Abstract
Alzheimer's disease (AD), the most common age-dependent neurodegenerative disorder, produces a progressive decline in cognitive function. The metabolic mechanism of AD has emerged in recent years. In this study, we used multivariate analyses of gas chromatography-mass spectrometry measurements to determine that learning and retention-related metabolic profiles are altered during aging in the hippocampus of the senescence-accelerated mouse prone 8 (SAMP8). Alterations in 17 metabolites were detected in mature and aged mice compared to young mice (13 decreased and 4 increased metabolites), including metabolites related to dysfunctional lipid metabolism (significantly increased cholesterol, oleic acid, and phosphoglyceride levels), decreased amino acid (alanine, serine, glycine, aspartic acid, glutamate, and gamma-aminobutyric acid), and energy-related metabolite levels (malic acid, butanedioic acid, fumaric acid, and citric acid), and other altered metabolites (increased N-acetyl-aspartic acid and decreased pyroglutamic acid, urea, and lactic acid) in the hippocampus. All of these alterations indicated that the metabolic mechanisms of age-related cognitive impairment in SAMP8 mice were related to multiple pathways and networks. Lipid metabolism, especially cholesterol metabolism, appears to play a distinct role in the hippocampus in AD.
Keywords: Aging, Alzheimer's disease, cholesterol, gas chromatography-mass spectrometry, hippocampus, metabolic profiles
Introduction
Alzheimer's disease (AD), the most common age-dependent neurodegenerative disorder, produces a progressive decline in cognitive function. The mechanistic determinants of AD consist of amyloid-β (Aβ) deposits, intracellular neurofibrillary tangles, disordered lipid metabolism, and energetic and mitochondrial dysfunction [1–3]. Senescence-accelerated mouse prone 8 (SAMP8), a spontaneous AD mouse model in which the aging processes are dominated by the above pathological mechanisms associated with cognitive dysfunction, appears to provide an excellent model to investigate the process of learning and memory decline in AD [4, 5].
Age is a major risk factor for AD. The abnormalities in cerebral metabolism in AD throughout aging have been gradually documented over recent years. Metabolic abnormalities in AD that have been described include abnormal lipid metabolism [6], such as cholesterol metabolic dysfunction [7, 8]. Significant deficits in mitochondrial bioenergetic pathways have also been reported [9, 10]. Studies reporting the dysfunction in amino acid metabolism have provided substantial evidence that these metabolites provide targets for AD [11, 12].
The hippocampus is one of the most important brain regions related to learning and memory and is also known as the most vulnerable region in AD pathology. In SAMP8 mice, a variety of cognitive techniques have demonstrated that hippocampus-related learning and retention deficits occur during aging [13, 14]. In our previous study of SAMP8 mice, we also detected impaired performance on the Morris water maze appearing at 7 months of age that deteriorated at 12 months of age [15].
To understand the biochemical basis of disease, metabonomics has been extensively applied using biological fluids and tissues [16]. Gas chromatography-mass spectrometry (GC-MS) has been proven to be a very suitable technique and is widely applied to metabolite analysis because of its high sensitivity, peak resolution, and reproducibility [17].
To search for biochemical markers for potential diagnosis and treatment of AD, we utilized a GC-MS-based metabonomics approach to screen for altered cognition-related metabolic profiles, and we investigated the differences in metabolite levels in the hippocampus of SAMP8 mice during the course of aging.
Materials and Methods
Animals
Male SAMP8 mice in three age groups [2 months old (young), 7 months old (mature), and 12 months old (aged)] were used. All mice were housed in a room at 20–22°C with a 12-hour light-dark cycle, and mice had access to food and water ad libitum. Animal care and use conformed to the guidelines for care and use of laboratory animals.
Tissue sample pretreatment
After mice were sacrificed by decapitation, the hippocampi were quickly dissected, placed in liquid nitrogen, and then stored at −80°C. For GC-MS analysis, 30 mg of mouse hippocampal tissue sample was transferred into a 2 mL centrifuge tube and submerged in a 1.0 mL solution of water-methanol-chloroform (2:5:2, v/v/v) dissolved in 30 μg heptadecanoic acid as an internal standard. The mixture was sonicated at 0°C and then centrifuged at 4°C at 14,000 rpm for 20 min. After centrifugation, 800 μL of supernatant was collected from each sample into a vial (5 mL) and evaporated with nitrogen gas at 50°C. Then, 100 μL of BSTFA with 1% TMCS was added to each sealed vial, and the derivatization reaction was carried out at 70°C for 60 min. After derivatization, the samples were ready to inject in the GC-MS for analysis.
GC-MS analysis
GC-MS analysis was performed using an Agilent 7890 Gas Chromatograph (Little Falls, DE, USA) equipped with an electronically controlled split/splitless injection port, an inert 5975 C mass selective detector with an electron impact (EI) ionization chamber, and a 7683B Series injector/autosampler. The GC separation was conducted using a HP-5MS 30 m × 0.25 mm I.D., 0.25 μm film thickness column (Agilent, CA, USA). Helium (99.999%) was used as the carrier gas with a flow rate of 1.0 mL min−1, and 1 μL of sample was injected in the splitless mode. The temperature of injection was set at 260°C. The column temperature was initially set at 50°C for 3 min, increased to 280°C at a rate of 8°C min–1, and maintained at 280°C for 10 min. The detector was a quadrupole mass spectrometer, and the temperatures of the quadrupole and the ion source were 150°C and 230°C, respectively, with a solvent delay of 6.5 min. Each sample was analyzed in duplicate.
The precision of the GC-MS method was evaluated by performing replicate determinations of internal standard and cholesterol as representative analytes in a hippocampal sample. The relative standard deviation (RSD) was determined as a measurement of the precision. The intra- and inter-day repeatability was determined on six replicates within one day and over three consecutive days, respectively. The intra-day precisions (%, RSD) of two representative analytes were 3.3% and 5.7%, and the inter-day precisions (%, RSD) of two representative analytes were 3.8% and 5.9%. The results showed the method had good precision (Supplementary Table 1).
GC-MS data processing
The total ion chromatograms were processed into a single data set from the data files of the samples using XCMS software. The parameters were default settings except for the following: fwhm = 5. The data set of processed mass ions was exported from XCMS to Excel and first treated to change all median retention times (MRTs) with units of seconds into values with units of minutes. The data were sorted in ascending order according to the MRTs. Integrated areas of ion peaks of adjacent MRTs belonging to the same compound peak were summed and considered as a single metabolite [18, 19]. Metabolite peaks were identified by comparing their mass-to-charge ratios and selected based on a standard mass chromatogram in the National Institute of Standards and Technology mass spectral library. Peaks with a similarity index greater than 70% were tentatively identified. The ratios of the intensities of the compound peak to the internal standard peak were calculated to construct the data matrix.
The data matrix was employed for multivariate analysis with principal components analysis (PCA) and partial least-squares discriminant analysis (PLS-DA) using the demo version of SIMCA-P software to determine the alteration of metabolic profiles during aging. For multivariate analysis, R2Y is a quality factor, while Q2 is a predictive factor. Typically, a robust model possesses a Q2 >0.4 and a R2 >0.5 [20]. Further comparisons of specific metabolites among groups were performed using one-way ANOVA, and p< 0.05 was considered as statistically significant.
Results
Altered metabolic profile during aging
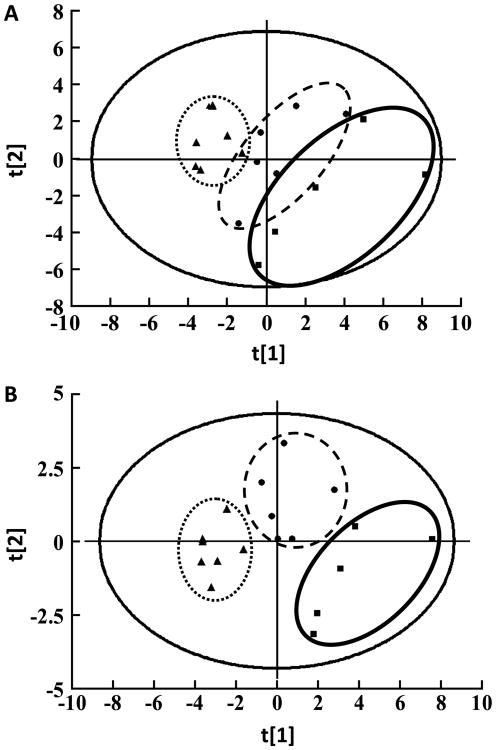
A typical total ion current chromatogram in the hippocampus of SAMP8 mice is shown in Fig. 1. Tested metabolites are listed in Table 1. The PCA score plot of the tested metabolites revealed that the samples from different groups were scattered into different regions and had a gathering trend during aging (Fig. 2A). The score plots of the first three principal components allowed for visualization of the data and comparison of the three sample groups, with an R2× and Q2 of 0.75 and 0.44, respectively. The score plot of the PLS-DA model indicated separation of the samples according to age (Fig. 2B). The model generated using three components had a cumulative R2Y of 0.81 and a cumulative Q2 of 0.58.
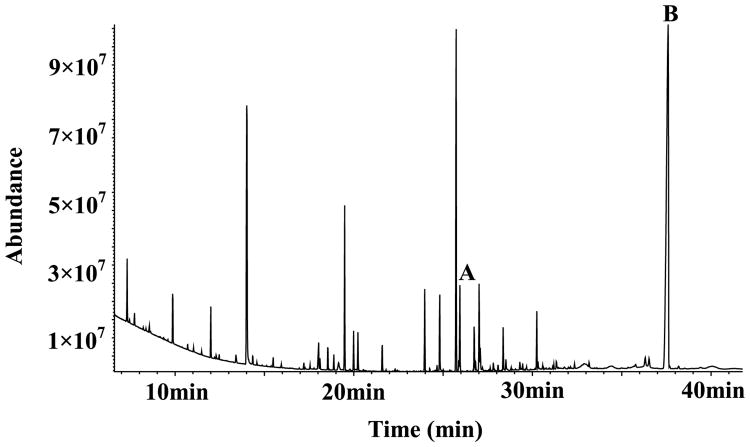
Fig. 1.
Typical GC-MS total ion chromatograms from the hippocampus after chemical derivatization. The internal standard of heptadecanoic acid peak (A) and the selected maker peak of cholesterol (B) were labeled above.
Table 1. The metabolites identified by GC-MS.
| Rt (min) | Metabolite | Relative peak to internal ion peak | ||
|---|---|---|---|---|
|
|
||||
| 2 month (mean ± SD) | 7 month (mean ± SD) | 12 month (mean ± SD) | ||
| 9.86 | Lactic acid (Lac) | 1.4807 ± 0.2681 | 1.1049 ± 0.2023 | 0.7882 ± 0.1326 |
| 10.70 | Alanine (Ala) | 0.0549 ± 0.0251 | 0.0376 ± 0.0082 | 0.0211 ± 0.0044 |
| 11.03 | Glycine (Gly) | 0.0314 ± 0.0084 | 0.0252 ± 0.0056 | 0.0184 ± 0.0034 |
| 13.40 | Urea | 0.1867 ± 0.0591 | 0.1166 ± 0.0252 | 0.0655 ± 0.0172 |
| 14.13 | Phosphoric acid (Pho) | 0.0070 ± 0.0016 | 0.0066 ± 0.0014 | 0.0057 ± 0.0012 |
| 14.35 | Gamma-aminobutyric acid (GABA) | 0.0492 ± 0.0152 | 0.0450 ± 0.0180 | 0.0304 ± 0.0040 |
| 14.58 | Butanedioic acid (But) | 0.0024 ± 0.0006 | 0.0021 ± 0.0005 | 0.0012 ± 0.0003 |
| 15.13 | Fumaric acid (Fum) | 0.0046 ± 0.0011 | 0.0037 ± 0.0005 | 0.0020 ± 0.0009 |
| 15.49 | Serine (Ser) | 0.1049 ± 0.0279 | 0.0820 ± 0.0159 | 0.0589 ± 0.0085 |
| 15.96 | Threonine (Thr) | 0.0186 ± 0.0033 | 0.0157 ± 0.0040 | 0.0156 ± 0.0021 |
| 17.55 | Malic acid (Mal) | 0.0101 ± 0.0036 | 0.0072 ± 0.0016 | 0.0050 ± 0.0009 |
| 18.00 | Pyroglutamic acid (P-Glu) | 0.2115 ± 0.0433 | 0.1480 ± 0.0424 | 0.0998 ± 0.0159 |
| 18.04 | Aspartic acid (Asp) | 0.3434 ± 0.0836 | 0.2356 ± 0.0393 | 0.2075 ± 0.0358 |
| 18.56 | Creatinine (Cr) | 0.2976 ± 0.1471 | 0.2764 ± 0.0360 | 0.2143 ± 0.1106 |
| 19.49 | Glutamate (Glu) | 2.6297 ± 0.2447 | 2.3631 ± 0.3003 | 2.1559 ± 0.0759 |
| 20.23 | N-acetyl-aspartic acid (NAA) | 0.0729 ± 0.0291 | 0.1211 ± 0.0479 | 0.1609 ± 0.0271 |
| 21.60 | Phosphoglyceride (P-Gly) | 0.2318 ± 0.0306 | 0.2763 ± 0.0417 | 0.3493 ± 0.0727 |
| 22.33 | Citric acid (Cit) | 0.0137 ± 0.0021 | 0.0139 ± 0.0013 | 0.0120 ± 0.0012 |
| 23.97 | Ascorbic acid (Asc) | 1.1793 ± 0.0633 | 1.0968 ± 0.0700 | 1.1481 ± 0.0899 |
| 24.82 | Hexadecanoic acid (Hex) | 0.9333 ± 0.1252 | 0.9793 ± 0.0591 | 0.8903 ± 0.0472 |
| 25.73 | Inositol (Ins) | 5.9575 ± 0.7579 | 5.8193 ± 0.3790 | 6.2595 ± 0.3988 |
| 26.74 | Oleic acid (Ole) | 0.1714 ± 0.0301 | 0.1957 ± 0.0097 | 0.1805 ± 0.0109 |
| 26.84 | Trans-9-octadecenoic acid (T-Oct) | 0.0072 ± 0.0006 | 0.0081 ± 0.0010 | 0.0071 ± 0.0010 |
| 27.01 | Octadecanoic acid (Oct) | 1.1772 ± 0.1547 | 1.2248 ± 0.0899 | 1.1905 ± 0.0441 |
| 29.30 | Phosphoinositol (P-Ins) | 0.0450 ± 0.0033 | 0.0467 ± 0.0027 | 0.0490 ± 0.0037 |
| 30.25 | 5,8,11,14,17-Eicosapentaenoic acid (Eic) | 0.5353 ± 0.0724 | 0.5416 ± 0.0450 | 0.5189 ± 0.0245 |
| 32.36 | Monostearin (M-Ste) | 0.0188 ± 0.0023 | 0.0197 ± 0.0017 | 0.0208 ± 0.0030 |
| 36.53 | Adenosine phosphate (P-Ade) | 0.0229 ± 0.0046 | 0.0271 ± 0.0067 | 0.0226 ± 0.0070 |
| 37.58 | Cholesterol (Cho) | 17.5943 ± 1.1256 | 19.9649 ± 0.6250 | 20.7026 ± 1.7915 |
Fig. 2.
The score plots from the PCA (A) and PLS-DA (B) analyses with the detected metabolites. Class 1 (■), Young mice group; Class 2 (●), Mature mice group; Class 3 (▲), Aged mice group. The PCA and PLS-DA score plots indicated that the samples from each group were scattered into three distinct regions (each sample represents one mouse, n = 5 in the young group; n = 6 in the mature group; n = 7 in the aged group).
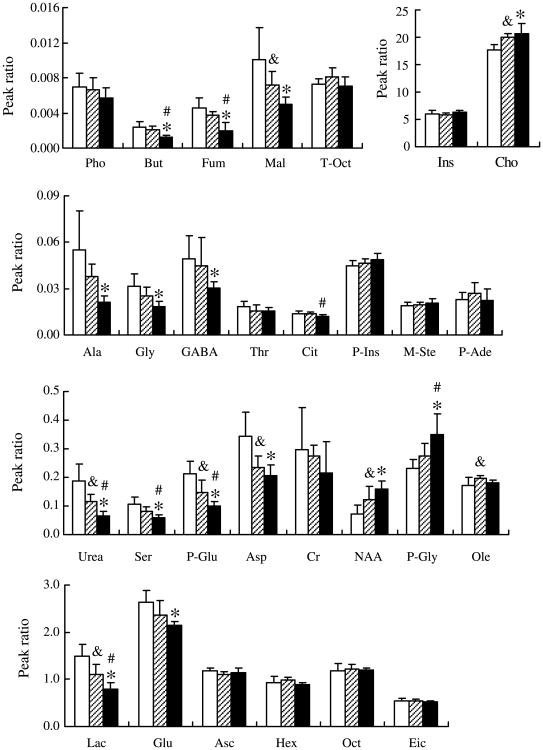
To accurately evaluate the metabolite alterations, one-way ANOVA was performed on these ratios, and significant differences were detected for 17 metabolites from the hippocampus of mice among three age groups, which were considered as potential distinct biomarkers (Fig. 3). Four metabolites were found to be significantly increased in the mouse hippocampus during aging. Compared to the levels in young mice, levels of N-acetyl-aspartic acid (NAA), oleic acid, and cholesterol increased significantly in mature mice, and NAA, phosphoglyceride, and cholesterol increased significantly in the aged mice. Compared to mature mice, in aged mice, only phosphoglyceride increased significantly. The levels of 13 metabolites decreased significantly in the hippocampus during aging. Compared to young mice, lactic acid, urea, malic acid, pyroglutamic acid, and aspartic acid decreased significantly in mature mice, and in aged mice, in addition to these 5 metabolites, alanine, glycine, gamma-aminobutyric acid (GABA), butanedioic acid, fumaric acid, serine, and glutamate decreased significantly. Compared to mature mice, the metabolites lactic acid, urea, butanedioic acid, fumaric acid, serine, pyroglutamic acid, and citric acid decreased significantly in aged mice.
Fig. 3.
Differences in metabolites were observed during aging in SAMP8 mice. Compared to the levels in young mice, NAA, oleic acid (Ole), and cholesterol (Cho) increased significantly in mature mice (&p < 0.05), and NAA, phosphoglyceride (P-Gly), and cholesterol increased significantly in aged mice (*p < 0.05). Compared to mature mice, in aged mice, only phosphoglyceride increased significantly (#p < 0.05). Compared to young mice, lactic acid (Lac), urea, malic acid (Mal), pyroglutamic acid (P-Glu), and aspartic acid (Asp) decreased significantly in mature mice (&p < 0.05), and in addition to these five metabolites, in aged mice, alanine (Ala), glycine (Gly), gamma-aminobutyric acid (GABA), butanedioic acid (But), fumaric acid (Fum), serine (Ser), and glutamate (Glu) decreased significantly (*p < 0.05). Compared to mature mice, the metabolites lactic acid, urea, butanedioic acid, fumaric acid, serine, pyroglutamic acid, and citric acid (Cit) decreased significantly in aged mice (#p < 0.05). The blank bars represented young mice, the shadow bars represented mature mice, and the dark bars represented aged mice. Data are presented as the mean ± SD (n = 5 in the young group; n = 6 in the mature group; n = 7 in the aged group).
Discussion
Metabolite abnormalities in the hippocampus have been investigated in the process of aging, and the score plots of multivariate analyses based on NMR spectra have demonstrated that the metabolic profiles in the hippocampus are altered in both normal [21] and AD aged rodents [22]. Using multivariate analysis of GC-MS results, we found that the score plot from PCA analysis with the metabolites of the hippocampus demonstrated that the samples from the different age groups were scattered into different regions such that they were combined according to age, indicating that the metabolite abnormalities in the hippocampus became dominant and homogeneous. The score plot from the PLS-DA analysis using all of the tested metabolites from hippocampus revealed that the samples from three age groups were scattered into three distinct regions, suggesting that their metabolic profiles were altered at 7 months of age (including 8 metabolites) and again transitioned to another distinct metabolic environment at 12 months of age (including 16 metabolites). The alterations in the metabolic profiles in the hippocampus coincided with the decline in cognitive performance of SAMP8 mice during aging in our previous study [15].
To observe the cognition-related metabolite alterations during aging, specific metabolites were further investigated. The present analyses of lipid metabolism detected abnormalities in the brain during both normal and AD aging. Although the weight of the brain is 2% of the total body weight, it contains 25% of the total cholesterol in the body [23]. In our GC-MS analysis, cholesterol also accounted for a large proportion of all tested compounds (Fig. 1). Cholesterol contained in membranes plays an important role in cytoskeletal function and various cellular signaling pathways. In cell culture systems, the production of Aβ is modulated by cholesterol, and studies using animal models have consistently demonstrated that hypercholesterolemia is associated with an increased deposition of cerebral Aβ peptides [24]. The link between cholesterol levels and AD is also amplified by the finding that many proteins involved in cholesterol metabolism, such as liver X receptor (LXR), have been shown to be associated with late onset AD [7, 25]. One study indicated that the application of a cholesterol-reducing LXR agonist improved the quantity of presynaptic boutons [7], regulated neuroinflammation, and decreased Aβ accumulation [26]. The administration of statins has also been demonstrated to protect against AD [27, 28]. Neural membranes contain several classes of phosphoglycerides, which not only constitute their backbone but also provide them with a suitable environment. Phosphoglycerides and phosphoglyceride-derived lipid mediators are suggested to be involved in AD pathology [29]. One study by Fraser et al. found that increased oleic acid in some regions of AD brains [30] affects Aβ production [31]. Several lines of evidence support protective as well as deleterious effects of oleic acid on AD; however, the bases for these effects are unclear [31]. In this study, we found that the lipid metabolites that were found to be altered all increased during aging. The levels of cholesterol increased significantly during the maturation period and continued through the aging period, but oleic acid only increased in mature mice, and phosphoglyceride only increased in aged mice, indicating that these metabolites, especially cholesterol, have distinct effects on AD pathology.
Reductions in cerebral metabolism that are sufficient to impair cognition in normal individuals also occur in AD. Amino acids play an important role in neurodegenerative diseases, including AD [11,32]. Amino acids are constituents of proteins, as well as neurotransmitters in the brain that exhibit regulating functions; glycine, aspartic acid, and glutamate play roles as excitatory neurotransmitters, while GABA is an inhibitory amino acid. We found that all of the amino acids that were found to be altered (alanine, serine, glycine, aspartic acid, glutamate, and GABA) decreased significantly in mature or aged mice. Overall, these results indicate that the composition and function of the hippocampus of SAMP8 mice were dysfunctional. Increasing evidence indicates a preceding and potentially causal role of mitochondrial bioenergetic deficits and brain hypometabolism coupled with increased mitochondrial oxidative stress in AD pathogenesis [9, 10]. We observed that the biochemistry of butanedioic acid, fumaric acid, malic acid, and citric acid, which are involved in the tricarboxylic acid cycle of mitochondria, the dysfunction of which is correlated with AD, decreased significantly during aging, such that at 7 months, malic acid decreased significantly, and three metabolites—butanedioic acid, fumaric acid, and citric acid—decreased significantly at 12 months of age. The energy metabolites declined in a manner similar to the amino acids, emerging in mature mice, deteriorating in aged mice, and coinciding with the decline in performance of SAMP8 mice. Abnormalities in other metabolites, including NAA, pyroglutamic acid, urea, and lactic acid, in the pathology of AD have been reported [33–35]. However, the cellular and molecular mechanisms of the involvement of these metabolites in AD remain to be explored.
A previous study indicated that a metabolic pathway including NAA, glutamate, and aspartic acid in the hippocampus assessed by proton magnetic resonance spectroscopy was found to be dysfunctional in AβPP/PS1 mice during aging [36,37]. In our study, we found, in addition to these three metabolites, another 14 lipid, amino acid, energetic, and other metabolites were altered in the hippocampus during aging in SAMP8 mice as determined using GC-MS. However, our results are similar to the recent findings of Trushina et al. [2], who revealed that, compared to wild type mice, in all three familial AD mouse models (AβPP, PS1, and AβPP/PS1 mice), there were significant alterations in the metabolite levels, including those involved in nucleotide metabolism, lipid metabolism, the mitochondrial Krebs cycle, and the carbohydrate, neurotransmitter, and amino acid metabolic pathways, indicating that multiple metabolic pathways were affected in both transgenic and spontaneous AD mice. These pathways were also significantly disturbed in AD patients, particularly the pathways involved in cholesterol biosynthesis and metabolism, transport, and lipid metabolism pathways, which were significantly altered in both cerebrospinal fluid and plasma [3, 38]. Taken together, our data are the first to demonstrate the presence of distinct changes in the levels of cholesterol in lipid metabolism dysfunction, along with multiple other metabolic pathways in the hippocampus of SAMP8 mice during aging. Further research in these pathways could shed light on the understanding of the disease mechanism and the development of biomarkers for AD.
In conclusion, the multivariate analysis using GC-MS performed in this study indicated that the metabolic mechanisms of age-related cognitive impairment in SAMP8 mice were related to multiple pathways and networks. The impairment in age-related cognitive performance was accompanied by homeostatic dysfunction of metabolite profiles, including alterations in levels of lipids, amino acids, energy-related metabolites, and other metabolites in the hippocampus of SAMP8 mice. Lipid metabolism, especially the cholesterol pathway, in the hippocampus appears to play an important role in AD.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81302471). This work was also supported by the Brain Aging and Cognitive Neuroscience Laboratory of Hebei Province. The authors wish to thank Prof. David Yew (Department of Anatomy, The Chinese University of Hong Kong) for providing the SAMP8 mice.
Footnotes
Authors' disclosures available online (http://www.j-alz.com/disclosures/view.php?id=1995).
Supplementary Material: The supplementary table is available in the electronic version of this article: http://dx.doi.org/10.3233/JAD-131463.
References
- 1.O'Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer's disease. Annu Rev Neurosci. 2011;34:185–204. doi: 10.1146/annurev-neuro-061010-113613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trushina E, Nemutlu E, Zhang S, Christensen T, Camp J, Mesa J, Siddiqui A, Tamura Y, Sesaki H, Wengenack TM, Dzeja PP, Poduslo JF. Defects in mitochondrial dynamics and metabolomic signatures of evolving energetic stress in mouse models of familial Alzheimer's disease. PLoS One. 2012;7:e32737. doi: 10.1371/journal.pone.0032737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trushina E, Dutta T, Persson XM, Mielke MM, Petersen RC. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer's disease using metabolomics. PLoS One. 2013;8:e63644. doi: 10.1371/journal.pone.0063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morley JE, Farr SA, Kumar VB, Armbrecht HJ. The SAMP8 mouse: A model to develop therapeutic interventions for Alzheimer's disease. Curr Pharm Des. 2012;18:1123–1130. doi: 10.2174/138161212799315795. [DOI] [PubMed] [Google Scholar]
- 5.Morley JE, Armbrecht HJ, Farr SA, Kumar VB. The senescence accelerated mouse (SAMP8) as a model for oxidative stress and Alzheimer's disease. Biochim Biophys Acta. 2012;1822:650–656. doi: 10.1016/j.bbadis.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Han X. Lipid alterations in the earliest clinically recognizable stage of Alzheimer's disease: Implication of the role of lipids in the pathogenesis of Alzheimer's disease. Curr Alzheimer Res. 2005;2:65–77. doi: 10.2174/1567205052772786. [DOI] [PubMed] [Google Scholar]
- 7.Jansen D, Janssen CI, Vanmierlo T, Dederen PJ, van Rooij D, Zinnhardt B, Nobelen CL, Janssen AL, Hafkemeijer A, Mutsaers MP, Doedee AM, Kuipers AA, Broersen LM, Mulder M, Kiliaan AJ. Cholesterol and synaptic compensatory mechanisms in Alzheimer's disease mice brain during aging. J Alzheimers Dis. 2012;31:813–826. doi: 10.3233/JAD-2012-120298. [DOI] [PubMed] [Google Scholar]
- 8.Maulik M, Westaway D, Jhamandas JH, Kar S. Role of cholesterol in APP metabolism and its significance in Alzheimer's disease pathogenesis. Mol Neurobiol. 2013;47:37–63. doi: 10.1007/s12035-012-8337-y. [DOI] [PubMed] [Google Scholar]
- 9.Bubber P, Haroutunian V, Fisch G, Blass JP, Gibson GE. Mitochondrial abnormalities in Alzheimer brain: Mechanistic implications. Ann Neurol. 2005;57:695–703. doi: 10.1002/ana.20474. [DOI] [PubMed] [Google Scholar]
- 10.Yao J, Brinton RD. Estrogen regulation of mitochondrial bioenergetics: Implications for prevention of Alzheimer's disease. Adv Pharmacol. 2012;64:327–371. doi: 10.1016/B978-0-12-394816-8.00010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison DW, Beal MF, Mazurek MF, Bird ED, Martin JB. A postmortem study of amino acid neurotransmitters in Alzheimer's disease. Ann Neurol. 1986;20:616–621. doi: 10.1002/ana.410200510. [DOI] [PubMed] [Google Scholar]
- 12.Nilsen LH, Melo TM, Saether O, Witter MP, Sonnewald U. Altered neurochemical profile in the McGill-R-Thy1-APP rat model of Alzheimer's disease: A longitudinal in vivo 1 H MRS study. J Neurochem. 2012;123:532–541. doi: 10.1111/jnc.12003. [DOI] [PubMed] [Google Scholar]
- 13.Flood JF, Morley JE. Learning and memory in the SAMP8 mouse. Neurosci Biobehav Rev. 1998;22:1–20. doi: 10.1016/s0149-7634(96)00063-2. [DOI] [PubMed] [Google Scholar]
- 14.Nomura Y, Okuma Y. Age-related defects in lifespan and learning ability in SAMP8 mice. Neurobiol Aging. 1999;20:111–115. doi: 10.1016/s0197-4580(99)00006-8. [DOI] [PubMed] [Google Scholar]
- 15.Ma Q, Qiang J, Gu P, Wang Y, Geng Y, Wang M. Age-related autophagy alterations in the brain of senescence accelerated mouse prone 8 (SAMP8) mice. Exp Gerontol. 2011;46:533–541. doi: 10.1016/j.exger.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- 17.Koek MM, Jellema RH, van der Greef J, Tas AC, Hankemeier T. Quantitative metabolomics based on gas chromatography mass spectrometry: Status and perspectives. Metabolomics. 2011;7:307–328. doi: 10.1007/s11306-010-0254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H, Xue R, Lu C, Deng C, Liu T, Zeng H, Wang Q, Shen X. Metabolomic study for diagnostic model of oesophageal cancer using gas chromatography/mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3111–3117. doi: 10.1016/j.jchromb.2009.07.039. [DOI] [PubMed] [Google Scholar]
- 19.Jonsson P, Bruce SJ, Moritz T, Trygg J, Sjostrom M, Plumb R, Granger J, Maibaum E, Nicholson JK, Holmes E, Antti H. Extraction, interpretation and validation of information for comparing samples in metabolic LC/MS data sets. Analyst. 2005;130:701–707. doi: 10.1039/b501890k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paban V, Fauvelle F, Alescio-Lautier B. Age-related changesinmetabolic profilesofrat hippocampus and cortices. Eur J Neurosci. 2010;31:1063–1073. doi: 10.1111/j.1460-9568.2010.07126.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Liu H, Wu J, Liu M, Wang Y. Metabonomic alterations in hippocampus, temporal and prefrontal cortex with age in rats. Neurochem Int. 2009;54:481–487. doi: 10.1016/j.neuint.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Salek RM, Xia J, Innes A, Sweatman BC, Adalbert R, Randle S, McGowan E, Emson PC, Griffin JL. A metabolomic study of the CRND8 transgenic mouse model of Alzheimer's disease. Neurochem Int. 2010;56:937–947. doi: 10.1016/j.neuint.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Curr Opin Lipidol. 2001;12:105–112. doi: 10.1097/00041433-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Ricciarelli R, Canepa E, Marengo B, Marinari UM, Poli G, Pronzato MA, Domenicotti C. Cholesterol and Alzheimer's disease: A still poorly understood correlation. IUBMB Life. 2012;64:931–935. doi: 10.1002/iub.1091. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Schuster GU, Hultenby K, Zhang Q, Andersson S, Gustafsson JA. Liver X receptors in the central nervous system: From lipid homeostasis to neuronal degeneration. Proc Natl Acad Sci U S A. 2002;99:13878–13883. doi: 10.1073/pnas.172510899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang J, Rivest S. Lipid metabolism and neuroinflammation in Alzheimer's disease: A role for liver X receptors. Endocr Rev. 2012;33:715–746. doi: 10.1210/er.2011-1049. [DOI] [PubMed] [Google Scholar]
- 27.Kurata T, Kawai H, Miyazaki K, Kozuki M, Morimoto N, Ohta Y, Ikeda Y, Abe K. Statins have therapeutic potential for the treatment of Alzheimer's disease, likely via protection of the neurovascular unit in the AD brain. J Neurol Sci. 2012;322:59–63. doi: 10.1016/j.jns.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 28.Pac-Soo C, Lloyd DG, Vizcaychipi MP, Ma D. Statins: The role in the treatment and prevention of Alzheimer's neurodegeneration. J Alzheimers Dis. 2011;27:1–10. doi: 10.3233/JAD-2011-110524. [DOI] [PubMed] [Google Scholar]
- 29.Frisardi V, Panza F, Seripa D, Farooqui T, Farooqui AA. Glycerophospholipids and glycerophospholipid-derived lipid mediators: A complex meshwork in Alzheimer's disease pathology. Prog Lipid Res. 2011;50:313–330. doi: 10.1016/j.plipres.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Fraser T, Tayler H, Love S. Fatty acid composition of frontal, temporal and parietal neocortex in the normal human brain and in Alzheimer's disease. Neurochem Res. 2010;35:503–513. doi: 10.1007/s11064-009-0087-5. [DOI] [PubMed] [Google Scholar]
- 31.Amtul Z, Westaway D, Cechetto DF, Rozmahel RF. Oleic acid ameliorates amyloidosis in cellular and mouse models of Alzheimer's disease. Brain Pathol. 2011;21:321–329. doi: 10.1111/j.1750-3639.2010.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura S. Amino acid metabolism in neurodegenerative diseases. Nihon Rinsho. 1992;50:1637–1642. [PubMed] [Google Scholar]
- 33.Sun N, Hartmann R, Lecher J, Stoldt M, Funke SA, Gremer L, Ludwig HH, Demuth HU, Kleinschmidt M, Willbold D. Structural analysis of the pyroglutamate-modified isoform of the Alzheimer's disease-related amyloid-beta using NMR spectroscopy. J Pept Sci. 2012;18:691–695. doi: 10.1002/psc.2456. [DOI] [PubMed] [Google Scholar]
- 34.Gueli MC, Taibi G. Alzheimer's disease: Amino acid levels and brain metabolic status. Neurol Sci. 2013;34:1575–1579. doi: 10.1007/s10072-013-1289-9. [DOI] [PubMed] [Google Scholar]
- 35.Trushina E, Mielke MM. Recent advances in the application of metabolomics to Alzheimer's disease. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbadis.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marjanska M, Curran GL, Wengenack TM, Henry PG, Bliss RL, Poduslo JF, Jack CR, Jr, Ugurbil K, Garwood M. Monitoring disease progression in transgenic mouse models of Alzheimer's disease with proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 2005;102:11906–11910. doi: 10.1073/pnas.0505513102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberg J, Spenger C, Wang FH, Andersson A, Westman E, Skoglund P, Sunnemark D, Norinder U, Klason T, Wahlund LO, Lindberg M. Age related changes in brain metabolites observed by 1H MRS in APP/PS1 mice. Neurobiol Aging. 2008;29:1423–1433. doi: 10.1016/j.neurobiolaging.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Oresic M, Hyotylainen T, Herukka SK, Sysi-Aho M, Mattila I, Seppanan-Laakso T, Julkunen V, Gopalacharyulu PV, Hallikainen M, Koikkalainen J, Kivipelto M, Helisalmi S, Lotjonen J, Soininen H. Metabolome in progression to Alzheimer's disease. Transl Psychiatry. 2011;1:e57. doi: 10.1038/tp.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.