Abstract
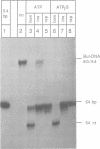
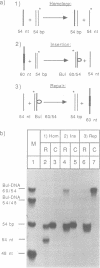
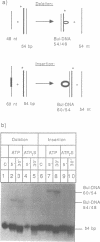
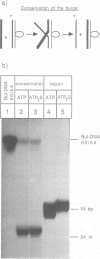
The role of ATP hydrolysis during the RecA-mediated recombination reaction is addressed in this paper. Recent studies indicated that the RecA-promoted DNA strand exchange between completely homologous double- and single-stranded DNA can be very efficient in the absence of ATP hydrolysis. In this work we demonstrate that the energy derived from the ATP hydrolysis is strictly needed to drive the DNA strand exchange through the regions where the interacting DNA molecules are not in a homologous register. Therefore, in addition to the role of the ATP hydrolysis in promoting the dissociation of RecA from the products of the recombination reaction, as described earlier, ATP hydrolysis also plays a crucial role in the actual process of strand exchange, provided that the lack of homologous register obstructs the process of branch migration.
Full text
PDF
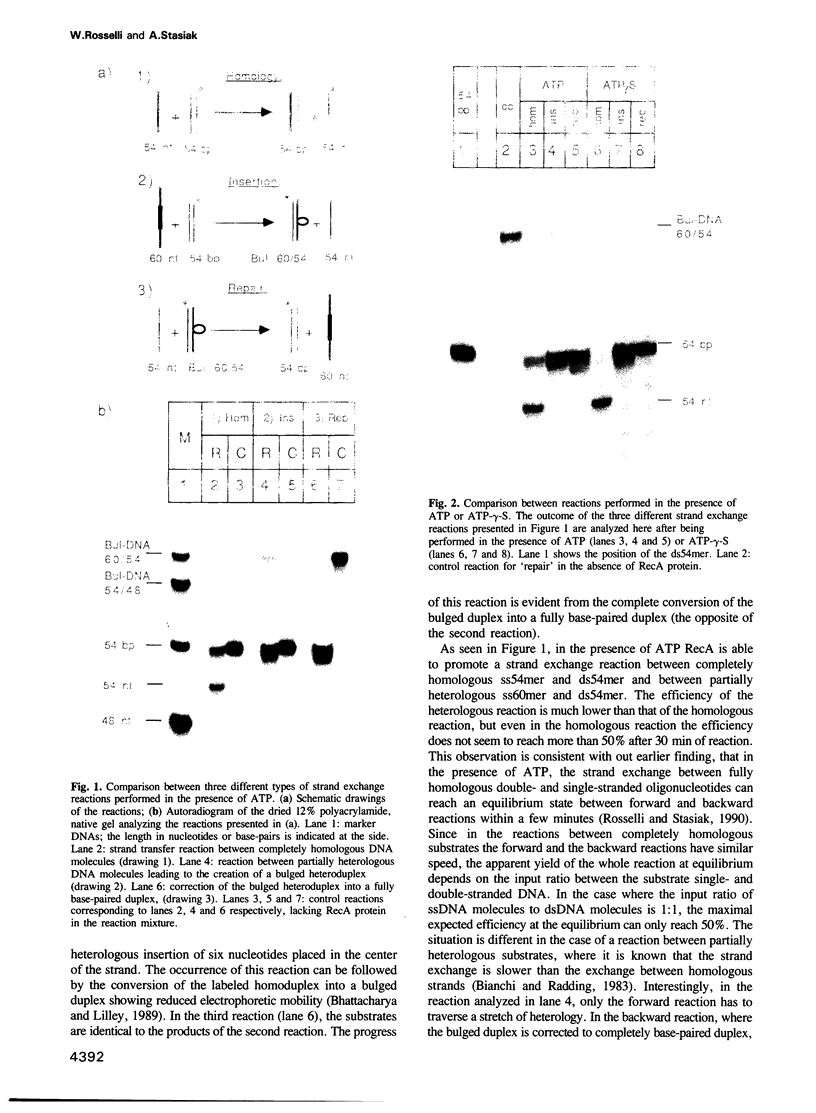
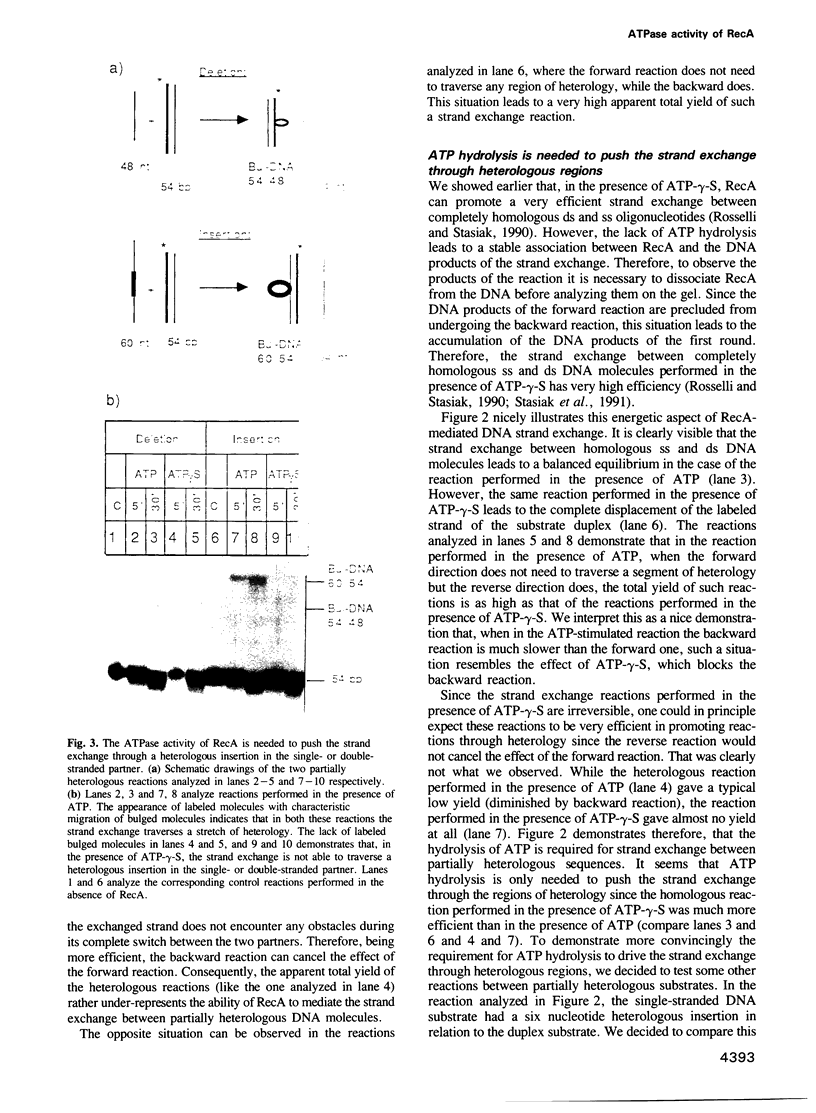



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhattacharyya A., Lilley D. M. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles). Nucleic Acids Res. 1989 Sep 12;17(17):6821–6840. doi: 10.1093/nar/17.17.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M. E., Radding C. M. Insertions, deletions and mismatches in heteroduplex DNA made by recA protein. Cell. 1983 Dec;35(2 Pt 1):511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Visualization of the paranemic joining of homologous DNA molecules catalyzed by the RecA protein of Escherichia coli. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2066–2070. doi: 10.1073/pnas.83.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. recA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., McEntee K., Lehman I. R. A simple and rapid procedure for the large scale purification of the recA protein of Escherichia coli. J Biol Chem. 1981 May 10;256(9):4676–4678. [PubMed] [Google Scholar]
- Honigberg S. M., Gonda D. K., Flory J., Radding C. M. The pairing activity of stable nucleoprotein filaments made from recA protein, single-stranded DNA, and adenosine 5'-(gamma-thio)triphosphate. J Biol Chem. 1985 Sep 25;260(21):11845–11851. [PubMed] [Google Scholar]
- Howard-Flanders P., West S. C., Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984 May 17;309(5965):215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu Rev Biophys Biophys Chem. 1991;20:539–575. doi: 10.1146/annurev.bb.20.060191.002543. [DOI] [PubMed] [Google Scholar]
- Menetski J. P., Bear D. G., Kowalczykowski S. C. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc Natl Acad Sci U S A. 1990 Jan;87(1):21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B., Koller T., Stasiak A. Characterization of the DNA binding activity of stable RecA-DNA complexes. Interaction between the two DNA binding sites within RecA helical filaments. J Mol Biol. 1990 Mar 5;212(1):97–112. doi: 10.1016/0022-2836(90)90307-8. [DOI] [PubMed] [Google Scholar]
- Register J. C., 3rd, Christiansen G., Griffith J. Electron microscopic visualization of the RecA protein-mediated pairing and branch migration phases of DNA strand exchange. J Biol Chem. 1987 Sep 15;262(26):12812–12820. [PubMed] [Google Scholar]
- Riddles P. W., Lehman I. R. The formation of plectonemic joints by the recA protein of Escherichia coli. Requirement for ATP hydrolysis. J Biol Chem. 1985 Jan 10;260(1):170–173. [PubMed] [Google Scholar]
- Roca A. I., Cox M. M. The RecA protein: structure and function. Crit Rev Biochem Mol Biol. 1990;25(6):415–456. doi: 10.3109/10409239009090617. [DOI] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. Physical map of the recA gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3144–3148. doi: 10.1073/pnas.76.7.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak A. Z., Rosselli W., Stasiak A. RecA-DNA helical filaments in genetic recombination. Biochimie. 1991 Feb-Mar;73(2-3):199–208. doi: 10.1016/0300-9084(91)90203-d. [DOI] [PubMed] [Google Scholar]
- Stasiak A., Stasiak A. Z., Koller T. Visualization of RecA-DNA complexes involved in consecutive stages of an in vitro strand exchange reaction. Cold Spring Harb Symp Quant Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- Tsang S. S., Chow S. A., Radding C. M. Networks of DNA and RecA protein are intermediates in homologous pairing. Biochemistry. 1985 Jun 18;24(13):3226–3232. doi: 10.1021/bi00334a023. [DOI] [PubMed] [Google Scholar]