Abstract
Lipomas are the most common soft tissue tumors in adults. They often carry chromosome aberrations involving 12q13~15 leading to rearrangements of the HMGA2 gene in 12q14.3, with breakpoints occurring within or outside of the gene. Here, we present eleven lipomas and one osteochondrolipoma with a novel recurrent chromosome aberration, t(12;18) (q14~15;q12~21). Molecular studies on eight of the tumors showed that full-length HMGA2 transcript was expressed in three and a chimeric HMGA2 transcript in five of them. In three lipomas and in the osteochondrolipoma, exons 1–3 of HMGA2 were fused to a sequence of SETBP1 on 18q12.3 or an intragenic sequence from 18q12.3 circa 10 kbp distal to SETBP1. In another lipoma, exons 1–4 of HMGA2 were fused to an intronic sequence of GRIP1 which maps to chromosome band 12q14.3, distal to HMGA2. The ensuing HMGA2 fusion transcripts code for putative proteins which contain amino acid residues of HMGA2 corresponding to exons 1–3 (or exons 1–4 in one case) followed by amino acid residues corresponding to the fused sequences. Thus, the pattern is similar to the rearrangements of HMGA2 found in other lipomas, i.e., disruption of the HMGA2 locus leaves intact exons 1–3 which encode the AT-hooks domains and separates them from the 3′-terminal part of the gene. The fact that the examined osteochondrolipoma had a t(12;18) and a HMGA2-SETBP1 fusion identical to the findings in the much more common ordinary lipomas, underscores the close developmental relationship between the two tumor types.
Keywords: lipoma, osteochondrolipoma, t(12;18)(q14~15;q12~21), recurrent chromosomal translocation, HMGA2-SETBP1, HMGA2 expression
Introduction
Lipomas are benign tumors composed of mature fat cells (1). It is the most common soft tissue tumor in adults with peak incidence between 40–60 years. They may appear at any site but with a broad distinction between subcutaneous (superficial) and deep-seated lesions. Although most lipomas are easily diagnosed, those occurring deep down (e.g., intramuscular lipoma, perineural lipoma) may be confused with liposarcomas. Though mitoses are rarely seen in histologic sections of lipomas, karyotypes can easily be obtained from short-term cultured tumor cells. In 1986, the first characteristic acquired chromosome aberration of lipomas was described, the translocation t(3;12)(q27~28;q13~15) (2,3). Since then, according to the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman database updated on February 13, 2015), 476 lipomas with chromosome aberrations have been reported, with involvement of chromosome bands 12q13~15 being seen in more than 300 of them. Recombination may take place with a wide variety of partners, but the by far most common is the translocation t(3;12)(q27~28;q14~15) which has been reported in 53 of the aberrant karyotypes registered in the Mitelman Database. Other recurrently involved chromosome segments found recombined with 12q13~15 in lipomas are 1p36 and 1p32~34 (each found in 29 cases), 2p22~24 (in 6 cases), 2q35~37 (10 cases), 5q33 (16 cases), 9p21~22 (7 cases) and 12p11~13 and 13q12~14 (8 cases each). The chromosome rearrangements often target the HMGA2 gene in 12q14.3 with breakpoints occurring both within and outside the locus; the essential outcome appears to be deregulation of HMGA2 with truncation of the gene as the critical event (4 and refs. therein). The most frequent translocation, t(3;12)(q27~28;q13~15), generates an HMGA2-LPP fusion gene coding for a transcription factor containing the AT-hook domain of HMGA2 and C-terminal LIM domains of LPP (5,6). HMGA2 has also been reported to form fusion genes with PPAP2 (at 1p32), ACKR3 (at 2q37), EBF1 (at 5q33), NFIB (at 9p22) and LHFP (at 13q12) (7–11). In the present study, we describe a novel recurrent chromosome translocation, t(12;18)(q14~15;q12~21) and its molecular consequences in 12 benign fat cell tumors.
Materials and methods
Ethics statement
The study was approved by the regional ethics committee (Regional komité for medisinsk forskning-setikk Sør-Øst, Norge, http://helseforskning.etikkom.no) and written informed consent was obtained from the patients.
Patients
Table I shows the patients' gender, age, diagnosis and the location of the tumor. All tumors were surgically removed.
Table I.
Clinical, cytogenetic and molecular data on the 12 benign fat cell tumors.
| Case no. | Gender/ age (years) | Diagnosis | Location | Karyotype | Cq, Ex1-Ex2 of HMGA2 | Cq, Ex4-Ex5 of HMGA2 | Cq of ACTB | HMGA2 fusion |
|---|---|---|---|---|---|---|---|---|
| 1 | M/58 | Lipoma | Intramuscular, left thigh | 46,XY,t(12;18)(q14~q15;q12~q21)[12]/46,XY[3] | 30.97 | 28.23 | 23.41 | Not done |
| 2 | F/44 | Lipoma | Intramuscular, right elbow | 46,XX,t(12;18)(q14~q15;q12~q21)[16] | 29.76 | 27.58 | 25.27 | Not done |
| 3 | M/54 | Lipoma | Intramuscular, right deltoid | 46,XY,t(12;18)(q14~q15;q12~q21)[12]/46,XY[3] | 28.58 | 36.84 | 24.33 | HMGA2-sequence from 18q12.3 |
| 4 | F/34 | Lipoma | Intramuscular, right deltoid | 46,XX,t(12;18)(q14~q15;q12~q21)[14] | 30.10 | 39.69 | 24.33 | HMGA2-SETBP1 |
| 5 | M/38 | Lipoma | Left thoracic wall | 46,XY,t(12;18)(q14~q15;q12~q21)[15] | 28.73 | 38.84 | 22.45 | HMGA2-GRIP1 |
| 6 | F/28 | Lipoma | Intramuscular, left thigh | 46,XX,t(12;18)(q14~q15;q12~q21)[14]/46,XX[1] | 32.57 | 30 | 22.96 | Not done |
| 7 | F/61 | Lipoma | Intramuscular, right splenius capitis muscle | 46,XX,t(12;18)(q14~q15;q12~q21)[12]/46,XX[3] | 26.28 | 39.36 | 23.67 | HMGA2-SETBP1 |
| 8 | M/55 | Osteochon-drolipoma | Intramuscular, subscapularis muscle | 46,XY,t(12;18)(q14~q15;q12~q21)[15] | 30.04 | 35.64 | 24.97 | HMGA2-SETBP1 |
| 9 | M/55 | Lipoma | Intramuscular, right infraspinatus muscle | 46,XY,t(2;18;12)(q37;q12~q21;q14~15)[9] | Not done | Not done | Not done | Not done |
| 10 | M/15 | Lipoma | Foot, right intrametatarsal | 46,XY,t(12;18)(q14~q15;q12~q21)[15] | Not done | Not done | Not done | Not done |
| 11 | M/64 | Lipoma | Right groin | 46,XY,t(8;9)(p21;q22),t(12;18)(q14~q15;q12~q21)[10] | Not done | Not done | Not done | Not done |
| 12 | M/56 | Lipoma | Intramuscular, left deltoid | 46,XY,t(12;18)(q14~q15;q12~q21)[5]/46,XY[5] | Not done | Not done | Not done | Not done |
Chromosome banding analysis
Samples from the operation specimens were mechanically and enzymatically disaggregated and short-term cultured as described elsewhere (12). The cultures were harvested and the chromosomes G-banded using Wright stain. The subsequent cytogenetic analysis and karyotype description followed the recommendations of the ISCN (13).
Total RNA isolation and cDNA synthesis
Tumor tissue adjacent to that used for cytogenetic analysis and histologic examination had been frozen and stored at −80°C from eight tumors (cases 1–8). Total RNA was extracted using miRNeasy kit, TissueLyser II homogenizer and Qiacube according to the manufacturer's recommendations (Qiagen Nordic, Stockholm, Sweden). For cDNA synthesis, 400–500 ng of total RNA were reverse-transcribed in a 20 μl reaction volume using iScript Advanced cDNA Synthesis kit for RT-qPCR according to the manufacturer's instructions (Bio-Rad Laboratories, Oslo, Norway). cDNA equivalent to 10 ng/μl of total RNA was used as template in subsequent real-time PCR assays. The Human Universal Reference Total RNA was used as control (Clontech Laboratories, Takara Bio Group, Saint-Germainen-Laye, France). According to the company's information, it is a mixture of total RNAs from a collection of adult human tissues, chosen to represent a broad range of expressed genes. Both male and female donors are represented.
Real-time PCR
Real-time PCR was carried out to determine the expression level of HMGA2. The TaqMan gene expression assays (Applied Biosystems, Foster City, CA, USA) Hs00171569_m1 (HMGA2 exons 1–2) and Hs00971725_m1 (HMGA2 exons 4–5) were used. The ACTB gene, assay Hs99999903_m1, was used as endogenous control. The 20 μl reaction volume contained 1× TaqMan Universal Master Mix II with UNG, lX of the 20× TaqMan Gene Expression Mix, and 1 μl cDNA (10 ng equivalent of RNA). Four replicates of each sample and the endogenous control were performed. Real-time PCR was run on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories). The thermal cycling included an initial step at 50°C for 2 min, followed by 10 min at 95°C, 40 cycles of 15 sec at 95°C and 1 min at 60°C. The data were analyzed using Bio-Rad CFX Manager software (Bio-Rad Laboratories).
3′-Rapid amplification cDNA ends (3′-RACE)
For 3′-RACE, 100 ng of total RNA were reverse-transcribed in a 20 μl reaction volume with the A3RNV-RACE primer (5′-ATC GTT GAG ACT CGT ACC AGC AGA GTC ACG AGA GAG ACT ACA CGG TAC TGG TTT TTT TTT TTT TTT-3′) using iScript Select cDNA Synthesis kit according to the manufacturer's instructions (Bio-Rad Laboratories). One microliter was used as template and amplified using the outer primer combination HMGA2-846F1 (5′-CCA CTT CAG CCC AGG GAC AAC CT-3′) and A3R-1New (5′-TCG TTG AGA CTC GTA CCA GCA GAG TCA C-3′). One microliter of the amplified products was used as template in nested PCR with the primers HMGA2-982F1 (5′-CAA GAG TCC CTC TAA AGC AGC TCA-3′) and A3R-3 (5′-CGA GAG AGA CTA CAC GGT ACT GGT-3′). For both PCRs, the 25 μl reaction volume contained 12.5 μl of Premix Taq (Takara Bio), template, and 0.4 μM of each of the forward and reverse primers. PCR cycling consisted of an initial step of denaturation at 94°C for 30 sec followed by 35 cycles of 7 sec at 98°C, 30 sec at 55°C, 90 sec at 72°C, and a final extension for 5 min at 72°C.
A total of 3 μl of the PCR products were stained with GelRed (Biotium, Hayward, CA, USA), analyzed by electrophoresis through 1.0% agarose gel and photographed. The rest of the amplified fragments were purified using the Thermo Scientific GeneJET PCR purification kit (Thermo Fisher Scientific, Oslo, Norway) and direct sequencing was performed using the light run sequencing service of GATC Biotech (http://www.gatc-biotech.com/en/sanger-services/lightrun-sequencing.html). The BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) programs were used for computer analysis of sequence data.
Reverse transcriptase PCR (RT-PCR)
To verify the results obtained by 3′-RACE, i.e., the presence of HMGA2-chimeric transcript (see below), PCRs were performed using the forward primer HMGA2-846F1 and the reverse primer SETBP1-5390R1 (5′-GCA GCG TGA GGT CAG GAG AGT GC-3′) for cases 4 and 7. The primers HMGA2-846F1 and SETBP1-5325F1 (5′-GGC GCT TCA GTA CGG CTG GAT C-3′) were used for case 8. For case 3, the primer HMGA2-846F1 and the reverse primer 18q21-Rev1 (5′-GCA TTG GCA GTC CCC TTG CAT T -3′) were used. For case 5, the primer HMGA2-846F1 and the reverse primer GRIP-intrR1 (5′-TTA AGG TGT GGC CTT TAG GCG TGA C-3′) were used. The 25 μl PCR volumes contained 12.5 μl of Premix Taq (Takara Bio), 1 μl of diluted cDNA (10 ng equivalent of RNA), and 0.4 μM of each of the forward and reverse primers. The PCRs were run on a C-1000 Thermal cycler (Bio-Rad Laboratories). The PCR conditions were: an initial denaturation at 94°C for 30 sec followed by 35 cycles of 7 sec at 98°C, 120 sec at 68°C, and a final extension for 5 min at 68°C.
Results
Pathology and cytogenetics
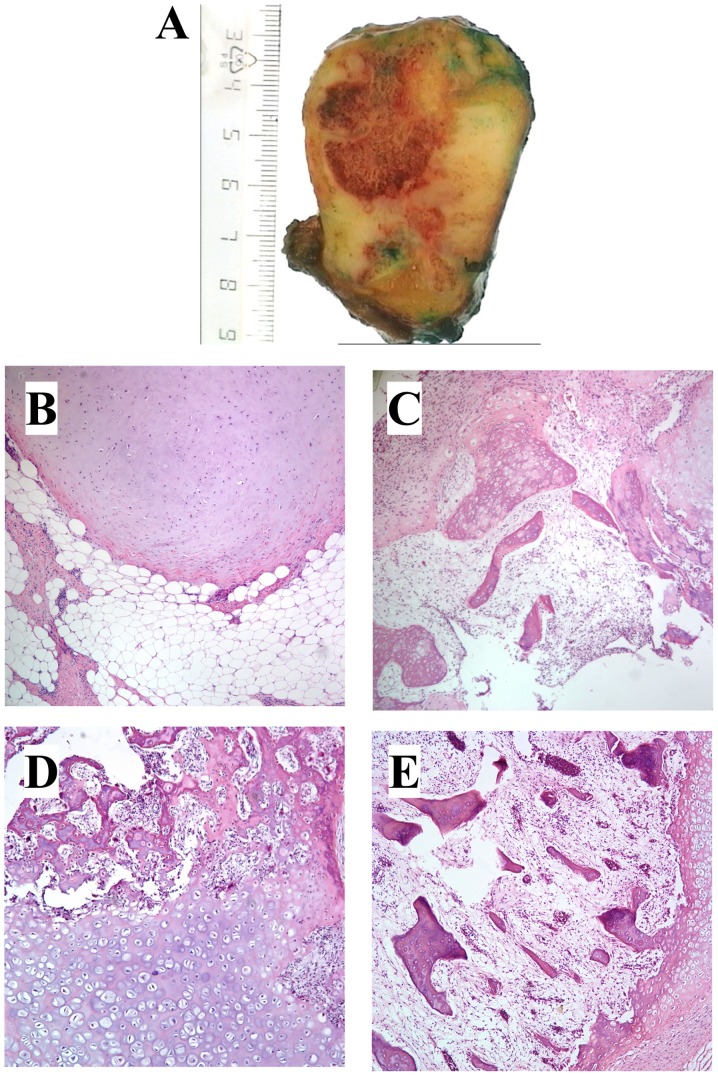
All tumors were ordinary lipomas except case 8, which was an osteochondrolipoma (Table I and Fig. 1), a rare variant of lipoma with metaplastic components of cartilage and bone within the fatty tissue. In this tumor, microscopic examination showed lobules of mature fat separated by strands of fibrous tissue and areas with hyaline cartilage. Other areas showed bony trabeculae with bone marrow between them (Fig. 1). There was no cellular atypia. Reactive changes with prominent lymphoid infiltrates in the bone marrow were seen, and in the fatty tissue and fibrous septa there were mature lymphocytes and some plasma cells (Fig. 1).
Figure 1.
Macroscopical and microscopical examination (H&E stained slides) of the osteochondrolipoma. (A) Cut surface of the osteochondrolipoma with macroscopically visible fatty tissue, cartilage and bone. (B) Mature cartilage nodule within fatty tissue. (C) Mature cartilage with bone formation and fatty tissue with lymphoid infiltration. (D) Mature cartilage and bone. (E) Mature cartilage and bone with bone marrow elements.
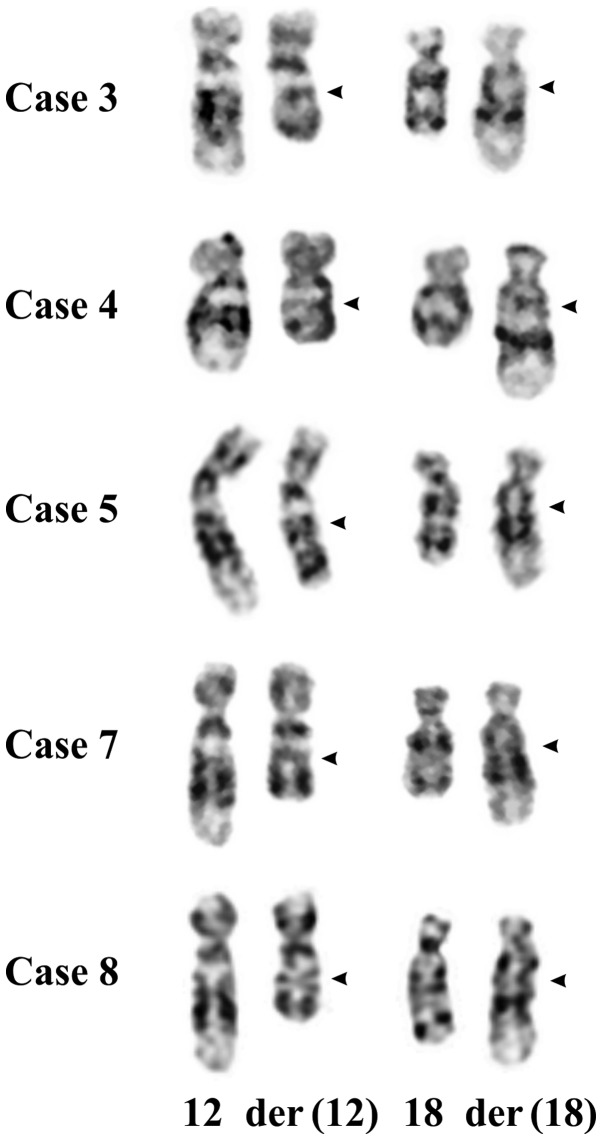
In all 12 cases, 8 males and 4 females (Table I), the tumor cells showed cytogenetic recombination between chromosome bands 12q14~15 and 18q12~21 (this was how the cases were selected). Nine lipomas and the osteochondrolipoma (case 8) had a reciprocal t(12;18)(q14~15;q12~21) as the sole karyotypic aberration, one lipoma carried a three-way translocation, t(2;18;12)(q37;q12~21;q14~15), and another tumor had a t(8;9) (p21;q22) in addition to t(12;18). Partial karyotypes of the t(12;18)(q14~15;q12~21) are shown in Fig. 2.
Figure 2.
Partial karyotypes of cases 3–5, 7 (lipomas), and 8 (osteochondrolipoma) showing the der(12)t(12;18)(q14~15;q12~21) and der(18)t(12;18) (q14~15;q12~21) together with the corresponding normal chromosome homologs; breakpoint positions are indicated by arrowheads.
Real-time PCR
Real-time PCR was performed with two commercially available TaqMan assays for the HMGA2 gene, one assay for exons 1–2 and the other for exons 4–5, in order to find out whether HMGA2 was rearranged. Because most HMGA2 rearrangements take place in intron 3 of the gene, the result is an overexpression of exons 1–3 of HMGA2. Thus, the mean quantification cycle (Cq) was compared between the two assays (Table I). Similar Cq values for both assays indicate that both 5′-end (exons 1–2) and 3′-end (exons 4–5) are equally expressed and the HMGA2 locus is most probably not rearranged. A difference in the Cq values between assays for exons 1–2 and exons 4–5, with the former having a lower Cq than the latter, indicates that 5′-end and 3′-end HMGA2 exons are unequally expressed and that HMGA2 is most probably rearranged. In three lipomas (cases 1, 2 and 6), similar Cq values between assays for exons 1–2 and exons 4–5 were found (Table I). In four lipomas and in the osteochondrolipoma (cases 3–5, 7 and 8), on the other hand, the Cq values for exons 4–5 were significantly lower than those for exons 1–2, indicating rearrangement of HMGA2 (Table I).
3′-RACE
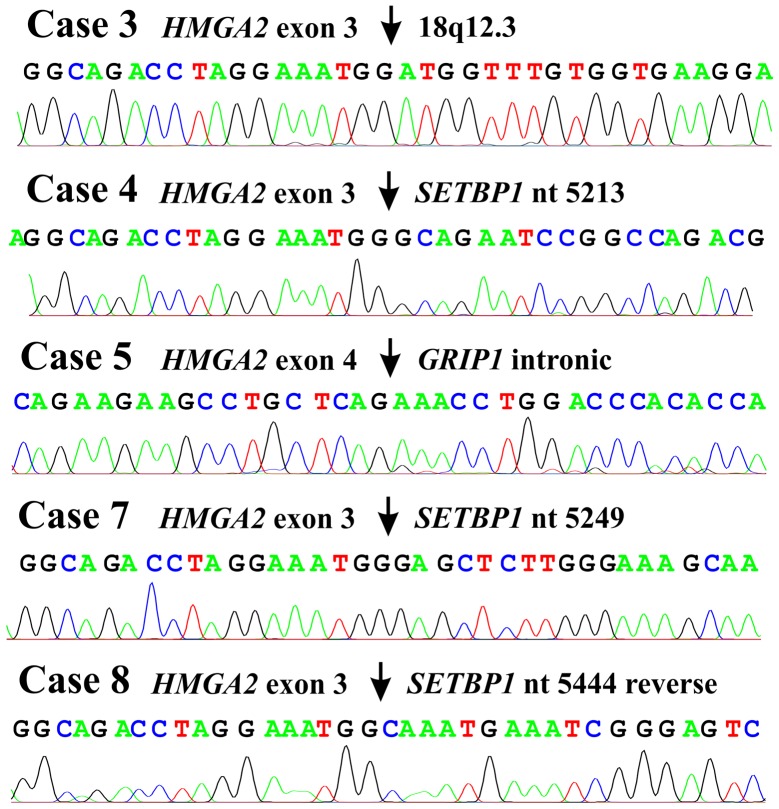
3′-RACE on lipomas (cases 3–5 and 7) and the osteochondrolipoma (case 8) (Table I) amplified fragments which by Sanger sequence analysis were found to be chimeric HMGA2-cDNA fragments. In lipomas 3, 4 and 7 as well as the osteochondrolipoma, exon 3 of HMGA2 was fused to sequences from 18q12.3 (Fig. 3) which in lipomas 4 and 7 and the osteochondrolipoma consisted of the 3′-untranslated region of the SETBP1 gene. In lipoma 3, exon 3 of HMGA2 was fused with a sequence 10 kbp downstream of the SETBP1 gene (Fig. 3). In lipoma 5, exon 4 of HMGA2 was fused to a sequence 500 Mbp distal to HMGA2 in an intron of GRIP1 in 12q14.3 (Fig. 3).
Figure 3.
Partial sequence chromatogram of 3′-RACE amplified cDNA fragment showing (arrow) the fusions of HMGA2.
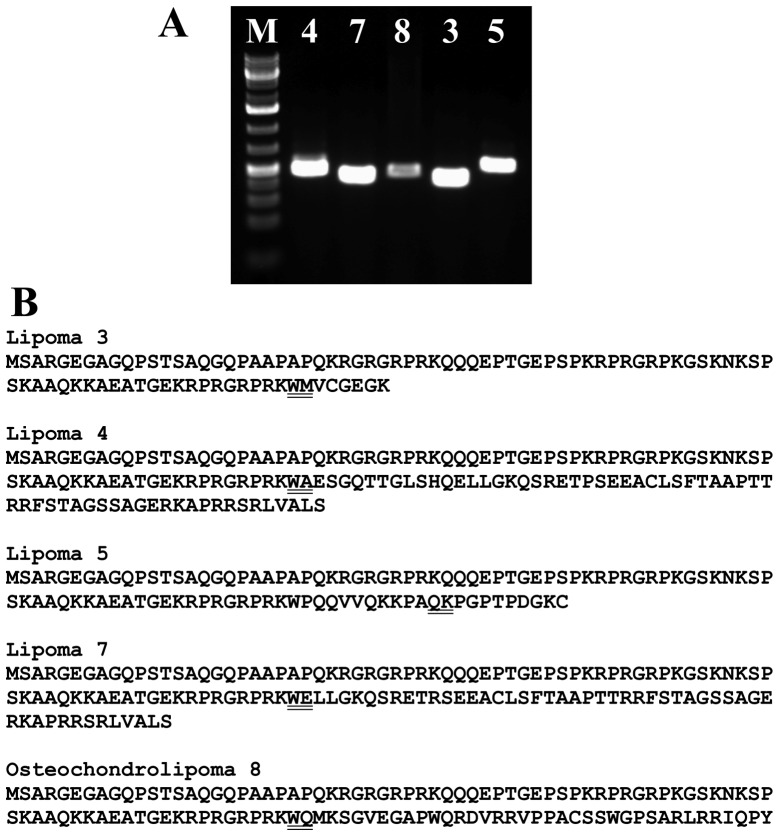
PCR with the primer HMGA2-846F1 and specific reverse primers amplified a cDNA fragment (Fig. 4A) which by direct sequencing was shown to display the same fusion point as the 3′-RACE amplified fragment (Fig. 3).
Figure 4.
(A) RT-PCR results for the expression of HMGA2-fusion in lipomas (cases 3–5 and 7) and osteochondrolipoma (case 8). PCRs were run using the forward primer HMGA2-846F1 and the reverse primer SETBP1-5390R1 for lipomas 4 and 7, primers HMGA2-846F1 and 18q21-Rev1 for case 3, primers HMGA2-846F1 and SETBP1-5325F1 for the osteochondrolipoma, and primers HMGA2-846F1 and GRIP-intrR1 for the lipoma of case 5. M, 1 kbp DNA ladder (GeneRuler; Fermentas, Vilnius, Lithuania). (B) The putative proteins encoded by the HMGA2-fusion transcripts.
Discussion
We describe here a new recurrent chromosome translocation, t(12;18)(q14~15;q12~21), in lipomas. According to the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman, Database last updated on February 13, 2015), there are 13 cases with aberration involving breakpoints on the long arm of chromosome 18 but none of them had t(12;18) (q14~15;q12~21). Whether some of these cases could nevertheless be cytogenetically similar to the tumors of our series, is a possibility we cannot test.
Because changes of chromosomal bands 12q13~15 in lipomas are almost always associated with rearrangement and/or activation of HMGA2 (14), we decided to investigate further the possible involvement of HMGA2 also in the present cases. The real-time PCR and 3′-RACE experiments showed that HMGA2 was expressed as a chimeric HMGA2 transcript in five cases. In four lipomas, exons 1–3 of HMGA2 were fused to a sequence of SETBP1 (cases 4, 7 and 8) or an intragenic sequence from 18q12.3 (case 3) 10 kbp distal to SETBP1. In one tumor (case 5), the translocation t(12;18) resulted in fusion of exons 1–4 of HMGA2 with an intronic sequence of GRIP1 which also maps to chromosome band 12q14.3. The ensuing HMGA2 fusion transcripts code for putative proteins which contain amino acid residues 1–83 of HMGA2 (accession number NP_003474.1) corresponding to exons 1–3 of the gene and amino acid residues from the fused sequences (cases 3, 4, 7 and 8) or, in the tumor of case 5, 94 amino acid residues are translated from exons 1–4 of HMGA2 and joined with residues from a fused sequence from chromosome 12 (Fig. 4B). Thus, the pattern is similar to that seen in other rearrangements of HMGA2 found in lipomas, i.e., disruption of the HMGA2 locus leaves intact exons 1–3 of the gene which encode the AT-hook domains and separates them from the 3′-terminal part of the gene (15). The biological activity of HMGA2 polypeptides in which the acidic tail has been replaced by a variable number of seemingly random amino acid residues, has not been studied in detail. The present data nevertheless add to the evidence that expression of truncated HMGA2 is involved in the development of lipomas. Fedele et al (16) showed that the expression of a truncated form of HMGA2 protein carrying only the three DNA-binding domains, or the expression of a fusion protein carrying the three DNA-binding domains of HMGA2 and the LIM domains of the lipoma preferred partner gene (LPP) protein, caused transformation of NIH3T3 cells. The acquisition of LPP ectopic sequences did not increase the transforming ability of the truncated form of HMGA2 (16). Moreover, transgenic mice expressing a truncated form of the HMGA2 protein develop obesity and an abnormally high prevalence of lipomas (17,18).
The real-time PCR results indicated that full length HMGA2 transcript was expressed in lipomas 1, 2 and 6 (Table I). In addition, FISH showed that the breakpoint was distal to the HMGA2 locus in lipomas 1 and 2 (data not shown). Similar data have been reported for lipomas carrying the t(5;12)(q32~33;q14~15) (9,19) where FISH studies (9) have shown that the genomic breakpoints usually lie outside the HMGA2 locus. Subsequently, Bartuma et al (19) showed that 4 out of 5 examined lipomas with t(5;12) had aberrant expression of the entire HMGA2 gene. Similar findings have also been reported for uterine leiomyomas with rearrangements involving chromosome band 12q15 (20): A study of 38 uterine leiomyomas showed that dysregulation of HMGA2 expression, not the formation of HMGA2 fusion transcripts, was the principal pathobiological mechanism in these tumors.
The recurrent HMGA2 partner gene, SETBP1, codes for a protein which contains six regions rich in proline (P), glutamine (E), serine (S), and threonine (T) residues (PEST sequences), three nuclear localization signals, three sequential proline-rich repeats PPLPPPPP at the carboxyl-terminal end, a region homologous to the SKI oncoprotein, and a SET binding region (21). The encoded protein has been shown to interact specifically with the SET protein both in a yeast two-hybrid system and in human cells (21). Constitutional mutations in this gene are associated with Schinzel-Giedion midface retraction syndrome (22). SETBP1 is otherwise involved in hematologic malignancies (23–25). The oncogenic function of SETBP1 was first reported in 2006 when a NUP98-SETBP1 fusion gene was identified in T cell acute lymphoblastic leukemia carrying a t(11;18)(p15;q12) (24). In 2010, SETBP1 was found to be overexpressed in a case of AML cytogenetically characterized by a t(12;18)(p13;q12) targeting ETV6 in the second breakpoint (23). The same study also showed that SETBP1 overexpression is a recurrent molecular event in AML (found in 53 of 192 patients) and is associated with shorter overall survival, especially in elderly patients (23). Recently, recurrent mutations in SETBP1, frequently targeting the SKI-homologous domain, have been identified in several types of myeloid malignancies, including chronic and acute myeloid leukemias (25 and refs. therein). In the three HMGA2-SETBP1 fusions described here, the breakpoint in SETBP1 occurred in the 3′-untranslated region (3′-UTR). This 3′-UTR is 4.8 kbp long, or almost half the size of the 9.9 kbp long transcript mRNA of SETBP1 (sequence with the accession number NM_015559 version 2). The function of this 3′-UTR is unknown but presumably it contains sequences that influence the expression of SETBP1 (26).
Case 8, which also had a HMGA2-SETBP1 fusion, was diagnosed as osteochondrolipoma (Fig. 1). This is a very rare type of tumor on which no prior cytogenetic or molecular genetic information exists. A search in ‘PubMed' using the term ‘osteochondrolipoma' yielded 6 articles (27–32). As the name indicates, the tumor is characterized by the presence of mature fatty tissue together with cartilage and bone formation with lipocytes, chondrocytes, and osteoblasts being the predominant cell types (31). The pathogenesis of osteochondrolipomas is unknown. Lin et al (33) reported that mesenchymal stem cells (MSCs) may be found in human lipomas and that they have characteristics similar to those of MSCs derived from adipose tissue. Thus, lipoma-derived MSCs can differentiate into adipocytes, osteoblasts, and chondrocytes after induction (33) and could be the source of the different cell types in these tumors. The present case with a t(12;18)(q14~15;q12~21) and an HMGA2-SETBP1 fusion identical to those found in the ordinary lipomas further supports the association both pathogenetically and otherwise between osteochondrolipomas and other lipoma subtypes. Worthy of mention is that the reciprocal translocation t(3;12)(q27~28;q13~15), i.e., the most common translocation in lipomas, has also been observed in three cases of osteolipoma (34). Evidently, the special phenotypes of these lipoma variants cannot be attributed to the tumor pathogenetic mechanism.
The present study provides yet another example of the fact that what at the cytogenetic level appears to be similar is in fact heterogeneous at the molecular level (9,19). This may reflect the intriguing pathogenetic role of HMGA2, which seems to be entirely different from the highly specific gene fusions present in, for example, myxoid liposarcomas (14).
Acknowledgements
The authors thank Hege Kilen Andersen and Kristin Andersen for technical help. The present study was supported by grants from the Norwegian Cancer Society and the Norwegian Radium Hospital Foundation.
References
- 1.Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F. WHO Classification of Tumours of Soft Tissue and Bone. 4th edition. IARC Press; Lyon: 2013. [Google Scholar]
- 2.Heim S, Mandahl N, Kristoffersson U, Mitelman F, Rööser B, Rydholm A, Willén H. Reciprocal translocation t(3;12) (q27;q13) in lipoma. Cancer Genet Cytogenet. 1986;23:301–304. doi: 10.1016/0165-4608(86)90012-9. [DOI] [PubMed] [Google Scholar]
- 3.Turc-Carel C, Dal Cin P, Rao U, Karakousis C, Sandberg AA. Cytogenetic studies of adipose tissue tumors. I. A benign lipoma with reciprocal translocation t(3;12)(q28;q14) Cancer Genet Cytogenet. 1986;23:283–289. doi: 10.1016/0165-4608(86)90010-5. [DOI] [PubMed] [Google Scholar]
- 4.Bartuma H, Hallor KH, Panagopoulos I, Collin A, Rydholm A, Gustafson P, Bauer HC, Brosjö O, Domanski HA, Mandahl N, et al. Assessment of the clinical and molecular impact of different cytogenetic subgroups in a series of 272 lipomas with abnormal karyotype. Genes Chromosomes Cancer. 2007;46:594–606. doi: 10.1002/gcc.20445. [DOI] [PubMed] [Google Scholar]
- 5.Petit MM, Mols R, Schoenmakers EF, Mandahl N, Van de Ven WJ. LPP, the preferred fusion partner gene of HMGIC in lipomas, is a novel member of the LIM protein gene family. Genomics. 1996;36:118–129. doi: 10.1006/geno.1996.0432. [DOI] [PubMed] [Google Scholar]
- 6.Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- 7.Bianchini L, Birtwisle L, Saâda E, Bazin A, Long E, Roussel JF, Michiels JF, Forest F, Dani C, Myklebost O, et al. Identification of PPAP2B as a novel recurrent translocation partner gene of HMGA2 in lipomas. Genes Chromosomes Cancer. 2013;52:580–590. doi: 10.1002/gcc.22055. [DOI] [PubMed] [Google Scholar]
- 8.Broberg K, Zhang M, Strömbeck B, Isaksson M, Nilsson M, Mertens F, Mandahl N, Panagopoulos I. Fusion of RDC1 with HMGA2 in lipomas as the result of chromosome aberrations involving 2q35–37 and 12q13–15. Int J Oncol. 2002;21:321–326. [PubMed] [Google Scholar]
- 9.Nilsson M, Mertens F, Höglund M, Mandahl N, Panagopoulos I. Truncation and fusion of HMGA2 in lipomas with rearrangements of 5q32→q33 and 12q14→q15. Cytogenet Genome Res. 2006;112:60–66. doi: 10.1159/000087514. [DOI] [PubMed] [Google Scholar]
- 10.Nilsson M, Panagopoulos I, Mertens F, Mandahl N. Fusion of the HMGA2 and NFIB genes in lipoma. Virchows Arch. 2005;447:855–858. doi: 10.1007/s00428-005-0037-9. [DOI] [PubMed] [Google Scholar]
- 11.Petit MM, Schoenmakers EF, Huysmans C, Geurts JM, Mandahl N, Van de Ven WJ. LHFP, a novel translocation partner gene of HMGIC in a lipoma, is a member of a new family of LHFP-like genes. Genomics. 1999;57:438–441. doi: 10.1006/geno.1999.5778. [DOI] [PubMed] [Google Scholar]
- 12.Mandahl N. Methods in solid tumour cytogenetics. In: Rooney DE, editor. Human Cytogenetics: Malignancy and Acquired Abnormalities. Oxford University Press; New York: 2001. pp. 165–203. [Google Scholar]
- 13.Schaffer LG, Slovak ML, Campbell LJ. ISCN 2009: An International System for Human Cytogenetic Nomenclature. Karger; Basel: 2009. [Google Scholar]
- 14.Heim S, Mitelman F. Cancer Cytogenetics. 3rd Edition. Wiley-Blackwell; New York, NY: 2009. [Google Scholar]
- 15.Cleynen I, Van de Ven WJ. The HMGA proteins: A myriad of functions (Review) Int J Oncol. 2008;32:289–305. [PubMed] [Google Scholar]
- 16.Fedele M, Berlingieri MT, Scala S, Chiariotti L, Viglietto G, Rippel V, Bullerdiek J, Santoro M, Fusco A. Truncated and chimeric HMGI-C genes induce neoplastic transformation of NIH3T3 murine fibroblasts. Oncogene. 1998;17:413–418. doi: 10.1038/sj.onc.1201952. [DOI] [PubMed] [Google Scholar]
- 17.Arlotta P, Tai AK, Manfioletti G, Clifford C, Jay G, Ono SJ. Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J Biol Chem. 2000;275:14394–14400. doi: 10.1074/jbc.M000564200. [DOI] [PubMed] [Google Scholar]
- 18.Battista S, Fidanza V, Fedele M, Klein-Szanto AJ, Outwater E, Brunner H, Santoro M, Croce CM, Fusco A. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 1999;59:4793–4797. [PubMed] [Google Scholar]
- 19.Bartuma H, Panagopoulos I, Collin A, Trombetta D, Domanski HA, Mandahl N, Mertens F. Expression levels of HMGA2 in adipocytic tumors correlate with morphologic and cytogenetic subgroups. Mol Cancer. 2009;8:36. doi: 10.1186/1476-4598-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quade BJ, Weremowicz S, Neskey DM, Vanni R, Ladd C, Dal Cin P, Morton CC. Fusion transcripts involving HMGA2 are not a common molecular mechanism in uterine leiomyomata with rearrangements in 12q15. Cancer Res. 2003;63:1351–1358. [PubMed] [Google Scholar]
- 21.Minakuchi M, Kakazu N, Gorrin-Rivas MJ, Abe T, Copeland TD, Ueda K, Adachi Y. Identification and characterization of SEB, a novel protein that binds to the acute undifferentiated leukemia-associated protein SET. Eur J Biochem. 2001;268:1340–1351. doi: 10.1046/j.1432-1327.2001.02000.x. [DOI] [PubMed] [Google Scholar]
- 22.Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, de Vries P, de Reuver R, Wieskamp N, Mortier G, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 23.Cristóbal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J, Novo FJ, Bandres E, Calasanz MJ, Bernabeu C, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115:615–625. doi: 10.1182/blood-2009-06-227363. [DOI] [PubMed] [Google Scholar]
- 24.Panagopoulos I, Kerndrup G, Carlsen N, Strömbeck B, Isaksson M, Johansson B. Fusion of NUP98 and the SET binding protein 1 (SETBP1) gene in a paediatric acute T cell lymphoblastic leukaemia with t(11;18)(p15;q12) Br J Haematol. 2007;136:294–296. doi: 10.1111/j.1365-2141.2006.06410.x. [DOI] [PubMed] [Google Scholar]
- 25.Trimarchi T, Ntziachristos P, Aifantis I. A new player SETs in myeloid malignancy. Nat Genet. 2013;45:846–847. doi: 10.1038/ng.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 27.Ahmadi SA, van Landeghem FK, Blechschmidt C, Lieber K, Haberl EJ, Thomale UW. Intratentorial osteochondrolipoma in a 9-year-old boy. J Neurosurg Pediatr. 2009;3:386–391. doi: 10.3171/2009.1.PEDS08237. [DOI] [PubMed] [Google Scholar]
- 28.Gru AA, Santa Cruz DJ. Osteochondrolipoma: A subcutaneous lipoma with chondroid and bone differentiation of the chest wall. J Cutan Pathol. 2012;39:461–463. doi: 10.1111/j.1600-0560.2011.01855.x. [DOI] [PubMed] [Google Scholar]
- 29.Gültekin SE, Kahraman S, Karadayı K. Parosteal osteochondrolıpoma of the mandıble. J Oral Maxillofac Pathol. 2012;16:280–282. doi: 10.4103/0973-029X.99090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishio J, Ideta S, Iwasaki H, Naito M. Scapular osteochondrolipoma: Imaging features with pathological correlation. Oncol Lett. 2013;6:817–820. doi: 10.3892/ol.2013.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rau T, Soeder S, Olk A, Aigner T. Parosteal lipoma of the thigh with cartilaginous and osseous differentiation: An osteochondrolipoma. Ann Diagn Pathol. 2006;10:279–282. doi: 10.1016/j.anndiagpath.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Soulard R, Nguyen AT, Souraud JB, Oddon PA, Fouet B, Cathelinaud O. Osteochondrolipoma of the submandibular region: A case report and review of the literature. Head Neck Pathol. 2012;6:486–491. doi: 10.1007/s12105-012-0358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin TM, Chang HW, Wang KH, Kao AP, Chang CC, Wen CH, Lai CS, Lin SD. Isolation and identification of mesenchymal stem cells from human lipoma tissue. Biochem Biophys Res Commun. 2007;361:883–889. doi: 10.1016/j.bbrc.2007.07.116. [DOI] [PubMed] [Google Scholar]
- 34.Fritchie KJ, Renner JB, Rao KW, Esther RJ. Osteolipoma: Radiological, pathological, and cytogenetic analysis of three cases. Skeletal Radiol. 2012;41:237–244. doi: 10.1007/s00256-011-1241-0. [DOI] [PubMed] [Google Scholar]