Abstract
Adenosine to inosine RNA editing catalyzed by ADAR enzymes is common in humans and altered editing is associated with disease. Experiments using substrate RNAs with adenosine analogs at editing sites are useful for defining features of the ADAR reaction mechanism. Here we evaluated the reactivity of ADAR2 with RNA containing the emissive adenosine analog thieno[3,4-d]-6-aminopyrimidine (thA). This nucleoside was incorporated into a mimic of the glutamate receptor B (GluR B) mRNA R/G editing site. We found that thA is recognized by AMV reverse transcriptase as A, and is deaminated rapidly by human ADAR2 to give thI. Importantly, ADAR reaction progress can be monitored by following the deamination-induced change in fluorescence of the thA-modified RNA. The observed high thA reactivity adds to our understanding of the structural features that are necessary for an efficient hADAR2 reaction. In addition, the new fluorescent assay is expected to accelerate mechanistic studies of ADARs.
Keywords: fluorescent nucleosides, RNA editing, ADAR, adenosine deaminase
Adenosine deaminases acting on RNA (ADARs) are enzymes that deaminate adenosine to form inosine in the context of double-helical RNA structure. Inosine base pairs with cytosine, and so is recognized by the translation machinery and polymerases as guanosine, often leading to codon changes in mRNA. This phenomenon is widespread in the human transcriptome with thousands of inosine sites identified.[1] In addition, misregulation of A to I RNA editing is associated with human disease,[2] such as Dyschromatosis Symmetrica Hereditaria (DSH)[3] and the autoimmune disease Aicardi-Goutieres Syndrome (AGS).[4] Hyper editing has been observed at certain sites in cancer cells, such as in the mRNA for AZIN1 (antizyme inhibitor 1),[5] leading to more aggressive tumor behavior during esophageal squamous cell carcinoma progression.
Studies of the ADAR proteins are hampered by the fact that kinetic experiments typically require time consuming methods such as (a) reverse transcription followed by either gel electrophoresis or PCR and DNA sequencing for each assay time point[6] or (b) 32P labeling of the 5′ phosphate of the reactive adenosine followed by laborious nuclease digestion and thin layer chromatography resolution of labeled nucleotides for each time point.[7] Monitoring ADAR-mediated editing by fluorescence changes of the reactive A would greatly simplify data acquisition, which in turn could facilitate mechanistic studies. However, there are no reports of adenosine analogs that are ADAR-reactive while also having useful fluorescent properties. We therefore developed methods to incorporate into ADAR substrates thA and thI, members of a new class of fluorescent nucleoside analogs derived from thieno[3,4-d]-pyrimidine as the heterocyclic nucleus.[8] These analogs exhibit desirable properties for use in biophysical experiments, such as isomorphicity with natural bases, intact Watson-Crick hydrogen bonding faces, and intense visible emission.[8] Thieno[3,4-d]-6-aminopyrimidine (thA), the emissive analog for adenosine (Figure 1A), was recently shown to mimic adenosine in the reaction of adenosine deaminase (ADA), a metabolic enzyme that deaminates the free nucleoside.[9] thA is processed by ADA with a reduced Vmax (~ 3 fold) and increased KM (~ 20 fold) compared to the natural substrate, likely because of the carbon-for-nitrogen substitution at the purine 7-position,[9] resulting in a loss of an important hydrogen bonding contact to an aspartic acid residue in the ADA active site.[10] Nevertheless, since thA and the corresponding inosine analog (thI, Figure 1A) exhibit different fluorescence properties, thA enabled a fluorescence-based, high throughput screen for inhibitors of ADA.[9] We anticipated that thA could be an even better substrate for human ADAR2 because the reaction rate for this enzyme is less dependent on the presence N7 in the edited purine than is ADA.[11]
Figure 1.
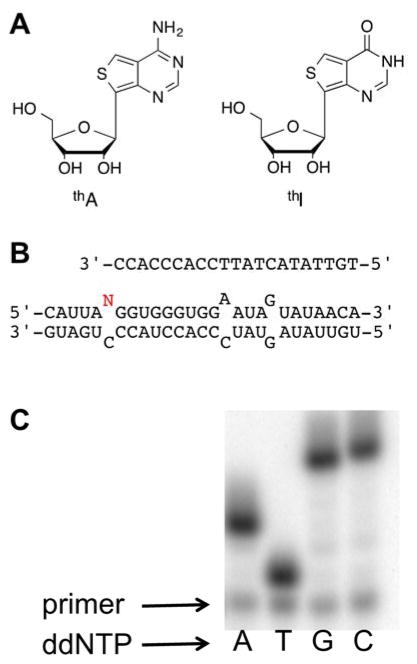
(A) Structures of emissive RNA nucleosides thA and thI. (B) Top: labeled primer used in the primer extension assay, Bottom: RNA duplex with editing site shown in red. (C) Only dideoxyT base pairs with thA, indicating that it is recognized by AMV RT as A.
Exploiting ADARs-mediated thA to thI fluorescence changes would be particularly useful given the complexities of measuring deamination rates for specific adenosines in RNA molecules.[7] However, the thA analog had not been previously incorporated into RNA and its reactivity in ADAR reactions was, therefore, unknown. The carbon-carbon bond linking the base to the sugar in thA and the sulfur atom corresponding to purine position 8 are structural features not previously tested in ADAR substrate analogs.
The proposed reaction mechanism for ADARs involves formation of a hydrated intermediate that lacks the aromaticity of adenosine or inosine. Modifications that decrease the aromaticity of adenosine facilitate hydration and increase the reaction rate.[11b, 12] We have previously used quantum mechanical calculations to predict the propensity for hydration of various purine analogs relative to 9-methylpurine.[13] These generally correlate well to observed ADAR2-catalyzed deamination rates for the corresponding adenosine analogs in RNA, particularly when the substrates match the shape of adenosine.[11b] Using these same methods, we calculated the covalent hydration free energy for thieno[3,4-d]-pyrimidine to be 8.5 kcal/mol more favorable than for 9-methylpurine (Figure 2). Given our previous studies, this difference suggested thA would be an excellent ADAR2 substrate.[11b, 13]
Figure 2.
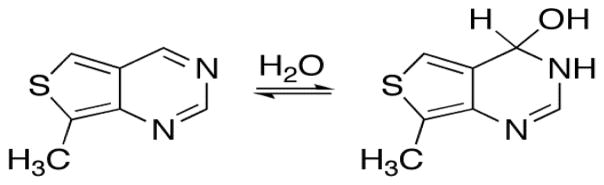
Thieno[3,4-d]-pyrimidine covalent hydration. The calculated hydration free energy compared to that of 9-methylpurine predicts high ADAR reactivity.[13]
The fluorescence and predicted ADAR reactivity of the thieno analogs led us to synthesize modified RNA, which required the preparation of the corresponding phosphoramidites (Figure 3). For thA, regioselective protection of the 2′-hydroxyl group to give the corresponding 2′-O-TBDMS via the 3′,5′-O-di-tert-butylsilyl-protected free nucleoside,[8] was followed by protection of the N6 amine with the N,N-diisobutylformamidine group (Supplementary Scheme 1). Tritylation of the 5′-hydroxyl group and phosphitylation at the 3′-hydroxyl group provided the thA phosphoramidite, suitable for automated RNA synthesis. The phosphoramidite of thI was synthesized in a similar manner (Supplementary Scheme 2). The free thI nucleoside synthesis was reported in our previous publication.[9] These phosphoramidites were used to incorporate the thieno analogs into a previously characterized mimic of the GluR B R/G editing site (Figure 1B) that is efficiently deaminated by human ADAR2 and various mutants of this enzyme.[7]
Figure 3.
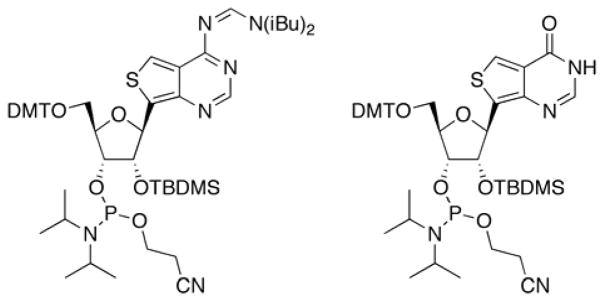
Structures of phosphoramidites for thA (left) and thI (right) used to prepare analog-containing RNA.
One method used for measuring ADAR activity consists of following the deamination reaction progress by a primer extension reaction.[14] We found both that thA is recognized by AMV reverse transcriptase like A, with only ddTTP incorporated at the complementary position (Figure 1C), and that thA is a substrate for ADAR2 (Table 1). Under single turnover conditions at saturating concentration of human ADAR2, we measured a kobs for the thA deamination reaction approximately two-fold higher than that observed for the reaction with adenosine (Table 1). The rate of ADAR2 editing of adenosine in this substrate using this assay was comparable to that found using the AMP/IMP thin layer chromatography assay.[15] Thus, the thA analog is an excellent substrate for human ADAR2 consistent with the prediction based on calculated hydration free energies. The carbon-carbon bond linking the base analog to ribose and the thieno heterocycle are structural features that appear to be well tolerated by the ADAR2 active site.
Table 1.
Rate of deamination of A and thA by ADAR2 in the primer extension assay
| substrate (N) | kobs, min−1 | krel |
|---|---|---|
| A | 0.87 ± 0.42 | 1 |
| thA | 1.83 ± 0.66 | 2.1 |
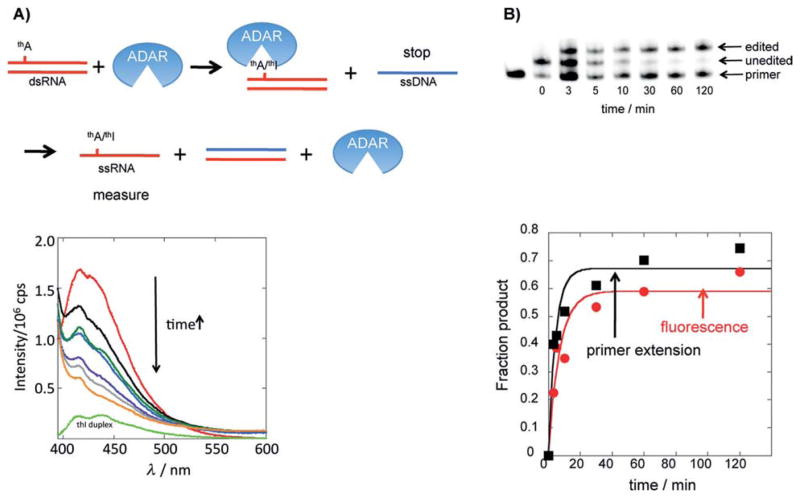
We recorded fluorescence spectra for thA- and thI-modified GluR B R/G site RNAs either in the single stranded form or in a duplex (see Figure 1B for duplex structure) in the presence of ADAR assay buffer components (Figure 4).[7] Excitation at 380 nm of the thA-containing single strand showed strong emission with a maximum at 417 nm (Figure 4A). Interestingly, the thI analog emission is quenched to a greater degree in the context of the oligonucleotide than is the emission for the thA analog such that, in contrast to the case of the free nucleosides, emission of thI-containing RNA is substantially lower than for the thA-containing RNA (Figure 4A). While the precise origin of this difference is difficult to define, it is likely a combination of factors such as different ground state and excited state interactions, particularly with neighboring nucleobases, and differences in solvation. While fluorescence quenching is also observed upon duplex formation, the emission of thA and thI differs by approximately twofold and remains distinct (Figure 4B).
Figure 4.
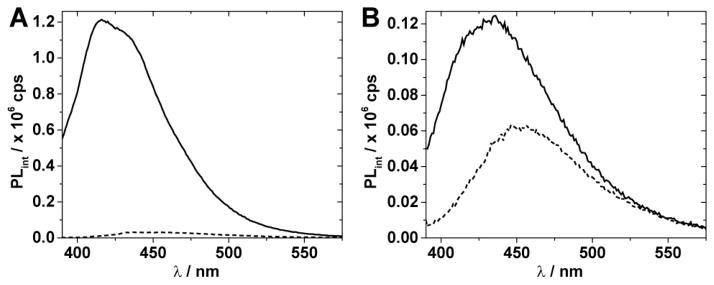
Emission spectra of 1 μM GluR B R/G site single stranded RNA (A) and duplex RNA (B) with incorporated thA (solid line), and thI (dashed line) in ADAR2 buffer at 30 °C containing DTT (0.5 mM), yeast tRNA (1 μg/μL), and RNasin (0.16 U/μL). Spectra were recorded on a Horiba Fluoromax-3 after excitation at 380 nm using slit-widths of 5 nm. Spectra are corrected for the blank fluorescence signal measured using the same instrument settings.
The current in vitro assays for ADAR deamination are not amenable to high throughput formats, and often necessitate the use of radiolabeled substrates. Since we have shown that RNAs containing thA or thI exhibit different fluorescence properties, we endeavored to develop a fluorescence assay for the ADAR reaction. The quenching of the fluorophores when in the context of an RNA duplex, not uncommon for fluorescent nucleosides, complicated direct measurement of fluorescence changes induced by ADAR (Figure 4B).[16] In addition, given the high enzyme/RNA ratios typical of in vitro ADAR reactions, real time fluorescence changes observed during an ADAR reaction are influenced by base flipping, a phenomenon known to alter the observed emission of the fluorescent base due to unstacking.[15] To overcome these complications, we devised a protocol to stop the reaction at specified time points while rendering the thA/thI-containing oligonucleotide single stranded by adding an excess of a complementary unlabeled single stranded DNA and heating the sample (Figure 5A (top)). The progress of the reaction could then be monitored by measuring the fluorescence decrease at 417 nm (380 nm excitation) since thA in single stranded RNA is substantially more emissive under these conditions than is thI in single stranded RNA (see Figure 4A above). In addition, since ADAR has low affinity for single stranded RNA and the heat denatures and inactivates the enzyme, ADAR-induced base flipping would not affect the fluorescence of the sample. Using this protocol, we observed the expected decrease in fluorescence as a function of time for the reaction of the thA-modified GluR B R/G site duplex with an active N-terminal deletion mutant of human ADAR2 (hADAR2-RD)[17] (Figure 5A (bottom). To confirm that the changes in fluorescence were indeed due to the deamination reaction, we carried out a parallel analysis of the reaction using the primer extension assay described above, finding good agreement between the two (Figure 5B).
Figure 5.
Fluorescence-based assay for an ADAR reaction. A) Top: The procedure used to stop the reaction such that thA/thI fluorescence can be measured in ssRNA form. Bottom: Plot of change in fluorescence as a function of time for the reaction of 1 mm thA-modified GluR B R/G site substrate and 1 mm hADAR2-RD. Red: 0 min, black: 3 min, blue: 5 min, dark green, 10 min, purple: 30 min, gray: 60 min, orange: 120 min, light green: thI-containing duplex. B) Top: Image of gel used to resolve products for primer extension editing assay for reaction evaluated by changes in fluorescence in (A). Bottom: Fraction product vs time for primer extension assay (black squares) and fluorescence assay (red circles) (kobs=0.14 0.01 min¢1 (fluorescence); kobs=0.15 0.07 min¢1 (primer extension).
We considered the use of time resolved fluorescence measurements to follow the ADAR reaction with the emissive RNA constructs in real time. Such an approach could have potentially distinguished between different helical or flipped out states of thA and thI in the ADAR–RNA complex. However, in addition to practical considerations, the excited state lifetimes of the two emissive nucleoside analogs are too close to one another to facilitate an unambiguous detection (see Figures S1–S3, Table S2). We therefore resorted to the use of indirect but assured fluorescence discrimination between substrate and product as described above.
Current estimates have the number of A to I editing sites in the human transcriptome at > 15,000 and our understanding of how the surrounding RNA sequence and structure controls editing efficiency is limited.[18] New assays that allow mechanistic data to be acquired more rapidly, particularly with short, synthetic RNA substrates, will accelerate efforts necessary to advance this field. The new assay described here is fast, does not require the generation of radiolabelled substrates with short half-lives and should be adaptable to a multi-well plate format.
In conclusion, we have evaluated the reactivity of the novel fluorescent adenosine analog thA with an RNA editing adenosine deaminase. This analog is an excellent substrate for human ADAR2 with a deamination rate comparable to the enzyme’s natural substrate. thA-modified single stranded RNA is substantially more fluorescent than the same RNA modified with the deamination product, thI. This difference facilitated the development of a new ADAR assay based on monitoring the deamination-induced reduction in fluorescence of the thA/thI-containing RNA. This work will significantly accelerate mechanistic studies of ADARs and set the stage for high throughput screens for ADAR inhibitors.
Supplementary Material
Acknowledgments
P.A.B. acknowledges the National Institutes of Health for financial support in the form of grant GM061115. R.A.M was supported by a Graduate Research Fellowship from the National Science Foundation (1148897). Y.T. acknowledges the National Institutes of Health for support in the form of grant GM069773. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Footnotes
SUPPORTING INFORMATION AVAILABLE: Methods for phosphoramidite synthesis, modified RNA purification and characterization, primer extension and fluorescence assays for ADAR are available online.
References
- 1.Pullirsch D, Jantsch MF. RNA Biology. 2010;7:1. doi: 10.4161/rna.7.2.11286. [DOI] [PubMed] [Google Scholar]
- 2.a) Morabito MV, Ulbricht RJ, O’Neil RT, Airey DC, Lu P, Zhang B, Wand L, Emeson RB. Mol Pharmacol. 2010;77:895. doi: 10.1124/mol.109.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]; c) Maas S, Kawahara Y, Tamburro KM, Nishikura K. RNA Biology. 2006;3:1. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Miyamura Y, Suzuki T, Kono M, Inagaki K, Ito S, Suzuki N, Tomita Y. Am J Hum Genet. 2003;73:693. doi: 10.1086/378209. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang XJ, He PP, Li M, He CD, Yan KL, Cui Y, Yang S, Zhang KY, Gao M, Chen JJ, Li CR, Jin L, Chen HD, Xu SJ, Huang W. Hum Mutat. 2004;23:629. doi: 10.1002/humu.9246. [DOI] [PubMed] [Google Scholar]
- 4.Rice GI, Kasher PR, Forte GMA, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanielli M, De-Laet C, de Lonay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin J-PS-M, Lourenco CM, Male AM, Marques W, Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe W, Vanderver A, Vassalo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O’Connell M, Lovell SC, Crow YJ. Nat Genetics. 2012;44:1243. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, Liu M, Yuan YF, Fu L, Kong KL, Qi L, Li Y, Zhang N, Tong AH, Kwong DL, Man K, Lo CM, Lok S, Tenen DG, Guan XY. Nat Med. 2013;19:209. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Mizrahi RA, Phelps K, Ching A, Beal PA. Nucleic Acids Res. 2012;40:9825. doi: 10.1093/nar/gks752. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lehmann KA, Bass BL. J Mol Biol. 1999;291:1. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 7.Maydanovych O, Easterwood LM, Cui T, Veliz EA, Pokharel S, Beal PA. Methods Enzymol. 2007;424:369. doi: 10.1016/S0076-6879(07)24017-0. [DOI] [PubMed] [Google Scholar]
- 8.Shin D, Sinkeldam RW, Tor Y. J Am Chem Soc. 2011;133:14912. doi: 10.1021/ja206095a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinkeldam RW, McCoy LS, Shin D, Tor Y. Angew Chem Int Ed. 2013;52:14026. doi: 10.1002/anie.201307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson DK, Rudolph FB, Quiocho FA. Science. 1991;252:1278. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]
- 11.a) Easterwood LM, Veliz EA, Beal PA. J Am Chem Soc. 2000;122:11537. doi: 10.1021/ja029742d. [DOI] [PubMed] [Google Scholar]; b) Pokharel S, Jayalath P, Maydanovych O, Goodman RA, Wang SC, Tantillo DJ, Beal PA. J Am Chem Soc. 2009;131:11882. doi: 10.1021/ja9034076. [DOI] [PubMed] [Google Scholar]
- 12.a) Veliz EA, Easterwood LM, Beal PA. J Am Chem Soc. 2003;125:10867. doi: 10.1021/ja029742d. [DOI] [PubMed] [Google Scholar]; b) Erion MD, Reddy MR. J Am Chem Soc. 1998;120:3295. [Google Scholar]
- 13.Wang SC, Beal PA, Tantillo DJ. J Comput Chem. 2010;31:721. doi: 10.1002/jcc.21364. [DOI] [PubMed] [Google Scholar]
- 14.Phelps K, Ibarra-Sosa J, Tran K, Fisher AJ, Beal PA. ACS Chem Biol. 2014;9:1780. doi: 10.1021/cb500270x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens OM, Yi-Brunozzi H-Y, Beal PA. Biochemistry. 2000;39:12243. doi: 10.1021/bi0011577. [DOI] [PubMed] [Google Scholar]
- 16.a) Sinkeldam RW, Greco NJ, Tor Y. Chem Rev. 2010;110:2579. doi: 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wilhelmsson LM. Q Rev Biophys. 2010;43:159. doi: 10.1017/S0033583510000090. [DOI] [PubMed] [Google Scholar]
- 17.Macbeth MR, Lingam AT, Bass BL. RNA. 2004;10:1563. doi: 10.1261/rna.7920904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Science. 2009;324:1210. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]; b) Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, Steidl C, Holt R, Jones S, Sun M, Leung G, Moore R, Severson T, Taylor GA, Teschendorff AE, Tse K, Turashvili G, Varhol R, Warren RL, Watson P, Zhao Y, Caldas C, Huntsman D, Hirst M, Marra MA, Aparicio S. Nature. 2009;461:809. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.