Abstract
In the presence of emetine, an inhibitor of protein synthesis, nascent DNA on forward arms of replication forks in hamster cell lines containing either single or amplified copies of the DHFR gene region was enriched 5- to 7-fold over nascent DNA on retrograde arms. This forward arm bias was observed on both sides of the specific origin of bidirectional DNA replication located 17 kb downstream of the hamster DHFR gene (OBR-1), consistent with at least 85% of replication forks within this region emanating from OBR-1. However, the replication fork asymmetry induced by emetine does not result from conservative nucleosome segregation, as previously believed, but from preferentially inhibiting Okazaki fragment synthesis on retrograde arms of forks to produce 'imbalanced DNA synthesis'. Three lines of evidence support this conclusion. First, the bias existed in long nascent DNA strands prior to nuclease digestion of non-nucleosomal DNA. Second, the fraction of RNA-primed Okazaki fragments was rapidly diminished. Third, electron microscopic analysis of SV40 DNA replicating in the presence of emetine revealed forks with single-stranded DNA on one arm, and nucleosomes randomly distributed to both arms. Thus, as with cycloheximide, nucleosome segregation in the presence of emetine was distributive.
Full text
PDF





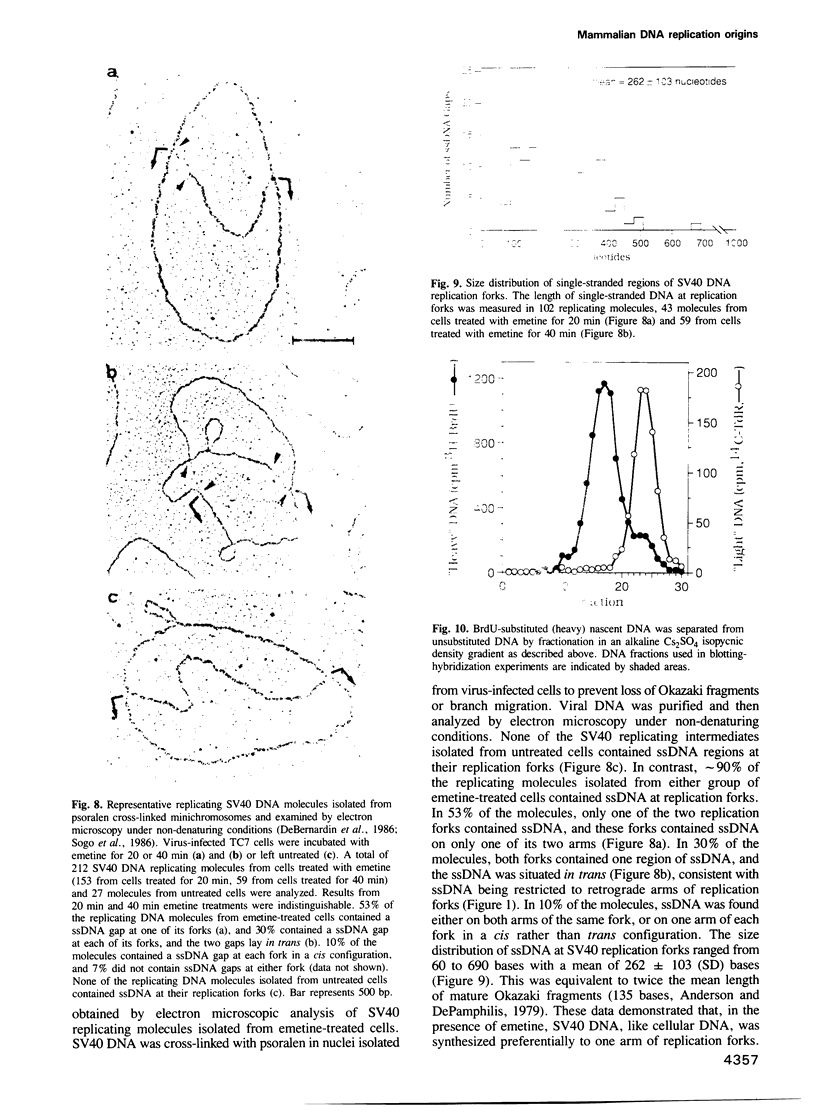



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almouzni G., Clark D. J., Méchali M., Wolffe A. P. Chromatin assembly on replicating DNA in vitro. Nucleic Acids Res. 1990 Oct 11;18(19):5767–5774. doi: 10.1093/nar/18.19.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anachkova B., Hamlin J. L. Replication in the amplified dihydrofolate reductase domain in CHO cells may initiate at two distinct sites, one of which is a repetitive sequence element. Mol Cell Biol. 1989 Feb;9(2):532–540. doi: 10.1128/mcb.9.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S., DePamphilis M. L. Metabolism of Okazaki fragments during simian virus 40 DNA replication. J Biol Chem. 1979 Nov 25;254(22):11495–11504. [PubMed] [Google Scholar]
- Bonne-Andrea C., Wong M. L., Alberts B. M. In vitro replication through nucleosomes without histone displacement. Nature. 1990 Feb 22;343(6260):719–726. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Isolation of the origin of replication associated with the amplified Chinese hamster dihydrofolate reductase domain. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7790–7794. doi: 10.1073/pnas.83.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W. C., Selegue J. E., Heintz N. H. Replication intermediates formed during initiation of DNA synthesis in methotrexate-resistant CHOC 400 cells are enriched for sequences derived from a specific, amplified restriction fragment. Biochemistry. 1986 Jan 28;25(2):441–449. doi: 10.1021/bi00350a025. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- Cusick M. E., DePamphilis M. L., Wassarman P. M. Dispersive segregation of nucleosomes during replication of simian virus 40 chromosomes. J Mol Biol. 1984 Sep 15;178(2):249–271. doi: 10.1016/0022-2836(84)90143-8. [DOI] [PubMed] [Google Scholar]
- Cusick M. E., Wassarman P. M., DePamphilis M. L. Application of nucleases to visualizing chromatin organization at replication forks. Methods Enzymol. 1989;170:290–316. doi: 10.1016/0076-6879(89)70053-7. [DOI] [PubMed] [Google Scholar]
- De Bernardin W., Koller T., Sogo J. M. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J Mol Biol. 1986 Oct 5;191(3):469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L., Wassarman P. M. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu Rev Biochem. 1980;49:627–666. doi: 10.1146/annurev.bi.49.070180.003211. [DOI] [PubMed] [Google Scholar]
- Dijkwel P. A., Vaughn J. P., Hamlin J. L. Mapping of replication initiation sites in mammalian genomes by two-dimensional gel analysis: stabilization and enrichment of replication intermediates by isolation on the nuclear matrix. Mol Cell Biol. 1991 Aug;11(8):3850–3859. doi: 10.1128/mcb.11.8.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handeli S., Klar A., Meuth M., Cedar H. Mapping replication units in animal cells. Cell. 1989 Jun 16;57(6):909–920. doi: 10.1016/0092-8674(89)90329-2. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Hamlin J. L. An amplified chromosomal sequence that includes the gene for dihydrofolate reductase initiates replication within specific restriction fragments. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4083–4087. doi: 10.1073/pnas.79.13.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. H., Stillman B. W. Nuclear DNA synthesis in vitro is mediated via stable replication forks assembled in a temporally specific fashion in vivo. Mol Cell Biol. 1988 May;8(5):1923–1931. doi: 10.1128/mcb.8.5.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V., Chalkley R. Histone segregation on replicating chromatin. Biochemistry. 1985 Nov 19;24(24):6930–6938. doi: 10.1021/bi00345a027. [DOI] [PubMed] [Google Scholar]
- Jackson V. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry. 1988 Mar 22;27(6):2109–2120. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- Jackson V. Deposition of newly synthesized histones: misinterpretations due to cross-linking density-labeled proteins with Lomant's reagent. Biochemistry. 1987 Apr 21;26(8):2325–2334. doi: 10.1021/bi00382a038. [DOI] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990 Jan 23;29(3):719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Kaufmann G., Anderson S., DePamphilis M. L. RNA primers in Simian virus 40 DNA replication. II. Distribution of 5' terminal oligoribonucleotides in nascent DNA. J Mol Biol. 1977 Nov 5;116(3):549–567. doi: 10.1016/0022-2836(77)90083-3. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Seiter A., Zentgraf H. Nucleosome assembly in vitro: separate histone transfer and synergistic interaction of native histone complexes purified from nuclei of Xenopus laevis oocytes. EMBO J. 1990 Apr;9(4):1309–1318. doi: 10.1002/j.1460-2075.1990.tb08240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffak I. M. Conservative segregation of nucleosome core histones. Nature. 1984 Jan 5;307(5946):82–85. doi: 10.1038/307082a0. [DOI] [PubMed] [Google Scholar]
- Leffak I. M., Grainger R., Weintraub H. Conservative assembly and segregation of nucleosomal histones. Cell. 1977 Nov;12(3):837–845. doi: 10.1016/0092-8674(77)90282-3. [DOI] [PubMed] [Google Scholar]
- Leffak I. M. Stability of the conservative mode of nucleosome assembly. Nucleic Acids Res. 1983 May 11;11(9):2717–2732. doi: 10.1093/nar/11.9.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu T. H., Hamlin J. L. High-resolution mapping of replication fork movement through the amplified dihydrofolate reductase domain in CHO cells by in-gel renaturation analysis. Mol Cell Biol. 1989 Feb;9(2):523–531. doi: 10.1128/mcb.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M. H., Huberman J. A. The two faces of higher eukaryotic DNA replication origins. Cell. 1990 Sep 7;62(5):845–847. doi: 10.1016/0092-8674(90)90258-g. [DOI] [PubMed] [Google Scholar]
- Ma C., Leu T. H., Hamlin J. L. Multiple origins of replication in the dihydrofolate reductase amplicons of a methotrexate-resistant chinese hamster cell line. Mol Cell Biol. 1990 Apr;10(4):1338–1346. doi: 10.1128/mcb.10.4.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli G., Baldari C. T., Carri M. T., Di Cello G., Buongiorno-Nardelli M. An electron microscope study of chromosomal DNA replication in different eukaryotic systems. Exp Cell Res. 1982 Jan;137(1):127–140. doi: 10.1016/0014-4827(82)90015-5. [DOI] [PubMed] [Google Scholar]
- Pospelov V., Russev G., Vassilev L., Tsanev R. Nucleosome segregation in chromatin replicated in the presence of cycloheximide. J Mol Biol. 1982 Mar 25;156(1):79–91. doi: 10.1016/0022-2836(82)90460-0. [DOI] [PubMed] [Google Scholar]
- Riley D., Weintraub H. Conservative segregation of parental histones during replication in the presence of cycloheximide. Proc Natl Acad Sci U S A. 1979 Jan;76(1):328–332. doi: 10.1073/pnas.76.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roufa D. J., Marchionni M. A. Nucleosome segregation at a defined mammalian chromosomal site. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1810–1814. doi: 10.1073/pnas.79.6.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roufa D. J. Replication of a mammalian genome: the role of de novo protein biosynthesis during S phase. Cell. 1978 Jan;13(1):129–138. doi: 10.1016/0092-8674(78)90144-7. [DOI] [PubMed] [Google Scholar]
- Sapp M., Worcel A. Purification and mechanism of action of a nucleosome assembly factor from Xenopus oocytes. J Biol Chem. 1990 Jun 5;265(16):9357–9365. [PubMed] [Google Scholar]
- Seidman M. M., Levine A. J., Weintraub H. The asymmetric segregation of parental nucleosomes during chrosome replication. Cell. 1979 Oct;18(2):439–449. doi: 10.1016/0092-8674(79)90063-1. [DOI] [PubMed] [Google Scholar]
- Smith S., Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989 Jul 14;58(1):15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Stahl H., Koller T., Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986 May 5;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- Vassilev L. T., Burhans W. C., DePamphilis M. L. Mapping an origin of DNA replication at a single-copy locus in exponentially proliferating mammalian cells. Mol Cell Biol. 1990 Sep;10(9):4685–4689. doi: 10.1128/mcb.10.9.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn J. P., Dijkwel P. A., Hamlin J. L. Replication initiates in a broad zone in the amplified CHO dihydrofolate reductase domain. Cell. 1990 Jun 15;61(6):1075–1087. doi: 10.1016/0092-8674(90)90071-l. [DOI] [PubMed] [Google Scholar]













