Abstract
DNA encapsulates silver clusters, and these hybrid nanomaterials form molecular sensors. We discuss a silver cluster-oligonucleotide sensor with four characteristics. First, a specific reporting cluster forms within a single-stranded DNA. This template uses the 5’ cluster domain CCCCAACTCCTT with different 3’ recognition sites for complementary oligonucleotides. The modular composite strand exclusively forms a cluster with λmax = 400 nm and with low emission. Conjugates were chromatographically purified, and their elemental analysis measured a cluster adduct with ~11 silver atoms. Second, hybridization transforms the cluster. Size exclusion chromatography shows that the 3’ recognition sites of the single-stranded conjugates hybridize with their complements. This secondary structural change both shifts cluster absorption from 400 to 490 nm and develops emission at 550 nm. Third, cluster size remains intact. Like their violet predecessors, purified blue-green clusters have ~11 silver atoms. Cluster integrity is further supported by extracting the complement from the blue-green conjugate and reversing the spectral changes. Fourth, the cluster transformation is an equilibrium. Complementary strands generate an isosbestic point and thus directly link single-stranded hosts for the violet cluster and their hybridized analogs for the blue-green cluster. This equilibrium shifts with temperature. A van’t Hoff analysis shows that longer and more stable duplexes favor the blue-green cluster. However, hybridized cluster hosts are less stable than their native DNA counterparts, and stability further degrades when short complements expose nucleobases within S1-S2. Duplex instability suggests that unpaired nucleobases coordinate the violet cluster and favor the single-stranded sensor. A balance between innate hybridization and exogenous folding highlights a distinct feature of silver clusters for sensing – they are both chromophoric reporters and ligands that modulate analyte-sensor interactions.
Noble metal clusters are molecules, and one distinguishing characteristic is their electronic structure.1–3 Gold and silver clusters with ≲100 atoms exhibit structured absorption spectra, and these spectral signatures vary with cluster stoichiometry, structure, and oxidation state.4–6 In addition, radiative relaxation via fluorescence is facilitated by sparsely organized electronic states.7,8 Thus, cluster absorption and emission spectra provide fingerprints, so specific species can be spectroscopic labels.9 Particular clusters are synthesized with ligands that not only constrain their growth but also inhibit their reactivity.4 We focus on silver clusters formed within DNA templates.8,10–12 These hybrid chromophores have four distinguishing characteristics. First, fluorescence quantum yields (ϕf) range from 10 – 65%, and extinction coefficients (ε) range from 120,000 −350,000 M−1cm−1.8,13,14 This level of molecular brightness (ε × ϕf) efficiency is comparable to organic chromophores and semiconductor nanocrystals and thus allows sensitive, fluorescence-based detection.15 Second, short ns fluorescence lifetimes and robust photostabilities enable laser-driven excitation, which produces high integrated emission signals.8,16 Third, weakly coupled dark states minimize intermittent fluorescence and thus increase net signal.17,18 Furthermore, small steady state populations reside in these dark states and can be optically redirected to produce more emission.19 Fourth, specific chromophores are encoded by the sequence of their DNA templates and are conveniently synthesized by reducing DNA-bound Ag+ .8,13,20
In this paper, we explore another molecular characteristic of silver clusters as DNA ligands. These adducts coordinate nucleobases and occupy specific DNA binding sites.8,13,21–25 Furthermore, silver clusters bind multiple nucleobases and thus crosslink and assemble DNA strands.23,26–28 Few-atom silver clusters are relatively small in relation to their DNA hosts. Thus, they are distinguished from larger nanoparticle-based sensors with dense coverages of hundreds to thousands of oligonucleotides.29 Silver clusters are also chromophoric reporters that recognize DNA through innate fluorescence switching. This again contrasts with nanoparticle- based detection that uses energy transfer with an exogenous dye or plasmon coupling.29,30 Our earlier studies considered an ~11 silver atom cluster with violet absorption and low emission.27 This cluster forms within a single-stranded DNA that has a cytosine-rich cluster domain and a recognition site for complementary oligonucleotides. Complements hybridize with this recognition site, and cluster absorption shifts from violet to near-infrared and near-infrared emission develops. We followed a similar strategy in this paper. A single-stranded DNA template again forms the ~11 silver atom violet cluster. However, this strand uses a different cluster domain, so hybridization shifts cluster absorption from the violet to the blue-green and green emission develops. A key finding is that this hybridized blue-green cluster conjugate equilibrates with its single-stranded violet predecessor. We address the structural, spectral, stoichiometric, and thermodynamic aspects of this cluster-DNA equilibrium.
Experimental
The clusters were synthesized using oligonucleotides (Integrated DNA Technologies), and these were purified by standard desalting by the manufacturer. An HPLC sample was also used (see S1-S212c:S2C12c/Ag11 in Table 1). Lyophilized oligonucleotides were dissolved in deionized water, and their concentrations were spectroscopically measured using extinction coefficients based on the nearest neighbor approximation.31 Sensor strands had a common 5’ component CCCCAACTCCTT (S1) and different 3’ recognition sites (S2). Subscripts for the following oligonucleotides designate the length and sequence of the recognition site and their complements (S2C). Strand polarity is 5’ – 3’:
S1-S212a: CCCCAACTCCTTCCCGCCGCTGGA; S2C12a: TCCAGCGGCGGG;
S1-S212b: CCCCAACTCCTTCCCGCCGCTAGA; S2C12b: TCTAGCGGCGGG;
S1-S212c: CCCCAACTCCTTCCCGCCTAGCTA; S2C12c: TAGCTAGGCGGG;
S1-S214: CCCCAACTCCTTCCCGCCTAGCTAGA; S2C14: TCTAGCTAGGCGGG;
S1-S216: CCCCAACTCCTTCCCGCCTAGCTAGAGG, S2C16: CCTCTAGCTAGGCGGG;
S1-S28: CCCCAACTCCTTCCCGCCTA; and S2C14a: CCTCTAGCTAGGCG. Silver nitrate (Acros) and sodium borohydride (Aldrich) were used as received. Samples were prepared in 10 mM citrate/citric acid buffers at pH = 6.5. The oligonucleotide concentration was 90 µM with 8 Ag+:oligonucleotide. The samples were heated to 80 °C for 5 mins to disrupt DNA aggregates that are favored by cytosine-rich DNA strands.32 The reducing agent was added at 4 BH4−:oligonucleotide, and its relative concentration did not significantly change the cluster yield.33 The samples were subsequently transferred to a high pressure reactor (Parr) with oxygen at 500 psi and were removed after 3 hours.
Table 1.
Comparison of Enthalpy and Entropy Changes for Dissociation of the Blue-Green Conjugates with Different S1-S2:S2C Hosts
| Entrya | Duplex | ΔH (kcal)b | ΔS (cal/K)b |
|---|---|---|---|
| A | S1-S212a:S2C12a/Ag11 | 46 ± 4 | 125 ± 12 |
| A’ | S1-S212a:S2C12a | 92 ± 6 | 252 ± 17 |
| B | S1-S212b:S2C12b/Ag11 | 55 ± 6 | 153 ± 19 |
| B4/2c | S1-S212b:S2C12b/Ag11 | 50 ± 2 | 135 ± 5 |
| B2/1d | S1-S212b:S2C12b/Ag11 | 52 ± 2 | 140 ± 4 |
| B’ | S1-S212b:S2C12b | 98 ± 5 | 266 ± 16 |
| Ce | S1-S212c:S2C12c/Ag11 | 51 ± 6 | 139 ± 18 |
| C’ | S1-S212c:S2C12c | 94 ± 7 | 262 ± 21 |
| Ccf | S1-S212c:S2C12c/Ag11 | 50 ± 4 | 135 ± 10 |
| D | S1-S214:S2C14/Ag11 | 72 ± 3 | 199 ± 9 |
| D’ | S1-S214:S2C14 | 105 ± 9 | 287 ± 31 |
| E | S1-S216:S2C16/Ag11 | 93 ± 4 | 258 ± 10 |
| E’ | S1-S216:S2C16 | 119 ± 11 | 328 ± 32 |
| F | S1-S216:S2C14/Ag11 | 68 ± 7 | 186 ± 23 |
| F’ | S1-S216:S2C14 | 102 ± 4 | 285 ± 10 |
| G | S1-S216:S2C14a/Ag11 | 27 ± 1 | 66 ± 4 |
| G’ | S1-S216:S2C14a | 109 ± 10 | 306 ± 28 |
Entries without a prime correspond to the cluster conjugates. Entries with a prime designation designate native hybridized DNA. Unless otherwise noted the relative concentrations were 4 Ag+:2 BH4−:S1-S2
Uncertainties were derived from a minimum of four measurements.
Parameters were derived using 4 Ag+:2 BH4−:S1-S212b. The overall concentration of S1-S212b was 15 µM with 3 equivalents of S2C12b.
Parameters were derived using 2 Ag+:1 BH4−:S1-S212b. The overall concentration of S1-S212b was 15 µM with 3 equivalents of S2C12b.
A HPLC purified sample had ΔH = 52 ± 2 kcal and ΔS = 141 ± 5 cal/K.
Parameters were derived from the concentration dependence of the melting temperatures (Fig. S8).
The blue-green conjugate was prepared by increasing the Na+ concentration to 100 mM. NaClO4 was used to maintain silver solubility.34 S2C complement was added, and the resulting sample was heated to 50°C prior to thermal analysis. Absorption spectra were collected on a Cary 50 spectrometer (Varian) equipped with a temperature controller (Quantum Northwest). Absorption spectra were collected in 2°C increments and the temperature equilibrated for 15 mins at each temperature increment. The samples were evaluated in quartz cells with pathlengths of 1 cm.
Thermodynamic measurements were based on absorbance changes at 490 nm.35 Absorbances were measured in 2°C increments and with 15 min equilibration times. Fractional conversion of the blue-green conjugate was based on limiting baseline absorbances at low and high temperatures. Temperature-dependent equilibrium constants were based on duplex dissociation, i.e. blue-green conjugates dissociate to single-stranded violet conjugates and their complementary strands (Eq. 2). Van’t Hoff analysis was confined to 20%-80% fractional conversion and yielded enthalpy and entropy changes for dissociation. The thermodynamic parameters did not change when equilibration times were increased to 30 min and when the direction of the temperature change was reversed. Uncertainties in these measurements were established from four or more measurements.
Emission spectra were acquired on a Fluoromax-3 (Jobin Yvon Horiba). Circular dichroism spectra were acquired on a Jasco J-810. Fluorescence correlation spectroscopy (FCS) studies were conducted using apparatus similar to that described earlier.36 Sample fluorescence was excited using a frequency-doubled diode laser operating at 488 nm. Autocorrelation analysis of the fluorescence fluctuations (G(τ)) was resolvable into diffusive (gD(τ)) and excited state (gES(τ)) contributions:18,37
| Eq. 1 |
where gD(τ) is based on a 3-D Gaussian model:
and gES(τ) is:
τ is the lag time, N is the average number of fluorescent species in the probe volume, ω is the transverse radius of the probe volume, z is the height of the probe volume, τd is the lag time at which the autocorrelation amplitude has decayed to approximately one half of its maximum value, G(0), F is the fractional occupancy of the dark state, and τes is the net correlation time for dark state shelving. The aspect ratio z/ω was determined using Rhodamine B (Rh B) and set to 6.7 for fitting. We used the occupancy (N) of the probe volume to determine the extinction coefficient of the blue-green conjugate. The size of the FCS probe volume was determined to be 1 fL from the fluorescence autocorrelation recorded for Rh B dissolved in water and the known diffusion constant of Rh B in water (420 µm2/s).38 When samples were diluted ten-fold, resulting concentrations scaled by approximately the same factor. A total of five samples were analyzed: S1-S212a:S2C12a, S1-S212b:S2C12b, S1-S212c:S2C12c, S1-S214:S2C14, and S1-S216:S2C16. Size exclusion chromatography studies were conducted as described earlier.27 We used a 300 × 7.8 mm i.d. column (BioSep, Phenomenex) on a HPLC system (Prominence, Shimadzu) using a 10 mM citrate buffer at pH = 7 with 300 mM NaClO4 to minimize matrix adsorption.39 After HPLC purification, the quantity of silver in the resulting DNA-conjugate was determined by inductively coupled plasma-atomic emission spectroscopy (Optima 7300 DV, Perkin Elmer).27 Control samples containing oligonucleotide and Ag+ were used to account for matrix effects.14
Results
Cluster Formation and Transformation
Our sensor was based on a specific silver cluster with a single violet absorption band and with low emission (Fig. 1). Three factors controlled this selective synthesis. First, the cluster formed within single-stranded DNA, and this strand had two components (Fig. 1). The 3’ recognition site (S2) hybridized with complementary DNA strands. The 5’ cluster domain CCCCAACTCCTT (S1) was originally chosen because it produced a strongly red-emitting cluster (Fig. S1A). Like other short oligonucleotides, this isolated 12-nucleobase sequence was indiscriminate and formed several types of clusters with overlapping absorption bands (Fig. S1B).21 However, composite S1-S2 strands produced only the violet absorbing cluster (Fig. 1). Thus, both components defined this specific cluster environment. Second, ratios from 2 to 8 Ag+:S1-S2 yielded a single absorption band with both constant λmax ≈ 400 nm and progressively increasing absorbances (Fig. 2A). Higher relative Ag+ concentrations produced additional transitions with λmax = 435 and 530 nm. These spectral trends supported a specific DNA binding site for the violet cluster. This site was first exclusively favored at lower Ag+ stoichiometries but then saturated as the concentration of Ag+ increased. Third, oxygen eliminated red-shifted absorptions but preserved the violet band (Fig. 2B). This change also occurs for samples exposed to air but is accelerated by higher pressures of oxygen.33 Similar reactivities of oxygen and hydrogen peroxide suggested that competing clusters were selectively oxidized.33,40 In summary, we synthesized a specific violet-absorbing silver cluster by combining a composite strand S1-S2 with 8 equivalents of Ag+, then reducing with BH4−, and finally exposing to oxygen.
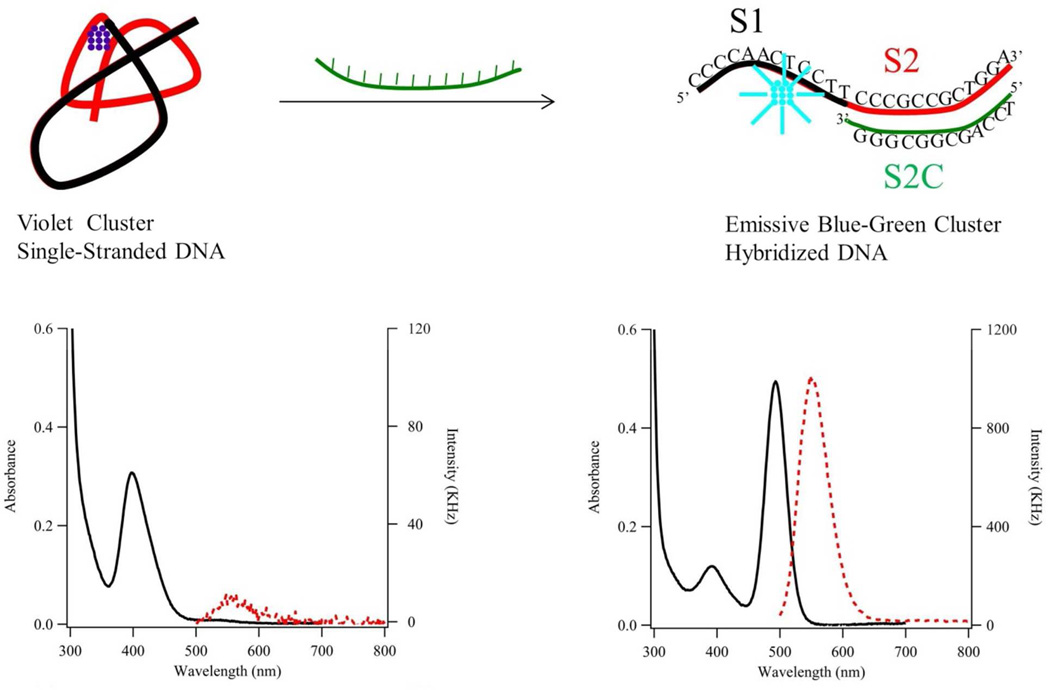
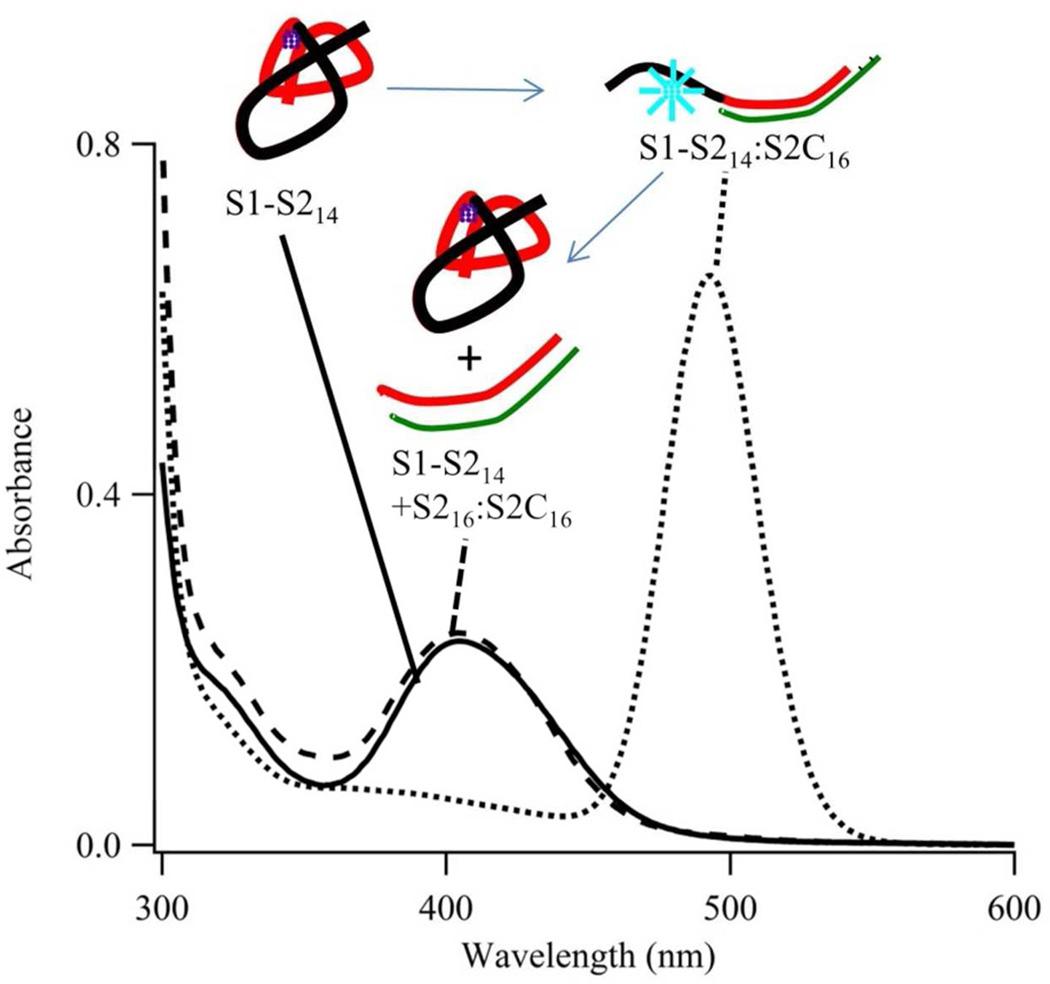
Figure 1. Reaction Scheme.
Our proposed reaction scheme describes how hybridization changes both DNA structure and silver cluster spectra. (Left) The composite strand has the 5’ cluster domain (S1 = CCCCAACTCCTT (black segment)) and one of five recognition sites (S212a = CCCGCCGCTGGA (red segment)). This single-stranded oligonucleotide hosts an ~11 silver atom with λmax = 400 nm (solid black line – left axis) and low emission (dashed red line – right axis). The cluster condenses its host strand. (Right) This cluster-laden strand hybridizes with its complementary strand (S2C12a = TCCAGCGGCGGG (green segment)). Cluster size is conserved but λmax shifts to 490 nm (solid black line – left axis) and strong green emission develops (dashed red line – right axis with 10X scale difference).
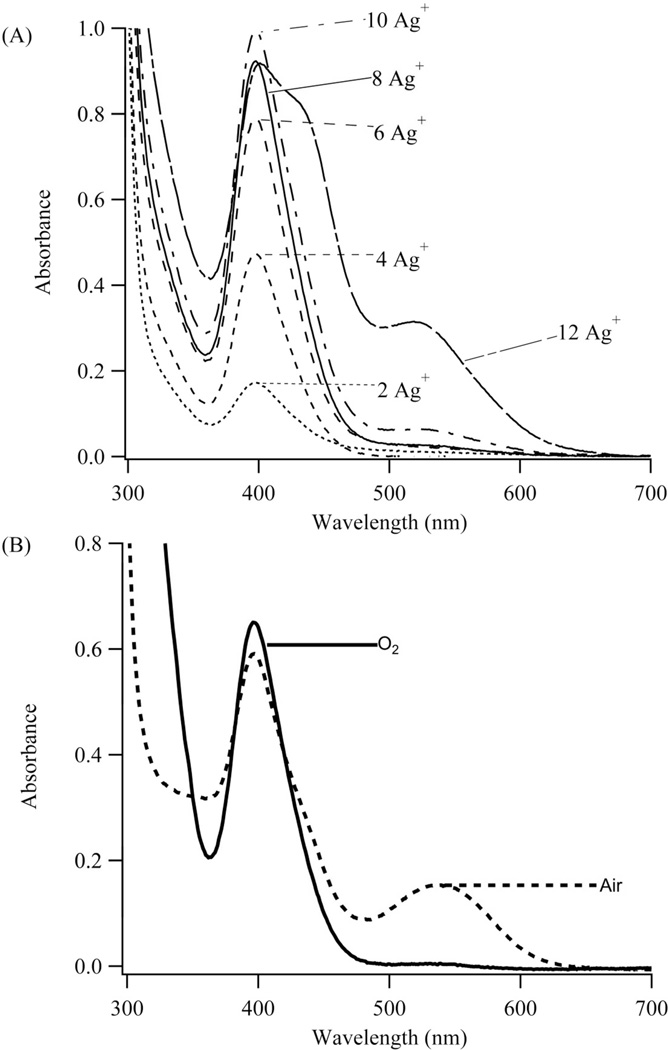
Figure 2. Cluster Synthesis.
(A) Absorption spectra of 45 µM S1-S212a with varying amounts of Ag+. In relation to the oligonucleotide concentration, the relative concentrations are 2 Ag+, 4 Ag+, 6 Ag+, 8 Ag+, 10 Ag+, and 12 Ag+. Up to 8 Ag+, absorption increases at 400 nm, which supports population of a specific cluster binding site. Beyond 8Ag+, new species with longer wavelength absorbances develop. (B) Absorption spectra of 30 µM S1-S212a with 8 Ag+ and 4 BH4- acquired with exposure to oxygen (solid line) and to air (dashed line). Oxygen eliminates alternate species and favors the violet absorbing species.
Three experiments demonstrated that DNA encapsulated the violet cluster. First, size exclusion chromatography isolated a species with absorptions both at 400 and 260 nm (Fig. 3A). Coincident cluster- and DNA-specific absorptions, respectively, supported a single type of cluster adduct with DNA. The cluster moiety condensed the native DNA strand and slowed conjugate elution. Second, circular dichroism accompanied the violet absorption and thus supported a chiral DNA host for the cluster (Fig. S2). Third, atomic emission measurements of purified species identified both silver and phosphorus. This latter element was derived only from DNA and thus provided the oligonucleotide concentration. From our chromatographically purified sample, elemental analysis measured in situ cluster sizes.12 Cluster sizes can also be determined from electrospray ionization mass spectrometry.25,41 However, these measurements of desolvated gas-phase species can be distorted by fragmentation and by favoring weakly bound adducts.42 In our investigation, we used four strands that all produced the violet cluster. These DNA hosts had the common 12 nucleotide cluster domain (S1) and different 8 – 16 nucleotide recognition sites (S2). These yielded the following relative stoichiometries: 11.0 ± 1.0 Ag:S1-S28, 11.1 ± 1.1 Ag:S1-S212a, 11.5 ± 1.5 Ag:S1-S212b, and 12.2 ± 1.0 Ag:S1-S216. The four strands varied almost two-fold in the amount of phosphorus because of the different S2 sequences. Thus, their similar silver stoichiometries supported a single cluster with ~11 atoms. Other studies substantiate this assignment. A cluster with ~11 atoms and violet absorption also formed when not only S2 but also S1 sequences vary.27 Undecanuclear silver also forms with alternate ligands and has violet absorption.43–45
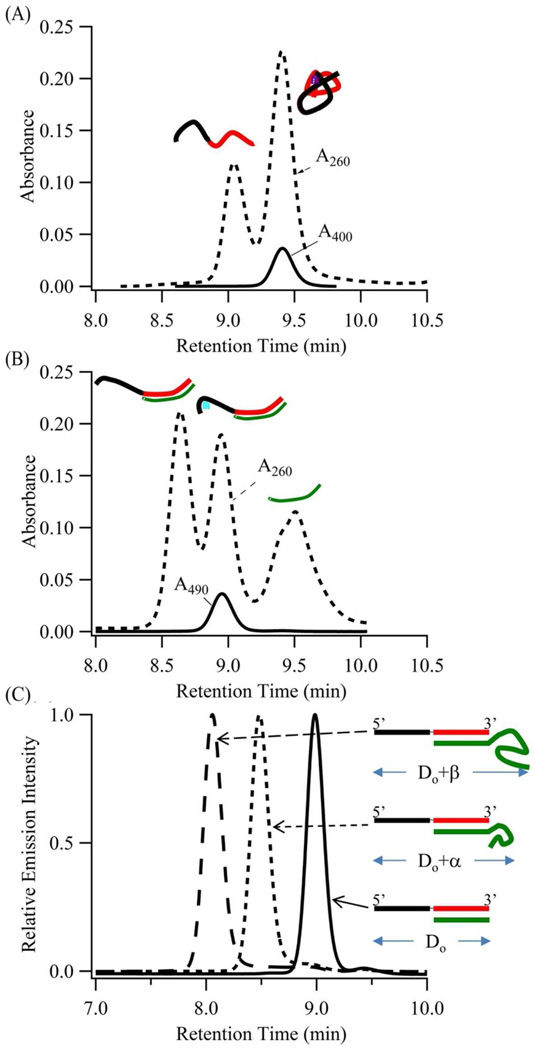
Figure 3. Secondary Structure.
Chromatograms derived for (A) S1-S212a with the violet cluster and for (B) S1-S212a :S2C12a with the blue-green cluster. Absorbances at 260 (dotted lines), 400 (solid line – A), and 490 (solid line – B) nm correspond to the DNA host, the violet cluster, and the blue-green cluster, respectively. Based on the absorbance at 260 nm in (A), the leading peak is native S1-S212a. The lagging peak is the cluster conjugate. Based on the absorbance at 260 nm in (B), the leading peak is due to native duplex S1-S212a:S2C12a, the intermediate peak is the blue-green cluster conjugate, and the lagging peak is the complement S2C12a. (C) Size exclusion chromatograms of blue-green conjugates in which S1-S212a (black and red strands) hybridizes with complements S2C12a, dT10-S2C12a, and dT20-S2C12a (green strands). These produce a blue-green conjugates with a hydrodynamic diameter of Do, Do + α, and Do + β, respectively. Progressively shorter retention times support hybridization of the S212a component and its S2C12a complement.
Complements to the S2 recognition site (S2C) transformed the encapsulated violet cluster -absorption shifted from 400 to 490 nm and emission developed at 550 nm (Fig. 1). Three factors promoted this reaction. First, higher complement concentrations produced more blue-green absorbing cluster, and this shift supported intermolecular association of S2 and S2C. An isosbestic point between the two absorption bands established that the complement linked the violet and blue-green species (Fig. S3A).46 Second, higher Na+ concentrations favored the blue-green species. This trend suggests that this cation alleviated electrostatic repulsion between the DNA polyelectrolytes S1-S2 and S2C and thereby promoted their intermolecular association (Fig. S3B).47 Third, lower temperatures increased blue-green absorption, while higher temperatures impeded complement association and recovered violet absorption (Figs. 5A and S4). Similar absorption spectra at higher and lower temperatures suggested that the violet cluster remained bound to single-stranded S1-S2 over our temperature range (Fig. S4). This temperature response supported base stacking and pairing that accompanies hybridization of two single-stranded oligonucleotides.31 We next evaluate how complements altered the cluster environment.
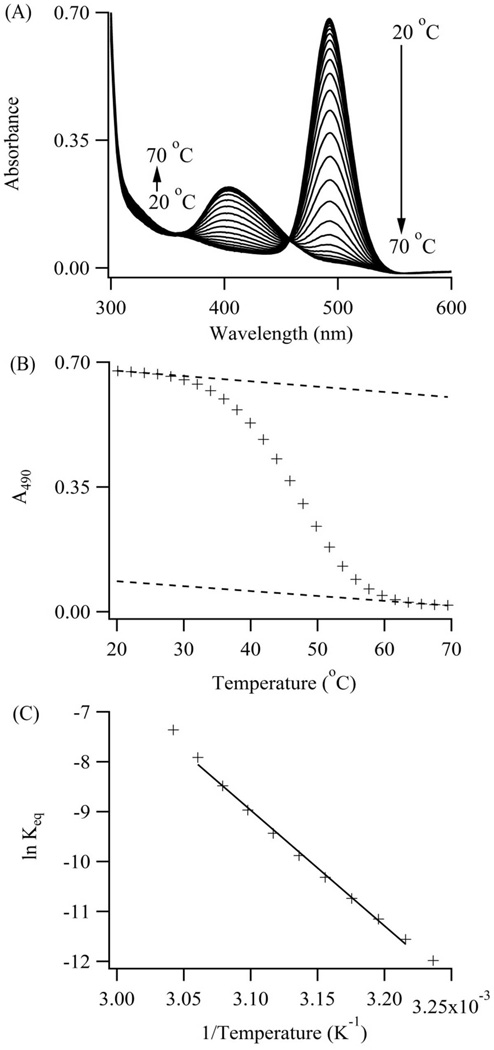
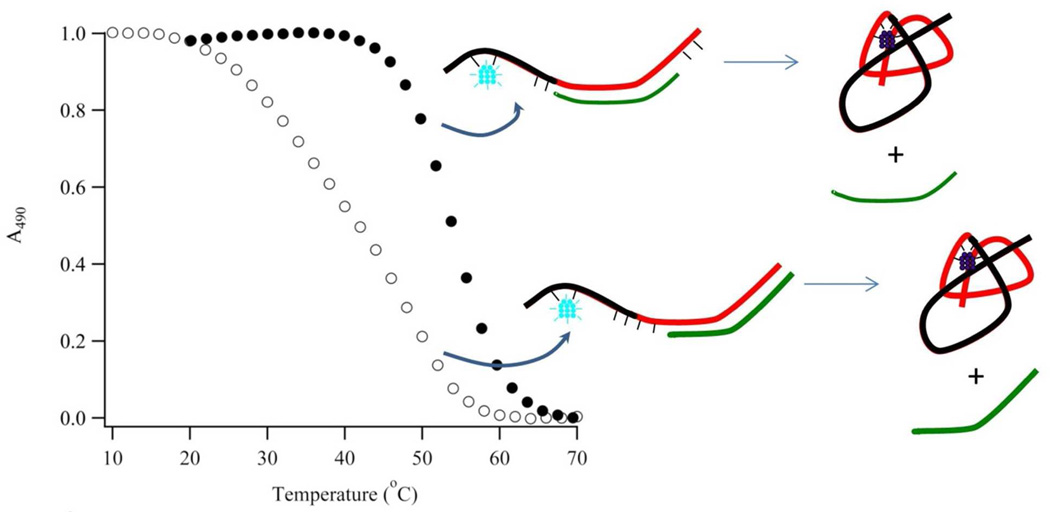
Figure 5. Cluster-DNA Equilibrium.
(A) Higher temperatures favor the violet over the blue-green absorption. An isosbestic point develops between the absorption bands and supports direct conversion of the violet to the blue-green conjugate via the complement S2C12c. (B) Blue-green cluster absorbances at 490 nm were extracted from the spectra and track the reaction progress. Extrapolated upper and lower baselines (dotted lines) define the reactant and product states over the full temperature range. These baselines yield fractional conversion of the blue-green conjugates and equilibrium constants for its dissociation. (C) A van’t Hoff representation for dissociation of the blue-green conjugate with S1-S212c:S2C12c yields the enthalpy and entropy changes for denaturation and hence cluster transformation.
Cluster Transformation and Hybridization
Complements S2C orchestrated a DNA structural transformation. Using size exclusion chromatography, blue-green conjugates eluted earlier than their violet predecessors and thus had larger hydrodynamic radii (Fig. 3A and B). We attributed this increased size to hybridization because complement S2C also reduced elution time for native S1-S2 (Fig. 3A and B). Modified complements substantiated formation of S2-S2C duplexes within the blue-green conjugates (Fig. 3C). Two 5’ sequences, dT10− and dT20−, were appended to S2C to indirectly speed cluster elution. These modifications were chemically innocuous because they project away from the S1 cluster domain. Furthermore, the N3 cluster binding sites in thymine are protonated and thus blocked at neutral pH because their pKa is 9.7.31,33,36 Complements S2C12a, dT10-S2C12a, and dT20-S2C12a progressively reduced blue-green cluster elution times, and this trend qualitatively established that S2C hybridized with the S1-S2-cluster complex. We also quantitatively evaluated these changes. Shifts in retention time empirically predict changes in hydrodynamic radii for structurally homologous biopolymers (Supporting Information – Section B).48,49 Based on the hydrodynamic radius of the blue-green conjugate with S1-S212a:S2C12a, we measured size changes due to the two homooligonucleotide thymine appendages - α for dT10 and β for dT20. Cluster conjugates and their native analogs have β/α = 2.08 ± 0.10 and 2.06 ± 0.10, respectively, and thus both experience a two-fold change from dT10 to dT20. A mixture of S2C12a and dT10−S2C12a was also reacted with the S1-S212a:silver cluster conjugate. Two chromatographic peaks suggested that only monomeric duplex-cluster conjugates formed (Fig. S5).27 Based on these chromatographic studies, we concluded that association of complement S2C with S1-S2 caused the violet to blue-green cluster conversion. This new environment was spectroscopically distinct not only because of red-shifted absorption and enhanced emission but also because of a weaker circular dichroism response at 490 nm (Fig. S2).
Cluster integrity
We also examined the spectral changes from a cluster perspective. Two experiments evaluated cluster stoichiometry. First, like their violet predecessors, blue-green conjugates were isolated by size exclusion chromatography and then analyzed by atomic emission. Resulting concentrations of silver and phosphorus provided the following Ag:DNA stoichiometries: 11.2 ± 0.5 Ag:S1-S212a:S2C12a, 10.7 ± 1.7 Ag:S1-S212c:S2C12c, and 10.4 ± 1.0 Ag:S1-S216:S2C16. These DNA hosts vary in the length and sequence, yet consistent silver stoichiometries supported a single ~11 atom cluster. This assignment is supported by mass spectrometry studies that identified an 11 silver atom DNA-adduct with similar blue-green electronic spectra.25,41 Thus, within our experimental error, the violet and blue-green clusters had the same stoichiometry.
Second, to further probe cluster integrity, we extracted the complement from a blue-green cluster conjugate (Fig. 4). Initially, a composite strand with a 14-nucleobase recognition site, S1-S214, formed the violet cluster. Then, 2.0 equivalents of the longer complement S2C16 generated the blue-green cluster, and the resulting S1-S214:S2C16 host had a two-nucleobase overhang on its terminus. In the last step, 2.1 equivalents of the perfectly-matched 16-nucleobase complement S216 extracted the hybridized stand and regenerated single-stranded S1-S214. The original violet absorption correspondingly recovered. Spectra reversed in both concentrated and dilute solutions, and this concentration-independence supports intrastrand transfer between two cluster binding sites (Fig. S6). Collectively, elemental analysis and reversible structural/spectral changes support the following cluster transformation reaction:
| (Eq. 2) |
Figure 4. Cluster Integrity.
Absorption spectra acquired for S1-S214 that is fully cycled from its single-stranded to its hybridized and back to its single-stranded state. First, single-stranded S1-S214 forms the violet cluster (solid line). Second, this conjugate hybridizes with its longer complement S2C16 (dotted line). Third, this conjugate reacts with the isolated recognition site S216 (dashed line). The first and third spectra match, and this spectral reversibility is also accomplished in diluted solutions (Fig. S6). These results support transfer of an intact cluster between two distinct DNA binding sites.
Cluster-DNA Equilibrium
This cluster transformation reversibly shifted with temperature, and van’t Hoff analysis revealed underlying thermodynamic and structural factors (Fig. 5). We measured equilibrium constants for thermal dissociation of the blue-green conjugates (Eq. 2). Equilibrium concentrations were based on blue-green cluster absorbances at 490 nm, and the extinction coefficient for this chromophore was derived from fluorescence correlation spectroscopy (Fig. S7). This technique spectrally eliminated unlabeled DNA and enumerated the green emissive species within an optically-defined volume. Cluster concentrations and absorbances were measured for five S1-S2:S2C constructs with 12, 14, and 16 nucleobase recognition sites (Table 1 – Entries A–E) and for samples with different degrees of dilution. These measurements produced similar extinction coefficients, so we used a common value of 31.7 (± 5.4) × 103 M−1cm−1 . Using material balance based on Eq. 2, blue-green cluster concentrations also provided equilibrium concentrations of the violet cluster and its complement. Equilibrium constants were measured at different temperatures, and van’t Hoff analysis yielded the enthalpy and entropy changes for dissociation of the blue-green conjugates.35
Three concentration studies supported our thermodynamic analysis. First, dilution diminished the temperatures needed to dissociate blue-green conjugates (Fig. S8). Melting temperatures corresponded to ΔG = 0, and the concentration dependence of the melting temperatures also yielded enthalpy and entropy changes.35 These thermodynamic parameters agree with those derived from van’t Hoff analysis (Table 1 – Entries C and Cc). Second, complement S2C concentrations were varied between two and four fold excess relative to S1-S2. This range ensured conversion of the violet and blue-green clusters. Third, samples were prepared with the normal concentration of 8 Ag+:4 BH4− :S1-S2 and also with 4 Ag+:2 BH4−:S1-S2 and 2 Ag+:1 BH4−:S1-S2. These amounts adjusted the relative proportion of labeled and unlabeled DNA (Fig. 3A). All three samples had the same overall concentrations of S1-S2 and S2C, yet enthalpy and entropy changes for dissociation of the blue-green conjugate were similar (Table 1 – Entries B, B4/2, and B2/1). This consistency indicated that the cluster and complement concentrations Eq. 2 tracked the overall oligonucleotide concentrations.
We combined thermodynamic studies with DNA structural changes and identified two factors that shifted the cluster equilibrium. First, hybridization was investigated using five composite strands with 12 to 16 nucleotide recognition sites were used (Table 1 – Entries A–E). These S2 recognition sites were spectrally interchangeable because their S1-S2 composite strands formed both the violet precursor and its blue-green product. However, cluster conjugates with these five strands were thermodynamically distinct. Enthalpies and entropies for dissociation were similar for three different 12-nucleotide recognition sites but progressively increased with 14 to 16 nucleotide recognition sites (Table 1 - Entries A-C, D, and E). In a similar manner, enthalpy and entropy changes for denaturation of the native S1-S2:S2C analogs also increased with duplex length (Table 1 - Entries A'-C', D', and E'). Thus, accumulation of base stacking and pairing interactions stabilized duplex appendages and consequently favored the blue-green cluster.31
Second, an opposing factor disfavored the blue-green cluster because its DNA hosts had lower enthalpy and entropy changes for denaturation than their native DNA counterparts. Two perspectives suggested that the violet cluster coordinated single-stranded S1-S2 and thereby promoted duplex denaturation. From a structural standpoint, this cluster coordinated multiple nucleobases because it condensed the native S1-S2 conformation and produced a relatively strong circular dichroism response (Figs. 3A and S2). From a thermodynamic standpoint, violet cluster coordination with S1-S2 could have reduced the enthalpic penalty of duplex denaturation in S1-S2:S2C. In addition, folding could have tempered the entropic advantage of producing an unstacked, single-stranded DNA.
This thermodynamic constraint was also supported by unpaired nucleobases within S1-S2:S2C. Experiments were based on a composite strand S1-S216 with a 16 nucleobase recognition site. It hybridized with two shorter 14-nucleobase complements, S2C14 and S2C14a (Fig. 6). These associated with the 5’ and 3’ regions of the recognition site, respectively. The native duplexes had similar enthalpy and entropy changes relative to each other and also relative to fully-complementary S1-S214:S2C14 (Table 1 – Entries D’, F’, and G’). Thus, inherent stabilities of these 14 nucleotide duplexes were insensitive to their position within S1-S216. However, the blue-green conjugate with S1-S216:S2C14a had lower enthalpy and entropy changes than its analogous complex with S1-S216:S2C14 (Fig. 6 and Table 1 – Entries F and G). To understand this difference, we compare these conjugates with the fully complementary analog S1-S214:S2C14 (Table 1 – Entry D). Enthalpy and entropy changes for denaturation of S1-S214:S2C14 and S1-S216:S2C14 conjugates had comparable values. This similarity suggested that the two overhanging nucleotides in S1-S216:S2C14 were thermodynamically innocuous because they were distant from the 5’ cluster domain. Lower enthalpy and entropy changes for the S1-S216:S2C14a conjugate suggested that the two unpaired nucleobases at the S1-S2 junction destabilized the duplex. Unpaired nucleobases coordinate silver clusters, so open coordination sites could have complexed with the violet cluster and facilitated folding and duplex denaturation.
Figure 6. Cluster-DNA Equilibrium.
Fractional conversion of the blue-green clusters conjugated with S1-S216:S2C14 (open circles) and S1-S216:S2C14a (closed circles). The slower rate of transition for the latter case suggests that the unpaired nucleobases in the interior of S1-S2 facilitate folding and denaturation of this duplex. Schematic representations of these two cases are included to emphasize the positions of the two unpaired nucleobases.
Discussion
We investigated a specific silver cluster-DNA conjugate that spectrally identifies oligonucleotides. This sensor has four characteristics: a single-stranded oligonucleotide forms a specific reporter cluster; hybridization distinguishes two optically-resolved cluster environments; cluster size is invariant; the cluster folds single-stranded DNA and impedes hybridization. These characteristics help explain how the sensor responds to oligonucleotide analytes.
The DNA-based sensor has a specific 5’ cluster domain (S1) and a general 3’ recognition site (S2) (Fig. 1). The 5’ leading sequence is largely comprised of cytosine because this nucleobase preferentially stabilizes silver clusters.20,22 The 3’ lagging sequence binds its complementary strand, and recognition sites with different lengths and sequences are used (Table 1). This design principle is used by other silver cluster-DNA conjugates and suggests that sequences for a suite of spectrally-diverse reporters could be coupled with sequences for a range of analytes.50–53 We are developing this general sensing strategy. Earlier studies used a composite S1-S2 strand that also forms an ~11 silver atom cluster with violet absorption and low emission.27 The S1 component favors a cluster with near-infrared absorption and emission, and this cluster forms after a complement hybridizes with its modular recognition site. The present studies used an alternate cluster domain. Again, complementary targets strands alter the violet cluster, but now the new cluster has blue-green absorption and strong green emission. We examine this spectral transformation from structural, stoichiometric, and thermodynamic perspectives.
The S2 component governs DNA secondary structure and the cluster environments. Complements to this recognition site create an isosbestic point between the cluster absorptions, and complements with thymine appendages speed blue-green cluster elution (Figs. S3A and 3C). These results establish that hybridization switches a single-stranded host for the violet cluster to a mixed single- and double-stranded host for the blue-green cluster (Figs. 1, 3, 4, S4, and S5). We propose that unpaired nucleobases distinguish these two binding sites. Single-stranded DNA exposes nucleobases and their electron-rich heteroatoms, such as the N3 of cytosine and thymine.21,36,54 However, these sites are acidic, and their protonation at low pH quenches cluster emission. Base pairing also protonates these sites and redefines cluster environments.20,24 To illustrate, hybridizing strands invade cluster binding sites and quench emission, while repositioned complements relieve static quenching.51 Thus, base pairing overrides cluster association, but the effect is reversible. Hybridization also regulates cluster environments within S1-S2. Single-stranded S1-S2 favors a violet cluster with low emission. Complement S2C hybridizes with S1-S2, and a different cluster with blue-green absorption and green emission develops (Fig. 1). The extent of base pairing within S2 controls the spectral switching because short complementary strands compromise cluster stability (Fig. 6). These results suggest that hybridization regulates cluster coordination by blocking association with the S2 component and thereby confining the cluster to the S1 component. Identifying the sites of cluster coordination would test this proposal.
DNA secondary structure also depends on the cluster adducts. The violet and blue-green clusters restrict the conformations of their single-stranded and double/single-stranded hosts respectively (Fig. 3A and B). This condensation suggests that the clusters coordinate multiple nucleobases within their monomeric DNA hosts. A near-infrared cluster forms within a related S1-S2 construct, but it coordinates multiple nucleobases through intermolecular DNA association.27 Two S1 components C3AC3AC3TC3A are recruited by two ~11 silver atom clusters and generate a dimeric assembly. We are addressing this stoichiometric and structural distinction between the monomeric blue-green and dimeric near-infrared conjugates by altering the sequence of the repeated motif in the latter case.
Hybridization defines two binding sites within S1-S2, and we measured the sizes of the cluster adducts. Violet and blue-green cluster adducts were chromatographically isolated, and elemental analysis revealed a common cluster with ~11 silver atoms (Fig. 3). Reversible spectral changes also support size conservation. Blue-green absorption bands revert to their original violet absorption bands when complement S2C is extracted from the S1-S2:S2C host (Figs. 4 and S6). We propose that these spectrally distinct forms are different isomers of the same cluster. Silver clusters with ≳5 atoms adopt three-dimensional structures, and these have two general characteristics. First, isomers have distinct electronic structures.5,6 The violet and blue-green clusters exhibit this characteristic because both their spectra and radiative relaxation rates differ. Second, cluster isomers are separated by shallow potential wells, and the distribution of cluster species depends on coordinating ligands.2,55,56 For example, thiols control the strength of chiroptical responses from gold clusters.57 Likewise, distinct circular dichroism spectra support different coordination environments for the violet and blue-green clusters (Fig. S2). Raman spectroscopy may distinguish such isomers.55,56
Hybridization and cluster-DNA coordination work in concert and control conjugate thermodynamics. Duplex length and hence base stacking/pairing within the S2:S2C duplexes control blue-green cluster stability (Table 1 – Entries A–E). However, these conjugates are less stable than their native DNA counterparts (Table 1 – Entries A’-E’). Our experiments suggest that this instability is driven by the violet cluster. Structural data support formation of a global complex with S1-S2. This cluster folds its host strand and has a strong circular response, and these observations suggest that this cluster binds multiple nucleobases within single-stranded S1-S2 (Figs. 3A and S2). The violet cluster also has a thermodynamic propensity to coordinate nucleobases because exposed nucleobases promote denaturation (Fig. S6). Thus, cluster-induced folding opposes innate DNA hybridization. These two factors highlight a distinctive feature of silver clusters – they are DNA ligands that both report and modulate analyte association. We continue to investigate this issue from structural and thermodynamic viewpoints. Prior studies established global structures of DNA-cluster conjugates, and finer structural details may be gleaned from fluorescence labeling.23,26,58,59 Silver cluster coordination supplements innate base stacking/pairing interactions, and we are working to balance these orthogonal interactions to more selectively identify closely-related targets.60–62
Conclusion
We have described a silver cluster that both reports and controls its DNA environment. The ~11 atom cluster moves between two spectroscopically distinct binding sites, and this transfer is accomplished by hybridization. However, the cluster tempers conversion by folding its single-stranded DNA host. This interplay between innate DNA-DNA interactions and silver cluster-DNA interactions offers the opportunity to both sensitively and selectively signal analyte recognition.
Supplementary Material
ACKNOWLEDGMENT
We thank the National Science Foundation (CBET-0853692), the National Institutes of Health (1R15GM102818), and the Beckman Foundation for support of this work. In addition, we thank the National Science Foundation (CHE-0718588 and CHE-0922834). D.N. and O.S. were supported by undergraduate research fellowships provided through the Furman Advantage program and a USE Award to Furman University from the Howard Hughes Medical Institute, respectively. J.P. is grateful for the support provided by the Henry Keith and Ellen Hard Townes Professorship. This work was performed, in part, at the Center for Integrated Nanotechnologies, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Los Alamos National Laboratory (Contract DE-AC52–06NA25396) and Sandia National Laboratories (Contract DE-AC04–94AL85000).
Footnotes
Supporting Information
The supporting information contains 8 figures describing spectra associated with the isolated S1 cluster domain, circular dichroism and corresponding absorption spectra of the violet and blue-green cluster conjugates, spectra associated with the complement and salt dependence of the cluster transformation, temperature dependence of the violet cluster spectra, chromatograms from a mixture of modified and unmodified complements, absorption spectra of the single-stranded, hybridized, and complement-deficient cluster conjugates in dilute solution, fluorescence correlation results, and concentration-dependent melting temperatures. In Appendix B, the effect of dT10 and dT20 appendages on S2C on the retention times of the blue-green cluster is described. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.de Heer WA. Rev. Mod. Phys. J1 - RMP. 1993;65:611–676. [Google Scholar]
- 2.Aikens CM. J. Phys. Chem. Lett. 2011;2:99–104. doi: 10.1021/jz101499g. [DOI] [PubMed] [Google Scholar]
- 3.Parker JF, Fields-Zinna CA, Murray RW. Acc. Chem. Res. 2010;43:1289–1296. doi: 10.1021/ar100048c. [DOI] [PubMed] [Google Scholar]
- 4.Qian H, Zhu M, Wu Z, Jin R. Acc. Chem. Res. 2012;45:1470–1479. doi: 10.1021/ar200331z. [DOI] [PubMed] [Google Scholar]
- 5.Bonacic-Koutecky V, Veyret V, Mitric R. J. Chem. Phys. 2001;115:10450–10460. [Google Scholar]
- 6.Schmidt M, Cahuzac P, Bréchignac C, Cheng H-P. J. Chem. Phys. 2003;118:10956. [Google Scholar]
- 7.Link S, El-Sayed MA. Annu. Rev. Phys. Chem. 2003;54:331–366. doi: 10.1146/annurev.physchem.54.011002.103759. [DOI] [PubMed] [Google Scholar]
- 8.Richards CI, Choi S, Hsiang J-C, Antoku Y, Vosch T, Bongiorno A, Tzeng Y-L, Dickson RM. J. Am. Chem. Soc. 2008;130:5038–5039. doi: 10.1021/ja8005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Patel SA, Dickson RM. Angew. Chem. Int. Ed. 2007;46:1927–2121. doi: 10.1002/anie.200123456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han BY, Wang EK. Analytical and Bioanalytical Chemistry. 2012;402:129–138. doi: 10.1007/s00216-011-5307-6. [DOI] [PubMed] [Google Scholar]
- 11.Obliosca JM, Liu C, Batson RA, Babin MC, Werner JH, Yeh H-C. Biosensors. 2013;3:185–200. doi: 10.3390/bios3020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petty JT, Story SP, Hsiang JC, Dickson RM. Journal of Physical Chemistry Letters. 2013;4:1148–1155. doi: 10.1021/jz4000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma J, Yeh HC, Yoo H, Werner JH, Martinez JS. Chem. Commun. 2010;46:3280–3282. doi: 10.1039/b927268b. [DOI] [PubMed] [Google Scholar]
- 14.Petty JT, Fan C, Story SP, Sengupta B, St. John Iyer A, Prudowsky Z, Dickson RM. J. Phys. Chem. Lett. 2010;1:2524–2529. doi: 10.1021/jz100817z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavis LD, Raines RT. ACS Chem. Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vosch T, Antoku Y, Hsiang J-C, Richards CI, Gonzalez JI, Dickson RM. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12616–12621. doi: 10.1073/pnas.0610677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patel SA, Cozzuol M, Hales JM, Richards CI, Sartin M, Hsiang J-C, Vosch T, Perry JW, Dickson RM. J. Phys. Chem. C. 2009;113:20264–20270. doi: 10.1021/jp9079537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petty JT, Fan C, Story SP, Sengupta B, Sartin M, Hsiang J-C, Perry JW, Dickson RM. J. Phys. Chem. B. 2011;115:7996–8003. doi: 10.1021/jp202024x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards CI, Hsiang J-C, Senapati D, Patel S, Yu J, Vosch T, Dickson RM. J. Am. Chem. Soc. 2009;131:4619–4621. doi: 10.1021/ja809785s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petty JT, Zheng J, Hud NV, Dickson RM. J. Am. Chem. Soc. 2004;126:5207–5212. doi: 10.1021/ja031931o. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie CM, Johnsen KR, Kiser JR, Antoku Y, Dickson RM, Petty JT. J. Phys. Chem. C. 2007;111:175–181. doi: 10.1021/jp0648487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soto-Verdugo V, Metiu H, Gwinn E. J. Chem. Phys. 2010;132:195102. doi: 10.1063/1.3419930. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta B, Springer K, Buckman JG, Story SP, Abe OH, Hasan ZW, Prudowsky ZD, Rudisill SE, Degtyareva NN, Petty JT. J. Phys. Chem. C. 2009;113:19518–19524. [Google Scholar]
- 24.Gwinn EG, O'Neill P, Guerrero AJ, Bouwmeester D, Fygenson DK. Adv. Mater. 2008;20:279–283. [Google Scholar]
- 25.O'Neill PR, Velazquez LR, Dunn DG, Gwinn EG, Fygenson DK. J. Phys. Chem. C. 2009;113:4229–4233. [Google Scholar]
- 26.Driehorst T, O’Neill P, Goodwin PM, Pennathur S, Fygenson DK. Langmuir. 2011;27:8923–8933. doi: 10.1021/la200837z. [DOI] [PubMed] [Google Scholar]
- 27.Petty JT, Giri B, Miller IC, Nicholson DA, Sergev OO, Banks TM, Story SP. Anal. Chem. 2013;85:2183–2190. doi: 10.1021/ac303531y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lippert B. Coord. Chem Rev. 2000;200–202:487–516. [Google Scholar]
- 29.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Angew. Chem. Int. Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. Nat. Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 31.Bloomfield VA, Crothers DM, Tinoco J. Ignacio Nucleic Acids: Structures, Properties, and Functions. Sausaltio, CA: University Science Books; 2000. [Google Scholar]
- 32.Gueron M, Leroy JL. Current Opinion in Structural Biology. 2000;10:326–331. doi: 10.1016/s0959-440x(00)00091-9. [DOI] [PubMed] [Google Scholar]
- 33.Petty JT, Story SP, Juarez S, Votto SS, Herbst AG, Degtyareva NN, Sengupta B. Anal. Chem. 2012;84:356–364. doi: 10.1021/ac202697d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen RH, Davidson N. Biopolymers. 1966;4:17–32. [Google Scholar]
- 35.Mergny J-L, Lacroix L. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 36.Sengupta B, Ritchie CM, Buckman JG, Johnsen KR, Goodwin PM, Petty JT. J. Phys. Chem. C. 2008;112:18776–18782. doi: 10.1021/jp804031v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Widengren J, Mets U, Rigler R. J. Phys. Chem. 1995;99:13368–13379. [Google Scholar]
- 38.Gendron PO, Avaltroni F, Wilkinson KJ. Journal of Fluorescence. 2008;18:1093–1101. doi: 10.1007/s10895-008-0357-7. [DOI] [PubMed] [Google Scholar]
- 39.Phan AT, Gueron M, Leroy JL. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 40.Jin R, Qian H, Wu Z, Zhu Y, Zhu M, Mohanty A, Garg N. J. Phys. Chem. Lett. 2010;1:2903–2910. [Google Scholar]
- 41.Schultz D, Gwinn EG. Chem. Commun. 2012;48:5748–5750. doi: 10.1039/c2cc17675k. [DOI] [PubMed] [Google Scholar]
- 42.Guo J, Kumar S, Bolan M, Desireddy A, Bigioni TP, Griffith WP. Anal. Chem. 2012;84:5304–5308. doi: 10.1021/ac300536j. [DOI] [PubMed] [Google Scholar]
- 43.Harb M, Rabilloud F, Simon D, Rydlo A, Lecoultre S, Conus F, Rodrigues V, Felix C. J. Chem. Phys. 2008;129:194108. doi: 10.1063/1.3013557. [DOI] [PubMed] [Google Scholar]
- 44.Martinez JI, Fernandez EM. European Physical Journal D. 2009;52:199–202. [Google Scholar]
- 45.Liu CW, Feng C-S, Fu R-J, Chang H-W, Saillard J-Y, Kahlal S, Wang J-C, Chang IJ. Inorganic Chemistry. 2010;49:4934–4941. doi: 10.1021/ic902571q. [DOI] [PubMed] [Google Scholar]
- 46.McNaught AD, Wilkinson A, editors. Second ed. Blackwell Scientific Publications: Oxford; 1997. [Google Scholar]
- 47.Manning GS. Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 48.Sim AYL, Lipfert J, Herschlag D, Doniach S. Phys. Rev. E. 2012;86:021901. doi: 10.1103/PhysRevE.86.021901. [DOI] [PubMed] [Google Scholar]
- 49.Akers GK. In: The Proteins. Neurath H, Hill RL, editors. Vol. 1. New York: Academic Press; 1975. p. 547. [Google Scholar]
- 50.Yeh HC, Sharma J, Han JJ, Martinez JS, Werner JH. Nano Lett. 2010;10:3106–3110. doi: 10.1021/nl101773c. [DOI] [PubMed] [Google Scholar]
- 51.Petty JT, Sengupta B, Story SP, Degtyareva NN. Anal. Chem. 2011;83:5957–5964. doi: 10.1021/ac201321m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang SW, Vosch T. Anal. Chem. 2011;83:6935–6939. doi: 10.1021/ac201903n. [DOI] [PubMed] [Google Scholar]
- 53.Lan G-Y, Chen W-Y, Chang H-T. Biosens. Bioelectron. 2011;26:2431–2435. doi: 10.1016/j.bios.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 54.Neidig ML, Sharma J, Yeh H-C, Martinez JS, Conradson SD, Shreve AP. J. Am. Chem. Soc. 2011;133:11837–11839. doi: 10.1021/ja202797w. [DOI] [PubMed] [Google Scholar]
- 55.Parker JF, Choi J-P, Wang W, Murray RW. J. Phys. Chem. C. 2008;112:13976–13981. [Google Scholar]
- 56.Tlahuice-Flores A, Whetten RL, Jose-Yacaman M. J. Phys. Chem C. 2013;117:12191–12198. doi: 10.1039/c3cp53837k. [DOI] [PubMed] [Google Scholar]
- 57.Provorse MR, Aikens CM. J. Am. Chem. Soc. 2010;132:1302–1310. doi: 10.1021/ja906884m. [DOI] [PubMed] [Google Scholar]
- 58.Rist MJ, Marino JP. Curr. Org. Chem. 2002;6:775. [Google Scholar]
- 59.Lilley DMJ. Q. Rev Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 60.Takezawa Y, Shionoya M. Acc. Chem. Res. 2012;45:2066–2076. doi: 10.1021/ar200313h. [DOI] [PubMed] [Google Scholar]
- 61.Sivakova S, Rowan SJ. Chem. Soc. Rev. 2005;34:9–21. doi: 10.1039/b304608g. [DOI] [PubMed] [Google Scholar]
- 62.Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Y, Medley CD, Cao Z, Li J, Colon P, Lin H, Tan W. Angew. Chem., Int. Ed. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.