aortic valves. J Anat. 2012;220:92–101. doi: 10.1111/j.1469-7580.2011.01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee TC, Zhao YD, Courtman DW, Stewart DJ. Abnormal aortic valve development in mice lacking endothelial nitric oxide synthase. Circulation. 2000;101:2345–8. doi: 10.1161/01.cir.101.20.2345. [DOI] [PubMed] [Google Scholar]
- 41•.Bosse K, Hans CP, Zhao N, Koenig SN, Huang N, Guggilam A, et al. Endothelial nitric oxide signaling regulates NOTCH1 in aortic valve disease. J Mol Cell Cardiol. 2013;60:27–35. doi: 10.1016/j.yjmcc.2013.04.001. The authors describe the first highly penetrant mouse model for bicuspid aortic valve by generating mice, which are haploinsufficient for NOTCH1 in a endothelial nitric oxide synthase (Nos3)-null background. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dupuis LE, Osinska H, Weinstein MB, Hinton RB, Kern CB. Insufficient versican cleavage and Smad2 phosphorylation results in bicuspid aortic and pulmonary valves. J Mol Cell Cardiol. 2013;60:50–9. doi: 10.1016/j.yjmcc.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas PS, Sridurongrit S, Ruiz-Lozano P, Kaartinen V. Deficient signaling via Alk2 (Acvr1) leads to bicuspid aortic valve development. PloS ONE. 2012;7:e35539. doi: 10.1371/journal.pone.0035539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolibash AJ, Kilman JW, Bush CA, Ryan JM, Fontana ME, Wooley CF. Evidence for progression from mild to severe mitral regurgitation in mitral valve prolapse. Am J Cardiol. 1986;58:762–7. doi: 10.1016/0002-9149(86)90352-8. [DOI] [PubMed] [Google Scholar]
- 45.Freed LA, Levy D, Levine RA, Larson MG, Evans JC, Fuller DL, et al. Prevalence and clinical outcome of mitral-valve prolapse. N Engl J Med. 1999;341:1–7. doi: 10.1056/NEJM199907013410101. [DOI] [PubMed] [Google Scholar]
- 46.Baddour LM, Bisno AL. Mitral valve prolapse: multifactorial etiologies and variable prognosis. Am Heart J. 1986;112:1359–62. doi: 10.1016/0002-8703(86)90408-4. [DOI] [PubMed] [Google Scholar]
- 47.Baddour LM, Phillips TN, Bisno AL. Coagulase-negative staphylococcal endocarditis. Occurrence in patients with mitral valve prolapse. Arch Intern Med. 1986;146:119–21. doi: 10.1001/archinte.146.1.119. [DOI] [PubMed] [Google Scholar]
- 48.Levine RA, Slaugenhaupt SA. Molecular genetics of mitral valve prolapse. Curr Opin Cardiol. 2007;22:171–5. doi: 10.1097/HCO.0b013e3280f3bfcd. [DOI] [PubMed] [Google Scholar]
- 49.Rabkin E, Aikawa M, Stone JR, Fukumoto Y, Libby P, Schoen FJ. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation. 2001;104:2525–32. doi: 10.1161/hc4601.099489. [DOI] [PubMed] [Google Scholar]
- 50.Gupta V, Barzilla JE, Mendez JS, Stephens EH, Lee EL, Collard CD, et al. Abundance and location of proteoglycans and hyaluronan within normal and myxomatous mitral valves. Cardiovasc Pathol. 2009;18:191–7. doi: 10.1016/j.carpath.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1994;352:337–9. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 52.Maslen CL, Corson GM, Maddox BK, Glanville RW, Sakai LY. Partial sequence of a candidate gene for the Marfan syndrome. Nature. 1991;352:334–7. doi: 10.1038/352334a0. [DOI] [PubMed] [Google Scholar]
- 53.Lee B, Godfrey M, Vitale E, Hori H, Mattei MG, Sarfarazi M, et al. Linkage of Marfan syndrome and a phenotypically related disorder to two different fibrillin genes. Nature. 1991;352:330–4. doi: 10.1038/352330a0. [DOI] [PubMed] [Google Scholar]
- 54.Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–11. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 55.Ng CM, Cheng A, Myers LA, Martinez-Murillo F, Jie C, Bedja D, et al. TGF-beta-dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J Clin Invest. 2004;114:1586–92. doi: 10.1172/JCI22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Habashi JP, Judge DP, Holm TM, Cohn RD, Loeys BL, Cooper TK, et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–21. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37:275–81. doi: 10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- 58.Liberfarb RM, Goldblatt A. Prevalence of mitral-valve prolapse in the Stickler syndrome. Am J Med Genet. 1986;24:387–92. doi: 10.1002/ajmg.1320240302. [DOI] [PubMed] [Google Scholar]
- 59.Najib MQ, Schaff HV, Ganji J, Lee HR, Click RL, Miller DC, et al. Valvular heart disease in patients with osteogenesis imperfecta. J Card Surg. 2013;28:139–43. doi: 10.1111/jocs.12064. [DOI] [PubMed] [Google Scholar]
- 60.Malfait F, Wenstrup RJ, De Paepe A. Clinical and genetic aspects of Ehlers-Danlos syndrome, classic type. Genet Med. 2010;12:597–605. doi: 10.1097/GIM.0b013e3181eed412. [DOI] [PubMed] [Google Scholar]
- 61.Lincoln J, Lange AW, Yutzey KE. Hearts and bones: shared regulatory mechanisms in heart valve, cartilage, tendon, and bone development. Dev Biol. 2006;294:292–302. doi: 10.1016/j.ydbio.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 62.Ritelli M, Dordoni C, Venturini M, Chiarelli N, Quinzani S, Traversa M, et al. Clinical and molecular characterization of 40 patients with classic Ehlers-Danlos syndrome: identification of 18 COL5A1 and 2 COL5A2 novel mutations. Orphanet J Rare. 2013;8:58. doi: 10.1186/1750-1172-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lincoln J, Florer JB, Deutsch GH, Wenstrup RJ, Yutzey KE. ColVa1 and ColXIa1 are required for myocardial morphogenesis and heart valve development. Dev Dyn. 2006;235:3295–305. doi: 10.1002/dvdy.20980. [DOI] [PubMed] [Google Scholar]
- 64.Henney AM, Tsipouras P, Schwartz RC, Child AH, Devereux RB, Leech GJ. Genetic evidence that mutations in the COL1A1, COL1A2, COL3A1, or COL5A2 collagen genes are not responsible for mitral valve prolapse. Br Heart J. 1989;61:292–9. doi: 10.1136/hrt.61.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wordsworth P, Ogilvie D, Akhras F, Jackson G, Sykes B. Genetic segregation analysis of familial mitral valve prolapse shows no linkage to fibrillar collagen genes. Br Heart J. 1989;61:300–6. doi: 10.1136/hrt.61.3.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Disse S, Abergel E, Berrebi A, Houot AM, Le Heuzey JY, Diebold B, et al. Mapping of a first locus for autosomal dominant myxomatous mitral-valve prolapse to chromosome 16p11.2-p12.1. Am J Hum Genet. 1999;65:1242–51. doi: 10.1086/302624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lardeux A, Kyndt F, Lecointe S, Marec HL, Merot J, Schott JJ, et al. Filamin-a-related myxomatous mitral valve dystrophy: genetic, echocardiographic and functional aspects. J Cardiovasc Transl Res. 2011;4:748–56. doi: 10.1007/s12265-011-9308-9. [DOI] [PubMed] [Google Scholar]
- 68.Kyndt F, Gueffet JP, Probst V, Jaafar P, Legendre A, Le Bouffant F, et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–9. doi: 10.1161/CIRCULATIONAHA.106.622621. [DOI] [PubMed] [Google Scholar]
- 69•.Sauls K, de Vlaming A, Harris BS, Williams K, Wessels A, Levine RA, et al. Developmental basis for filamin-A-associated myxomatous mitral valve disease. Cardiovasc Res. 2012;96:109–19. doi: 10.1093/cvr/cvs238. The authors describe the first mouse model of myxomatous mitral valve disease associated with mutation of Filamin A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sasaki A, Masuda Y, Ohta Y, Ikeda K, Watanabe K. Filamin associates with Smads and regulates transforming growth factor-beta signaling. J Biol Chem. 2001;276:17871–7. doi: 10.1074/jbc.M008422200. [DOI] [PubMed] [Google Scholar]
- 71.Feng Y, Chen MH, Moskowitz IP, Mendonza AM, Vidali L, Nakamura F, et al. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc Natl Acad Sci U S A. 2006;103:19836–41. doi: 10.1073/pnas.0609628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hart AW, Morgan JE, Schneider J, West K, McKie L, Bhattacharya S, et al. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum Mol Genet. 2006;15:2457–67. doi: 10.1093/hmg/ddl168. [DOI] [PubMed] [Google Scholar]
- 73.Kern CB, Wessels A, McGarity J, Dixon LJ, Alston E, Argraves WS, et al. Reduced versican cleavage due to Adamts9 haploinsufficiency is associated with cardiac and aortic anomalies. Matrix Biol. 2010;29:304–16. doi: 10.1016/j.matbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mahimkar R, Nguyen A, Mann M, Yeh CC, Zhu BQ, Karliner JS, et al. Cardiac transgenic matrix metalloproteinase-2 expression induces myxomatous valve degeneration: a potential model of mitral valve prolapse disease. Cardiovasc Pathol. 2009;18:253–61. doi: 10.1016/j.carpath.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barnette DN, Hulin A, Ahmed AS, Colige AC, Azhar M, Lincoln J. Tgfbeta-Smad and MAPK signaling mediate scleraxis and proteoglycan expression in heart valves. J Mol Cell Cardiol. 2013;65:137146. doi: 10.1016/j.yjmcc.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 77.Hatemi AC, Gursoy M, Tongut A, Bicakhan B, Guzeltas A, Cetin G, et al. Pulmonary stenosis as a predisposing factor for infective endocarditis in a patient with Noonan syndrome. Tex Heart Inst J. 2010;37:99–101. [PMC free article] [PubMed] [Google Scholar]
- 78.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- 79.Sznajer Y, Keren B, Baumann C, Pereira S, Alberti C, Elion J, et al. The spectrum of cardiac anomalies in Noonan syndrome as a result of mutations in the PTPN11 gene. Pediatrics. 2007;119:e1325–31. doi: 10.1542/peds.2006-0211. [DOI] [PubMed] [Google Scholar]
- 80.Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–57. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 81.Araki T, Chan G, Newbigging S, Morikawa L, Bronson RT, Neel BG. Noonan syndrome cardiac defects are caused by PTPN11 acting in endocardium to enhance endocardial-mesenchymal transformation. Proc Natl Acad Sci U S A. 2009;106:4736–41. doi: 10.1073/pnas.0810053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aoki Y, Niihori T, Narumi Y, Kure S, Matsubara Y. The RAS/MAPK syndromes: novel roles of the RAS pathway in human genetic disorders. Hum Mutat. 2008;29:992–1006. doi: 10.1002/humu.20748. [DOI] [PubMed] [Google Scholar]
- 83.Schubbert S, Bollag G, Shannon K. Deregulated Ras signaling in developmental disorders: new tricks for an old dog. Curr Opin Genet Dev. 2007;17:15–22. doi: 10.1016/j.gde.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 84.Tartaglia M, Gelb BD, Zenker M. Noonan syndrome and clinically related disorders. Best Pract Res Clin Endocrinol Metab. 2011;25:161–79. doi: 10.1016/j.beem.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–7. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 86.Okubo A, Miyoshi O, Baba K, Takagi M, Tsukamoto K, Kinoshita A, et al. A novel GATA4 mutation completely segregated with atrial septal defect in a large Japanese family. J Med Genet. 2004;41:e97. doi: 10.1136/jmg.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiang R, Fan LL, Huang H, Cao BB, Li XP, Peng DQ, et al. A novel mutation of GATA4 (K319E) is responsible for familial atrial septal defect and pulmonary valve stenosis. Gene. 2014;534:320–3. [PubMed] [Google Scholar]
- 88.Misra C, Sachan N, McNally CR, Koenig SN, Nichols HA, Guggilam A, et al. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS Genet. 2012;8:e1002690. doi: 10.1371/journal.pgen.1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maragh S, Miller RA, Bessling SL, McGaughey DM, Wessels MW, de Graaf B, et al. Identification of RNA binding motif proteins essential for cardiovascular development. BMC Dev Biol. 2011;11:62. doi: 10.1186/1471-213X-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90•.Postma AV, van Engelen K, van de Meerakker J, Rahman T, Probst S, Baars MJ, et al. Mutations in the sarcomere gene MYH7 in Ebstein anomaly. Circ Cardiovasc Genet. 2011;4:43–50. doi: 10.1161/CIRCGENETICS.110.957985. This study is the first to report mutations in MYH7 in individuals with Ebstein anomaly of the tricuspid valve. [DOI] [PubMed] [Google Scholar]
- 91.Bettinelli AL, Mulder TJ, Funke BH, Lafferty KA, Longo SA, Niyazov DM. Familial Ebstein anomaly, left ventricular hypertrabeculation, and ventricular septal defect associated with a MYH7 mutation. Am J Med Genet A. 2013;161:3187–90. doi: 10.1002/ajmg.a.36182. [DOI] [PubMed] [Google Scholar]
- 92.Horne BD, Camp NJ, Muhlestein JB, Cannon-Albright LA. Evidence for a heritable component in death resulting from aortic and mitral valve diseases. Circulation. 2004;110:3143–8. doi: 10.1161/01.CIR.0000147189.85636.C3. [DOI] [PubMed] [Google Scholar]
- 93.Lewin MB, Otto CM. The bicuspid aortic valve: adverse outcomes from infancy to old age. Circulation. 2005;111:832–4. doi: 10.1161/01.CIR.0000157137.59691.0B. [DOI] [PubMed] [Google Scholar]
- 94.Nigam V, Srivastava D. NOTCH1 represses osteogenic pathways in aortic valve cells. J Mol Cell Cardiol. 2009;47:828–34. doi: 10.1016/j.yjmcc.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lincoln J, Kist R, Scherer G, Yutzey KE. Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev Biol. 2007;305:120–32. doi: 10.1016/j.ydbio.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Acharya A, Hans CP, Koenig SN, Nichols HA, Galindo CL, Garner HR, et al. Inhibitory role of NOTCH1 in calcific aortic valve disease. PloS ONE. 2011;6:e27743. doi: 10.1371/journal.pone.0027743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Probst V, Le Scouarnec S, Legendre A, Jousseaume V, Jaafar P, Nguyen JM, et al. Familial aggregation of calcific aortic valve stenosis in the western part of France. Circulation. 2006;113:856–60. doi: 10.1161/CIRCULATIONAHA.105.569467. [DOI] [PubMed] [Google Scholar]
- 98.Bella JN, Tang W, Kraja A, Rao DC, Hunt SC, Miller MB, et al. Genome-wide linkage mapping for valve calcification susceptibility loci in hypertensive sibships: the Hypertension Genetic Epidemiology Network Study. Hypertension. 2007;49:453–60. doi: 10.1161/01.HYP.0000256957.10242.75. [DOI] [PubMed] [Google Scholar]
- 99••.Thanassoulis G, Campbell CY, Owens DS, Smith JG, Smith AV, Peloso GM, et al. Genetic associations with valvular calcification and aortic stenosis. N Engl J Med. 2013;368:503–12. doi: 10.1056/NEJMoa1109034. First large-scale genome-wide association study linking genetic variation in LPA (which encodes for lipoprotein A) locus with aortic valve calcification and aortic stenosis. [DOI] [PMC free article] [PubMed] [Google Scholar]