Abstract
Objectives:
There are regions in the core gene of hepatitis B virus (HBV) where missense mutations are clustered, and mutations in that region are related to severe liver disease. However, there were some differences of the major regions for mutation clustering among ethnic groups. To explore the phenomenon of clustering mutations in Korean patients with chronic HBV infection and to elucidate the correlation between clustering mutation region of the core gene and the severity of liver damage, we analyzed the precore/core gene sequence of HBV in the sera from fifteen chronic hepatitis B (CH-B) patients.
Methods:
We analysed the HBV precore and core sequences in the sera obtained from fifteen patients (14 males and 1 female, mean age 30.0 years) with biopsy-proven CH-B. The patients were divided into two groups according to the pathological severity of CH-B; namely, group I consisted of 8 patients with chronic persistent hepatitis (CPH), and group II included 7 patients with chronic active hepatitis (CAH). After extraction of HBV DNA from each serum by proteinase K and phenol-chloroform solution, the entire precore and core region of HBV was amplified by PCR, and then the PCR products were subjected to direct sequencing using thermostable DNA polymerase. Fisher’s exact test and Mann-Whitney U test were used for statistical analysis.
Results:
A total of 181 nucleotide substitutions were found in the HBV core gene from the 15 CH-B patients, of which 23 were missense and 158 were silent. The nucleotide and amino acid substitution rates were not significantly different between the two groups (p>0.05). Two mutational hot spots (MHS), codons 21–34 (MHS1) and codons 85–100 (MHS2), were found in the deduced amino acid alignment of the core gene. The alteration rate of amino acid residue in these regions were 2.857×10−2 and 5.000×10−2, respectively. Of 8 CPH patients, 5 showed missense mutations only in MHS2. In comparison, of 7 CAH patients, 3 showed them both in MHS1 and MHS2, 1 only in MHS1, and 1 only in MHS2; thus, missense mutation in MHS1 was exclusively found in patient with CAH.
Conclusions:
There were two mutation clusterings in the core region of adr subtype of HBV from Korean CH-B patients. Mutations in MHS1 (codon 21–34), but not in MHS2 (codon 85–100), are more likely to be related to the severity of CH-B. A longitudinal study using sequential samples is warranted to further clarify the role of MHS1 in the pathogenesis of more severe CH-B.
Keywords: Hepatitis B virus, Core region, Mutation clustering, Chronic hepatitis B
INTRODUCTION
Chronic hepatitis B virus (HBV) infection might lead to a wide spectrum of liver injury ranging from chronic healthy HBsAg carrier to rapidly progressive decompensated liver cirrhosis. With advances in molecular biology technology, the role of HBV variants in the pathogenesis of such varied liver damage has been investigated; precore mutants of the HBV were first found in patients with severe liver disease after seroconversion to anti-HBe1–6) and later also in those with normal transaminase levels7–9); therefore, precore variant is no longer considered to be associated with the severity of chronic hepatitis B (CH-B).
Hepatic injury due to HBV is thought to be immune-mediated10). The HBcAg is the most probable immunological target of cytotoxic T cell (CTL)11). Studies on endogenously-processed viral peptides demonstrated that a peptide as small as 8 to 9 amino acids could be recognized by CTL12,13). Thus, it could be supposed that if there is ‘pressure’ from CTL, one might expect to find a mutation of the amino acid sequence in a restricted segment of the core gene. The studies from Japan and Taiwan have shown the clustering of missense mutations in the core gene14,15), though there were some differences of the major regions for mutation clustering between the two studies. These differences might originate from differing distributions of HLA antigens or from the varying amino acid sequences among HBV subtypes between the two countries15), and this proposal remains to be confirmed by more studies in different countries.
Korea is a hyperendemic area of HBV16). However, the role of HBV variants in the severity of liver damage in Korean patients with chronic HBV infection has not been assessed. To explore the phenomenon of clustering mutations in patients with chronic HBV infection and to elucidate the correlation between clustering mutation region of the core gene and the severity of liver damage, we analyzed the precore/core gene sequence of HBV in the sera from fifteen Korean patients with CH-B.
METHODS
1. Patients
We analysed the HBV precore and core sequences in the sera obtained from fifteen patients (14 males and 1 female, mean age 30.0 years) with biopsy-proven CH-B. The patients were divided into two groups according to the pathological severity of CH-B; namely, group I consisted of 8 patients with chronic persistent hepatitis (CPH), and group II included 7 patients with chronic active hepatitis (CAH). Serologic markers for HBV (HBsAg and HBeAg/anti-HBe) were detected by commercially available radioimmunoassay kits (Ausria II and HBe Kit, respectively; Abbott laboratories, North Chicago, IL, USA).
2. Extraction of DNA from Serum
Serum (200–300μl) obtained from each patient was incubated at 70°C for 3 hours in a mixture of proteinase K (100μg/ml), 0.5% (wt/vol) SDS, 5mM EDTA, and 10 mM Tris-HCl, pH 8.0. The solution was subjected to phenol-chloroform extraction, and DNA was precipitated with ethanol.
3. Amplification and Direct Sequencing of the Precore and Core Region of HBV
To amplify a segment of HBV DNA that constitutes the entire precore and core region, we prepared a set of synthetic oligonucleotide primers according to the HBV sequence reported by Kim et al.17) : a sense primer (nt 1604–1623, 5′-CATGGAGACC ACCGTGAACG-3′) and a antisense primer (nt 2526–2547, 5′-GTG AGGAAAGGAGGGAGTTTGC-3′). By these primers, a segment of HBV DNA spanning 944 nucleotide from 1604 to 2547, which comprises the entire precore and core region, was amplified by the polymerase chain reaction (PCR). These primers were synthesized by the phosphoramidite method. The PCR reaction was conducted in a 100μl mixture containing 10μl of the specimen DNA, 2.5 U of Taq polymerase (Perkin-Elmer Cetus, Norwalk, CT, USA), 400 mM of each dNTP, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2 and 0.5μ M of each primer. The reaction mixture, except each primer, was preheated to 85°C and each primer was added to the mixture. The amplification reaction was cycled 35 times with 30 seconds of denaturation at 94°C, 30 seconds of annealing at 55°C, and 1 minute of extension at 72°C in a programmable thermal cycler (GeneAmp 9600, Perkin-Elmer/Cetus).
We carefully followed the recommendation of Kwok and Higuchi to avoid false-positive PCR18).
For direct sequencing of a portion of HBV DNA, each PCR product was eluted from the 0.8% agarose gel slice, excised under the indirect ultraviolet visualization, by using the Magic™ PCR Prep kit (Promega, Madison, WI, USA).
We prepared several sequencing primers to sequence bi-directionally: sense primers, P1 (nt 1741–1758, 5′-GGGAGGAGATTAGGTTAA-3′), P2 (nt 1843–1867, 5′-CATGTTCATGTCCTACTGTTCA-3′), P3 (nt 2047–2071, 5′-CTCATCATACAGCACTCAGGCAAGC-3′), P4 (nt 2292–2310, 5′-TACAGACCACCAAATGCCC-3′); antisense primers, M1 (nt 2472–2497, 5′-AGAATAAAGCCCAGTAAAGTTTCCC-3′), M2 (nt 2287–2310, 5′-GGGCATTTGGTGGTCTGTA-3′), M3 (nt 2114–2138, 5′-TGCTGGGTCTTCCAAATTACTTCCC-3′), M4 (nt 2047–2067, 5′-GCCTGAGTGCT GTATGATGAG-3′), M5 (nt 1910–1934, 5′-AGAAGCTCCAAATTCTTTATACG-3′). Each sequencing primer was radiolabeled with γ-32P-ATP using T4 polynucleotide kinase. From 40 to 100 fmol of purified PCR product and 1.5 pmol of each radiolabeled primer was used for cycle sequencing reaction19) using thermostable DNA polymerase (fmol™ sequencing system, Promega, Madison, WI, USA). The reaction products were analyzed on a 8% sequencing gel. For ease of detection of mutations, we used a nontraditional gel-loading format20) in which all G-terminated reaction products from a single primer are loaded side-by-side, followed by all the A-terminated reactions, etc. (GGGGAAAATTTTCCCC) (Fig. 1). This format allowed mutations to be detected by virtue of the differences in the banding patterns, rather than by reading each sequence and comparing it to the known sequence. HBsAg subtypes in the fifteen patients were all adr. When four previously reported amino acid residues of the HBV with adr subtype17,21–23) were aligned, the sequences showed well conserved amino acid residues of core peptides and the only difference was in the codon 169. Since, in all patients, the deduced amino acid residue in the codon 169 was consistent with all 3 sequences reported in Japan, we used that sequence as a prototype HBV.
Fig. 1.
The nonstandard gel-loading format. Using the nonstandard gel-loding format, all sequences generated from a single primer were run side-by-side (GGGAAATTTCCC). The mutations were detected based on differences in the banding patterns. The arrowheads in the figure denote mutations. The standard gel-loading format is displayed on the left side for comparison.
4. Statistical Analysis
Fisher’s exact test and Mann-Whitney U test were used for statistical analysis.
RESULTS
1. Nucleotide Sequence and Deduced Amino Acid Residue of Core Gene
The entire nucleotide sequence of the core gene of HBV was obtained in all 15 patients. A total of 181 nucleotide substitutions were found in the HBV core gene from the 15 CH-B patients (101 from 8 CPH patients, 80 from 7 CAH patients). Twenty-three of them were missense mutations and 158 were silent. The substitution rate at each nucleotide position was 2.198×10−2 (2.230×10−2 for CPH patients and 2.082×10−2 for CAH patients), and the occurrence of missense mutation at each codon was 0.838×10−2 (0.683×10−2 for CPH patients and 1.015×10−2 for CAH patients); therefore, the nucleotide and amino acid substitution rates were not significantly different between the two groups (p>0.05). Six of the 8 CPH (75.0%) and six of the 7 CAH (87.1%) patients had at least one missense mutation in the HBV core gene (Table 1).
Table 1.
Clinical and Laboratory Data on 15 Patients with Chronic Hepatitis B
| No. | Age | Sex | HBeAg | Anti-HBe | Precore stop codon | Core mutation (missense) | Pathology |
|---|---|---|---|---|---|---|---|
| CP1 | 22 | M | + | − | − | − | CPH |
| CP2 | 21 | M | + | − | − | + | CPH |
| CP3 | 21 | M | + | − | − | + | CPH |
| CP4 | 21 | M | + | − | − | + | CPH |
| CP5 | 19 | M | − | + | − | + | CPH |
| CP6 | 20 | M | + | − | − | + | CPH |
| CP7 | 30 | M | + | − | − | + | CPH |
| CP8 | 20 | M | + | − | − | − | CPH |
| CA1 | 55 | M | + | − | − | − | CAH |
| CA2 | 54 | M | + | − | − | + | CAH |
| CA3 | 31 | M | + | − | − | + | CAH |
| CA4 | 19 | M | + | − | − | + | CAH |
| CA5 | 21 | M | + | − | − | + | CAH |
| CA6 | 57 | F | + | − | − | + | CAH |
| CA7 | 36 | M | + | − | − | + | LC, active |
2. Missense Mutation Clustering in Different Regions of HBV Core Gene
Two mutational hot spots were found in codons 21–34 (MHS1) and codons 85–100 (MHS2) in the deduced amino acid alignment of the HBV core gene (Fig. 2). The alteration rate of amino acid residue in these regions were 2.857×10−2 and 5.000×10−2, respectively. Of 8 CPH patients, 5 showed missense mutations only in MHS2. In comparison, of 7 CAH patients, 3 showed them both in MHS1 and MHS2, 1 only in MHS1 and 1 only in MHS2; thus, missense mutation in MHS1 was exclusively found in patients with CAH.
Fig. 2.
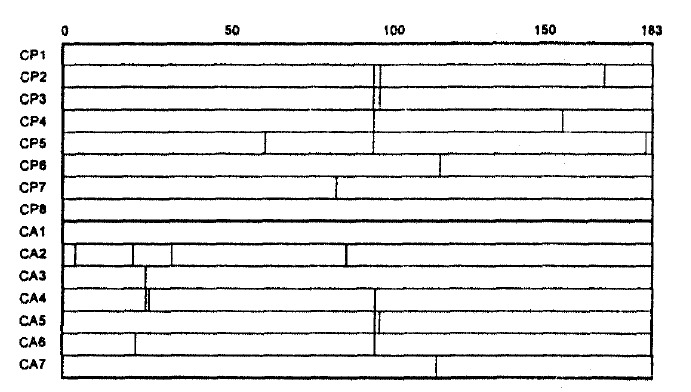
Deduced amino acid residues in 15 patients.
DISCUSSION
Viruses yield a high proportion of mutants on continuous replication in the absence of known mutagens. These spontaneous mutations accumulate in the genomes of the viruses, resulting in the variation of phenotypes (“viral quasispecies”)24,25), which subject the virus to selective pressure during evolution. When no variants with higher fitness are generated, the consensus (average) sequence of the population remains unchanged. By contrast, changes in environment of selective pressures lead to the emergence of variants with higher fitness for the new environment, which undergo further rounds of replication and become progeny virions. Such immune selection occurs not only with antibodies, but also with cytotoxic T cells26,27).
The in vivo mutation rate of hepadna virus was estimated to be lower than that of RNA viruses and higher than that of DNA viruses28). Presumably, such a high frequency of mutation is a result of HBV replication via reverse transcription of a RNA intermediate29). But there are constraints on the ability of HBV to accept mutations without becoming non-viable. The genome is only about 3200 bases long, the smallest of any DNA virus that infects man, and all the genetic material codes for proteins with the regulatory elements being found within these coding regions30).
It has been suggested that the core peptide is an immunological target of CTL in HBV infection11). Recent studies revealed that the endogenously processed viral peptides bound with the class 1 major histocompatibility complex (MHC) were recognized by CTL12). Indeed, the core gene of HBeAg-positive Japanese and Chinese patients with chronic hepatitis showed clustered amino acid substitutions or deletions14,15), probably as a result of immune pressure from CTL. However, the mutation clustering regions reported from different countries showed divergence. Mutation clusterings were found between amino acids 80 and 110 in Japanese patients14), whereas, in Chinese, those were found in codon 48–60, 84–101 and 147–15515). In the present study, we found that missense mutations in core gene of Korean patients with chronic active hepatitis were clustered in codon 21–34 and 85–100; therefore, we confirmed such an ethnic diversity of major regions of mutation clustering. It has not been defined what causes this diversity. A recent study of the amino acid sequences eluted from MHC molecules revealed that class I MHC had allele-specific binding motifs31). Thus, the major regions for mutation clustering could be different among chronic HBV-infected patients from different ethnic groups because of differing distributions of HLA antigens32–34) and the varying amino acids sequences among HBV subtypes.
It is not known why there are differences in the severity of liver disease among different HBV-infected individuals. In the present study, the number of missense mutations were not significantly different between the patients with CPH and CAH. Similarly, the occurrences of missense mutations in the MHS2 (codon 85–100), which is consistent with the MCR1 reported by Ehata et al.14), were not different between the two groups. In contrast, a mutation clustering region of 14 amino acid residues (MHS1; core codon from 21 to 34) was found in four of 7 patients (57.1%) with CAH, but in none of 8 (0.0%) CPH patients, suggesting that it is not the mutations in the MHS2 but those in the MHS1 that are more related to severity of CH-B. Actually, the mutation clustering region corresponding to MHS1 was also shown by Ehata et al.35) in four of seven (57.1%) Japanese patients with severe exacerbation of CH-B or fulminant hepatitis due to adr subtype of HBV.
Recently, Bertoletti et al. reported that the optimal amino acid sequence recognized by CTL from acute hepatitis B patients was a 10 mer (residues 18 to 27) containing the predicted peptide-binding motif for HLA-A2 and that this peptide could stimulate CTL that was able to recognize endogenously synthesized hepatitis B core antigen36). Interestingly, the MHS1 overlaps with the minimal optimal cytotoxic T-cell epitope. The missense mutations in MHS1 may result from the immune selection by CTL since the most frequent HLA-A phenotype in Korea is A2 (phenotype frequency=0.5392)32). However, it was also reported that patients with chronic HBV infection failed to mount an efficient CTL response to a synthetic HBV core antigen residue 11 to 2737). Furthermore, it is now clear that the different disease induced by HBV are caused by complex interaction between a particular viral genotype and the “quality” and perhaps “quantity” of the host’s immune response to that viral agent. Thus, to clarify the role of missense mutations in MHS1 in the pathogenesis of more severe CH-B, a longitudinal study with the use of sequential serum samples rather than a cross sectional study is warranted.
Footnotes
This work was supported in part by the Research Grant No. 1993-003 from Seoul National University Hospital and by a research grant from Liver Research Foundation of Korea (1993).
REFERENCES
- 1.Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, Thomas HC. Mutation preventing formation of hepatitis Be antigen in patients with CH-B infection. Lancet. 1989;2:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 2.Brunetto M, Stemler M, Schodel F, Bonino F. Identification of an HBV variant which cannot produce precore derived HBeAg and may be responsible for severe hepatitis. Ital J Gastroenterol. 1989;21:151–154. [Google Scholar]
- 3.Tong S, Li J, Vitvitski L, Trepo C. Active hepatitis B virus replication in the presence of anti-HBe is associated with viral variants containing an inactive pre-C region. Virology. 1990;176:596–603. doi: 10.1016/0042-6822(90)90030-u. [DOI] [PubMed] [Google Scholar]
- 4.Santantonio T, Jung M-C, Miska S, Pastore G, Pape GR, Will H. Prevalence and type of pre-C HBV mutants in anti-HBe positive carriers with chronic liver disease in a highly endemic area. Virology. 1991;183:840–844. doi: 10.1016/0042-6822(91)91022-9. [DOI] [PubMed] [Google Scholar]
- 5.Carman WF, Ferrao M, Lok ASF, Ma OCK, Lai CL, Thomas HC. Precore sequence variation in Chinese isolates of hepatitis B virus. J Infect Dis. 1991;165:127–133. doi: 10.1093/infdis/165.1.127. [DOI] [PubMed] [Google Scholar]
- 6.Naoumov NV, Schneider R, Grotzinger T, Jung MC, Miska S, Pape GR, Will H. Precore mutants hepatitis B virus infection and liver disease. Gastroenterology. 1992;102:538–543. doi: 10.1016/0016-5085(92)90101-4. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto H, Yotsumoto S, Akahane Y, Yamanaka T, Miyazaki Y, Sugai Y, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J Virol. 1990;64:1298–1303. doi: 10.1128/jvi.64.3.1298-1303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koh KC, Lee H-S, Kim CY. Universal emergence of precore mutant hepatitis B virus along with the seroconversion to anti-HBe irrespective of subsequent acitivity of chronic hepatitis B. Korean J Intern Med. 1994;9:61–66. doi: 10.3904/kjim.1994.9.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carman WF, McIntyre G, Hadziyannis S, Fattovich G, Alberti A, Thomas HC. Core protein evolution after selection of hepatitis B precore mutants and correlation with disease severity. In: Nishioka K, Suzuki H, Mishiro D, Oda T, editors. Viral Hepatitis and Liver Disease. Springer-Verlag; 1994. pp. 273–276. [Google Scholar]
- 10.Peters M, Vierling J, Gershwin ME, Milich D, Chisari FV, Hoofnagle JH. Immunology and the liver. Hepatology. 1991;13:977–994. [PubMed] [Google Scholar]
- 11.Ferrari C, Penna A, Glugerci T, Tong MJ, Ribera E, Fiaccadori F, Chisari FV. Intrahepatic, nucleocapsid antigen-specific T cell in chronic active hepatitis B. J Immunol. 1987;139:2050–2058. [PubMed] [Google Scholar]
- 12.Van Bleek GM, Nathenson SG. Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature. 1990;348:213–216. doi: 10.1038/348213a0. [DOI] [PubMed] [Google Scholar]
- 13.Rotzschke O, Falk K, Deres K, Shild H, Norda M, Metzger J, Jung G, Rammensee HG. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature. 1990;348:252–254. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- 14.Ehata T, Omata M, Yokosuka O, Hosoda K, Ohto M. Variations in codons 84–101 in the core nucleotide sequence correlate with hepatocellular injury in chronic hepatitis B virus infection. J Clin Invest. 1991;89:332–338. doi: 10.1172/JCI115581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuang W-L, Omata M, Ehata T, Yokosuka O, Ito Y, Imazeki F, Lu S-N, Chang W-Y, Ohto M. Precore mutations and core clustering mutations in chronic hepatitis B virus infection. Gastroenterology. 1993;104:263–271. doi: 10.1016/0016-5085(93)90861-6. [DOI] [PubMed] [Google Scholar]
- 16.Kim CY, Lee H-S, Yim KW. HBsAg+ and HBsAg- chronic active hepatitis, liver cirrhosis and hepatocellular carcinoma in Korea; Prevalence and difference in age distribution. Seoul J Med. 1992;33:127–131. [Google Scholar]
- 17.Kim KT, Hyun SW, Kim YS, Rho HM. Complete nucleotide sequence of hepatitis B virus (subtype adr) Korean J Biochem. 1988;21:319–331. [Google Scholar]
- 18.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 19.Carothers AM, Urlaub G, Mucha J, Grunberger D, Chasin LA. Point mutation analysis in a mammalian gene: Rapid preparation of total RNA. PCR amplfication of cDNA, and Taq sequencing by a novel method. Bio Techniques. 1989;7:494–499. [PubMed] [Google Scholar]
- 20.Hedden V, Callen W, Short JM, Kretz K. Improved sequence analysis of mutations identified with the Big Blue System: New protocol can be completed in 2 days. Strategies in Mol Biol. 1993;6:27–28. [Google Scholar]
- 21.Kobayashi M, Koike K. Complete nucleotide sequence of hepatitis B virus DNA of subtype adr and its conserved gene organization. Gene. 1984;30:227–232. doi: 10.1016/0378-1119(84)90124-0. [DOI] [PubMed] [Google Scholar]
- 22.Ono Y, Onda H, Sasada R, Igarachi K, Sugino Y, Nishioka K. The complete nucleotide sequences of the cloned hepatitis B virus DNA: subtype ade and adw. Nucleic Acids Res. 1983;11:1747–1757. doi: 10.1093/nar/11.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujiyama A, Miyanohara A, Nozaki C, Yoneyama T, Ohtomo N, Matsubara K. Cloning and structural analysis of hepatitis B virus DNAs, subtype adr. Nucleic Acids Res. 1983;11:4601–4610. doi: 10.1093/nar/11.13.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domingo E, Sabo D, Tanichi T, Weissmann C. Nuceotide sequence heterogeneity of a RNA phage population. Cell. 1978;13:735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- 25.Domingo E, Martinez-Salas E, Sobrino F, et al. The quasispecies (extremely heterogenous) nature of viral RNA genome populations: Biological relevance-a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 26.Pircher H, Moskophidis D, Rohrer U, Burki K, Hengartner H, Zinkernagel RM. Viral escape by selection of cytotoxic T cell-resistant virus variants in vivo. Nature. 1990;346:629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- 27.Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CRM, Rizza CR, McMichael AJ. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 28.Lee H-S, Ulrich P, Vyas GN. Mutations in the S-gene affecting the immunologic determinant of the envelope protein of hepatitis B virus. J Hepatol. 1991;13(Suppl 4):S97–101. doi: 10.1016/0168-8278(91)90035-a. [DOI] [PubMed] [Google Scholar]
- 29.Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 30.Miller RH, Kaneko S, Chung CT, Girones R, Purcell RH. Compact organization of the hepatitis B virus genome. Hepatology. 1989;9:322–327. doi: 10.1002/hep.1840090226. [DOI] [PubMed] [Google Scholar]
- 31.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motif revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 32.Kim T-G, Han H, Lim B-U, Kim W-I, Kim S-M. Distribution of HLA class I alleles and haplotypes in Korean. J Korean Med Sci. 1993;8:180–186. doi: 10.3346/jkms.1993.8.3.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumagai M. Genetic factors (HLA) and cellular immunity in the pathogenesis of asymptomatic HBsAg carrier. Act Hepatol Jpn. 1979;20:339–345. [Google Scholar]
- 34.Lin DY, Liaw YF, Chu CM, Chen GH. Histocompatibility profile of hepatoma in Taiwan. J Formosan Med Assoc (Taiwan I Hseuh Hui T sa Chih) 1982;81:1126–1131. [PubMed] [Google Scholar]
- 35.Ehata T, Omata M, Chuang W-L, Yokosuka O, Ito Y, Hosoda K, Ohto M. Mutations in the core nucleotide sequence of hepatitis B virus correlate with severe liver damage. In: Nishioka K, Suzuki H, Mishiro D, Oda T, editors. Viral Hepatitis and Liver Disease. Springer-Verlag; 1994. pp. 277–282. [Google Scholar]
- 36.Bertoletti A, Chisari FV, Penna A, Guilhot S, Galati L, Missale G, Fowler P, Schlicht H-J, Vitiello A, Chesnut RC, Fiaccadori F, Ferrari C. Definition of a minimal optimal cytotoxic T-cell epitope within the hepatitis B virus nucleocapsid protein. J Virol. 1993;67:2376–2380. doi: 10.1128/jvi.67.4.2376-2380.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penna A, Chisari FV, Bertoletti A, Missale G, Fowler P, Giuberti T, Fiaccadori F, Ferrari C. Cytotoxic T lymphocytes recognize an HLA-A2-restricted epitope within the hepatitis B virus nucleocapsid antigen. J Exp Med. 1991;174:1565–1570. doi: 10.1084/jem.174.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]


