Abstract
The bacterial GTPase FtsZ forms a cytokinetic ring at midcell, recruits the division machinery, and orchestrates membrane and peptidoglycan cell wall invagination. However, the mechanism for FtsZ regulation of peptidoglycan metabolism is unknown. The FtsZ GTPase domain is separated from its membrane-anchoring C-terminal conserved (CTC) peptide by a disordered C-terminal linker (CTL). Here, we investigate CTL function in Caulobacter crescentus. Strikingly, production of FtsZ lacking the CTL (ΔCTL) is lethal: cells become filamentous, form envelope bulges, and lyse, resembling treatment with β-lactam antibiotics. This phenotype is produced by FtsZ polymers bearing the CTC and a CTL shorter than 14 residues. Peptidoglycan synthesis still occurs downstream of ΔCTL, however cells expressing ΔCTL exhibit reduced peptidoglycan crosslinking and longer glycan strands than wildtype. Importantly, midcell proteins are still recruited to sites of ΔCTL assembly. We propose that FtsZ regulates peptidoglycan metabolism through a CTL-dependent mechanism that extends beyond simple protein recruitment.
Cell shape in bacteria is maintained by the peptidoglycan (PG) cell wall, a covalently linked network of glycan strands crosslinked by short peptides that provides mechanical integrity to the envelope and prevents cell lysis in the face of turgor pressure1. During growth and division, PG is remodeled by synthetic and hydrolytic enzymes whose activities must be precisely coordinated to preserve the integrity of the cell wall. Moreover, achieving specific cell shapes and forming new cell poles during division requires that PG remodeling be spatially organized. The cytoskeletal proteins FtsZ and MreB are dynamically associated with and direct localization of cell wall remodeling enzymes during cell division and elongation1,2. In addition, polymers of FtsZ, MreB, and the intermediate filament-like protein, crescentin, have been implicated in the mechanical regulation of cell shape2–5. However, it is unclear how the biochemical and biophysical properties of these cytoplasmic membrane-associated proteins influence peptidoglycan remodeling in the periplasm to define the shape of the cell1,2.
FtsZ polymerizes at midcell to form the Z-ring, a dynamic structure upon which the cell division machinery (divisome) assembles6. The divisome comprises dozens of proteins including factors that regulate FtsZ structure and membrane attachment, remodel the cell wall, and invaginate the membranes. In most bacteria, FtsZ is the first protein to arrive at the incipient division site, and it is required for recruitment of cell wall regulators and divisome proteins, which occurs in serial stages7–9. In addition to its primary role in cell division, in at least some organisms FtsZ directs PG synthesis for elongation of the cell prior to cell envelope constriction10,11. Apart from regulating PG remodeling and envelope invagination through protein recruitment, in vitro studies suggest that FtsZ is capable of generating force and deforming membranes4,12. Based on these findings, a mechanical role for FtsZ in inner membrane constriction has been proposed. However, it is not known if force generation by FtsZ is required in vivo, nor is it clear how FtsZ-mediated force might be coupled to PG remodeling to coordinately invaginate the multiple layers of the cell envelope during cytokinesis13.
FtsZ is structurally similar to eukaryotic tubulin14,15 and polymerizes in a GTP-dependent manner, forming linear protofilaments16,17. The FtsZ monomer can be divided into four regions: (i) a short N-terminal peptide, (ii) a conserved GTPase domain essential for polymerization, (iii) an intrinsically disordered C-terminal linker (CTL), and (iv) a C-terminal conserved peptide (CTC) that interacts with membrane-tethering proteins (Fig. 1a)18. A comparison of FtsZ across species reveals that the CTL is poorly conserved in sequence, and varies widely in length from approximately 9 to more than 300 residues. In Escherichia coli and Bacillus subtilis, the CTL was recently demonstrated to be essential for cell division, playing a sequence-independent role in executing cytokinesis19,20. Almost any intrinsically disordered sequence similar in length to the native CTL was sufficient to support normal cytokinesis, even if there was no detectable sequence similarity to the native CTL. The authors of these studies proposed that the CTL functions as an entropic spring20 or flexible linker19 that is crucial for Z-ring formation and constriction.
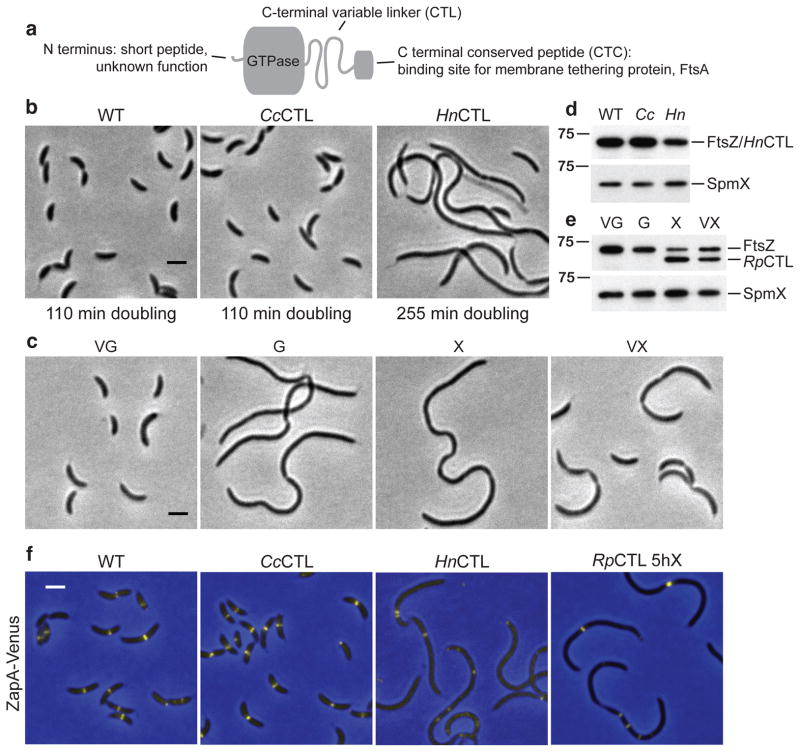
Figure 1. TheCaulobacter crescentus CTL fulfills an essential, sequence-dependent function.
(a) Domain organization of FtsZ. (b) Phase contrast images of strains expressing wildtype (WT, NA1000), CcCTL control (EG776), or HnCTL (EG769) ftsZ variants as the only copy of FtsZ in the cell and their corresponding doubling times in PYE medium. Bar = 2 μm. (c) Phase contrast images of strain EG1060, bearing vanillate (V)-induced expression of WT ftsZ and xylose (X)-induced expression of RpCTL ftsZ. Cells were grown in the presence of the indicated inducer or glucose (G) for 5 h prior to imaging. Bar = 2 μm. (d–e) Immunoblots against lysates from strains in (b–c) using FtsZ (top) or SpmX antisera (bottom). Owing to lack of a specific antibody raised against a loading control distinct enough in size from FtsZ and each of the variants used in this study, SpmX was probed against identical samples on a separate blot. Cc = CcCTL, Hn = HnCTL. FtsZ antiserum was raised against the entire WT FtsZ molecule. Some of the reduction in signal observed for HnCTL and RpCTL may be attributable to loss of CTL-derived epitopes. Positions of molecular weight markers (kDa) are indicated (left). (f) ZapA-Venus fluorescence (yellow) overlaid on phase contrast (blue) images of cells producing the indicated FtsZ CTL variants and ZapA-Venus. WT (EG941), CcCTL (EG1418), and HnCTL (EG1389) were produced from the ftsZ locus as the only FtsZ in the cell. RpCTL (EG1419) production was induced with xylose while depleting WT FtsZ for 5 h prior to imaging.
Here, we study FtsZ function in the α-proteobacterium Caulobacter crescentus, which has long been used as a model for investigating cell shape, development, and regulation of cell cycle-dependent events such as cytokinesis21. Among bacteria, α-proteobacteria stand out as having exceptionally long FtsZ CTLs18,22. Whereas the CTLs of E. coli and B. subtilis FtsZ are approximately 50 amino acids long, that of C. crescentus is 172 amino acids, and many α-proteobacteria have CTLs longer than 300 amino acids. In the present work, we set out to understand the physiological significance of the long CTL in C. crescentus FtsZ, and in the process uncovered a surprising requirement for the FtsZ CTL in regulating specific aspects of PG remodeling. From these findings, we propose that FtsZ regulates PG metabolism through a CTL-dependent mechanism that is in addition to its ability to recruit proteins to midcell.
RESULTS
The FtsZ CTL fulfills an essential function in C. crescentus
To investigate the function of the CTL in C. crescentus, we engineered a panel of C. crescentus FtsZ CTL variants bearing the wildtype (WT) C. crescentus CTL (CcCTL), three other α-proteobacterial CTLs (from Hyphomonas neptunium (HnCTL), Rickettsia parkeri (RpCTL), or Magnetospirillum magneticum (MmCTL)), or the much shorter E. coli CTL (EcCTL) in place of the C. crescentus CTL (Table 1, Supplementary Fig. 1). As primary sequence conservation is low in this region of FtsZ, the most similar CTL we tested, HnCTL, is only 18% identical and 35% similar to CcCTL. The other CTLs tested had no detectable sequence similarity (Supplementary Fig. 1). Using allele exchange, we asked if any of these variants could replace WT ftsZ and found that only CcCTL or HnCTL yielded viable C. crescentus cells (Table 1). Moreover, while the control CcCTL strain was morphologically indistinguishable from WT and grew at a similar rate, HnCTL cells were filamentous and slow growing (Fig. 1b).
Table 1. CTL variants used in this study.
The length, percent proline (% Pro) and theoretical isoelectric point (pI) listed for each linker do not include contributions from the four amino acids introduced at the junctions with the GTPase domain and CTC by addition of restriction enzyme recognition sites for cloning purposes.
| Name | Length (amino acids) | % Pro | pI | Complement? | Relative FtsZ levels |
|---|---|---|---|---|---|
| CcCTL | 172 | 16.3 | 4.35 | yes | +++ |
| ΔCTL | 0 | 0 | NA | no | +++ |
| HnCTL | 165 | 15.2 | 3.94 | yes | ++ |
| RpCTL | 116 | 6.9 | 4.46 | no | ++ |
| MmCTL | 224 | 16.1 | 10.4 | no | − |
| EcCTL | 51 | 9.8 | 10.0 | no | − |
| Nt34 | 34 | 20.6 | 6.23 | yes | ++/+ |
| Nt68 | 68 | 22.1 | 5.30 | yes | +++ |
| Nt102 | 102 | 19.6 | 4.64 | no | +++ |
| Nt136 | 136 | 17.6 | 4.51 | yes | +++ |
| Ct36 | 36 | 11.1 | 8.74 | yes | ++/+ |
| Ct70 | 70 | 11.4 | 4.73 | no | +++ |
| Ct104 | 104 | 12.5 | 4.45 | yes | +++ |
| Ct138 | 138 | 15.2 | 4.57 | yes | +++ |
To examine the phenotypes of cells producing non-complementing FtsZ CTL variants, we created strains in which the vanillate-inducible promoter (PvanA) drives expression of the only copy of WT ftsZ and the xylose-inducible promoter (PxylX) drives expression of a CTL variant (Supplementary Fig. 2a). Expressing WT ftsZ from either promoter using this strategy supports normal cytokinesis (Supplementary Fig. 2b, c). When RpCTL, MmCTL, or EcCTL was produced while depleting WT FtsZ, cells became filamentous, consistent with their inability to complement loss of ftsZ (Fig. 1c, Supplementary Fig. 2d). We examined steady-state levels of each variant by immunoblotting. While RpCTL and HnCTL proteins were detected at near-WT levels, MmCTL and EcCTL were undetectable (Fig. 1d, e, Supplementary Fig. 2e). WT FtsZ was depleted to 10–40% of normal levels in the absence of vanillate after 6.5 h (Fig. 1e, Supplementary Fig. 2c, e). We assessed Z-ring formation in cells producing WT FtsZ, CcCTL, HnCTL, or RpCTL by imaging a fluorescent fusion of the FtsZ-binding protein, ZapA (Fig. 1f). ZapA-Venus localized to midcell bands or rings in each. However, in HnCTL-producing cells, patches of ZapA-Venus were also observed, suggesting a reduced capacity to form Z-rings.
Length is not the primary determinant of CTL function
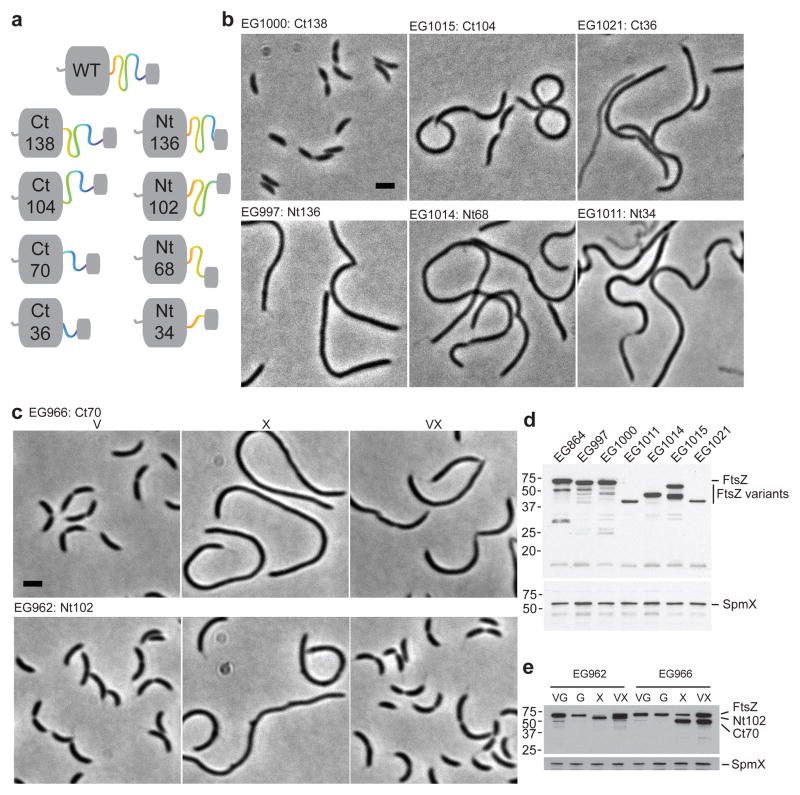
To further decipher the essential regions of the CTL, we asked if FtsZ variants lacking portions of the CcCTL support cytokinesis. We engineered CTL variants bearing only the N-terminal 34, 68, 102, or 136 residues (Nt34, Nt68, Nt102, and Nt136) or C-terminal 36, 70, 104, or 138 residues (Ct36, Ct70, Ct104, and Ct138) of the CTL (Fig. 2a). Surprisingly, most of these variants supported viability when expressed as the only copy of ftsZ (Fig. 2b, d). However, while the first 34 residues of the CTL were dispensable for full function (Fig. 2b, Ct138), all other deletion variants were filamentous. The two shortest variants tested contained only 34 or 36 residues yet supported cytokinesis, suggesting that C. crescentus tolerates large changes to CTL length while maintaining viability. Interestingly, two variants of intermediate length (Ct70 and Nt102) were non-functional and yielded filamentous cells when produced while depleting WT FtsZ (Fig. 2c, e). These data indicate that length is not the primary determinant of CTL function in C. crescentus, as the non-complementing variants are almost identical in length to two complementing variants. Similarly, Ct138 and Nt136 are almost identical in length, but Nt136-producing cells are filamentous while Ct138-producing cells divide normally (Fig. 2b).
Figure 2. C. crescentus tolerates large changes to the length of the CTL.
(a) Graphical representation of the CTL deletion variants analyzed. Ct138 = FtsZ with only the C-terminal 138 residues of the CTL, Nt136 = FtsZ with only the N-terminal 136 residues of the CTL, and so forth. (b) Phase contrast images of strains producing the indicated CTL variants as the only copy of FtsZ in the cell, expressed from the ftsZ locus. Bar = 2 μm. (c) Phase contrast images of the indicated strains, grown with vanillate (V), xylose (X), or both (VX) for 5 h prior to imaging. Bar = 2 μm. (d and e) Immunoblots against lysates from strains in (b–c) using FtsZ (top) or SpmX (bottom) antisera. Positions of molecular weight markers (kDa) are indicated (left).
The Nt34 and Ct36 variants were both present at low steady state protein levels (Fig. 2d), suggesting that composition of the CTL contributes to FtsZ levels. As proteolysis of FtsZ is a known point of regulation in C. crescentus23,24, we determined the half-lives of the HnCTL, Ct138, Nt34, and Ct36 variants and found that, indeed, Nt34 (t1/2 = ~27 min) and Ct36 (t1/2 = ~23 min) turned over substantially faster than WT FtsZ (t1/2 = ~48 min) (Supplementary Fig. 2f). Collectively, the above analyses indicate that the CTL in C. crescentus plays an essential, sequence-dependent role in cell division. Moreover, CTL composition contributes to post-transcriptional regulation of FtsZ levels, including affecting protein turnover.
FtsZ lacking the CTL induces bulging and rapid cell lysis
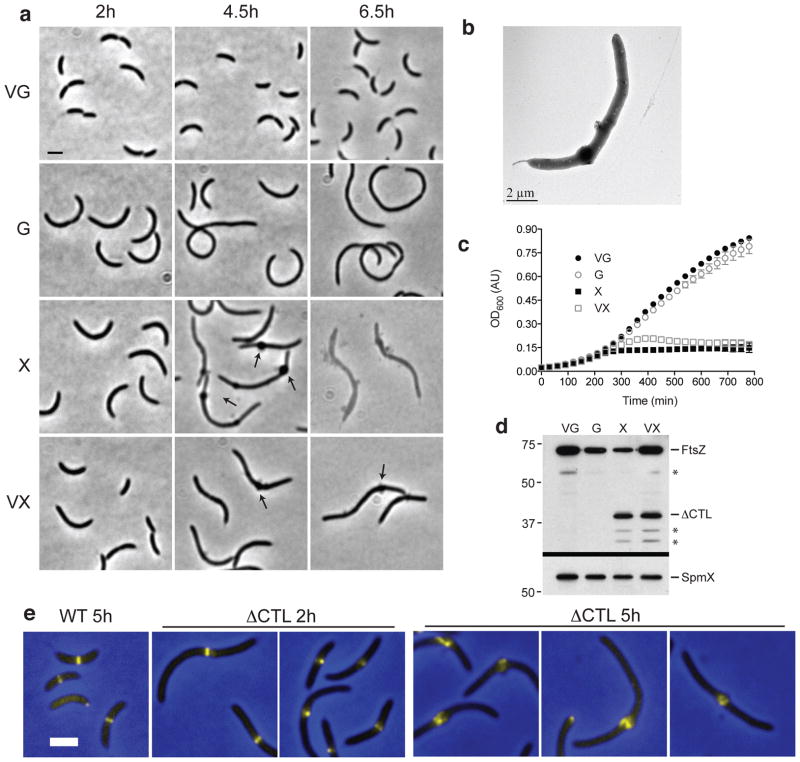
Having established that the CTL plays an important role in C. crescentus FtsZ function, we next examined the effects of producing FtsZ completely lacking the CTL (ΔCTL). To do this, we generated a strain bearing vanillate-induced WT ftsZ and xylose-induced ΔCTL. Unexpectedly, induction of ΔCTL caused cells to grow into filaments with localized envelope bulges and to rapidly lyse (Fig. 3a–c). In the absence of vanillate, bulges began to appear at 2 h post-induction of ΔCTL and were abundant at 4–5 h post-induction. Transmission electron microscopy (TEM) of ΔCTL-expressing cells confirmed that most bulges are roughly symmetrical expansions of the cell envelope (Fig. 3b). Cells began to lyse by 5 h post-induction of ΔCTL (Fig. 3a, c). The effects of ΔCTL expression were dominant lethal: bulging and lysis occurred in the presence of vanillate, but were delayed when compared to the phenotype in the absence of WT ftsZ induction (Fig. 3a, c). Using a second inducible expression system, with PxylX driving expression of ftsZ and the recently described myo-inositol-inducible PiolC25,26 driving expression of ΔCTL, we were able to deplete WT FtsZ to <5% of normal levels (Supplementary Fig. 4a–c). Bulging and lysis was observed under these conditions as well, indicating that significant amounts of WT FtsZ are not required for the ΔCTL phenotype. ΔCTL expression was also toxic to cells grown in minimal media, causing filamentation, rough cell envelope appearance, and lysis (Supplementary Fig. 4d, e). Unlike Nt34 or Ct36, deleting the CTL entirely did not have a pronounced effect on FtsZ steady-state protein levels (Fig. 3d, Supplementary Fig. 4f).
Figure 3. ΔCTL production causes dominant lethal envelope bulging and cell lysis.
(a) Phase contrast images of cells from strain EG852, with vanillate-induced expression of ftsZ and xylose-induced expression of ΔCTL. Cells were grown for the indicated amount of time in PYE medium with the indicated inducer (vanillate (V), xylose (X), and/or glucose (G)) prior to imaging. Arrows highlight envelope bulges. Bar = 2 μm. (b) Whole mount transmission electron micrograph of an EG852 cell grown in PYE xylose for 4 h. (c) Growth of strain EG852 in PYE with the indicated inducers added at time 0, as monitored by OD600. Average ± standard deviation (s.d.) from three technical replicates is plotted. (d) Immunoblot of lysates from cells in (a) after 7 h growth in PYE with the indicated inducers using FtsZ (top) or SpmX antisera (bottom). Asterisks indicate degradation products. Positions of molecular weight markers (kDa) are indicated (left). (e) Merged ZapA-Venus fluorescence (yellow) and phase contrast (blue) images of cells producing WT FtsZ (EG1396) for 5 h or ΔCTL (EG950) for 2 h or 5 h.
Using ZapA-Venus as a proxy for FtsZ localization, we found that bulges occur at sites of FtsZ assembly (Fig. 3e). ZapA-Venus localized to rings in cells producing WT FtsZ, as expected, and localized to the bulges of ΔCTL-producing cells. However, ZapA-Venus localization was more heterogeneous and less focused into a ring in cells producing ΔCTL as compared to WT; this effect was most apparent at 5 h post-induction. We verified that ZapA-Venus reported on the localization of FtsZ in each strain by producing an N-terminal CFP fusion to FtsZ or ΔCTL while depleting WT FtsZ (Supplementary Fig. 4g, h). Although these fusions were non-functional, CFP-FtsZ localized to midcell bands and CFP-ΔCTL was enriched in bulges. Collectively, these data indicate that FtsZ lacking the CTL induces local, toxic effects that lead to loss of envelope integrity and cell lysis.
Induction of bulging and lysis is CTL length-dependent
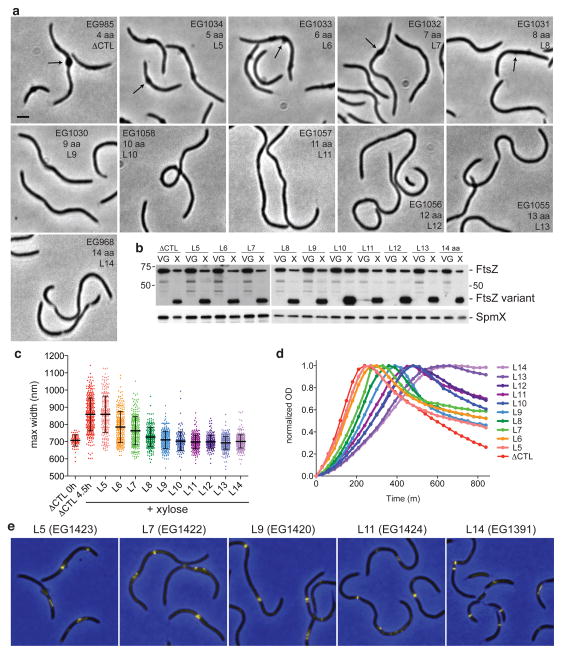
All organisms that possess an FtsZ homolog with a CTC bear a CTL of at least ~9 residues18. Moreover, expression of ftsZ lacking the CTL in B. subtilis or another CTL variant in E. coli was reported to cause cell lysis19,20. This led us to speculate that a minimal CTL might be a conserved requirement to prevent FtsZ-induced cell lysis. To test this, we added residues from the C-terminus of the CTL to the ΔCTL variant in single amino acid increments and examined the effects of expressing these variants while depleting WT FtsZ. Interestingly, we observed an exquisite CTL length-dependence for the induction of bulging and lysis (Fig. 4): expression of ftsZ with linkers up to 8 amino acids (including 4 residues introduced by restriction enzyme sites) induced bulging (Fig. 4a, b), with a length-dependent decrease in maximum cell width (used as a read-out for envelope bulging) (Fig. 4c). These variants also induced lysis, with the time to lysis increasing with linker length (Fig. 4d). FtsZ variants with linkers of 9 to 12 amino acids did not promote bulging, but induced lysis 6 to 8 h post-induction. 13 and 14 amino acid linker variants did not induce bulging or rapid lysis, but failed to support cell division. All short linker variants were produced at similar levels (Fig. 4b).
Figure 4. Bulging and lysis is CTL length-dependent.
(a) Phase contrast images of strains producing FtsZ with the indicated CTL lengths grown with xylose for 4.5 h prior to imaging. The number of amino acids (aa) in the linker includes 4 residues added from restriction sites introduced for cloning purposes. Thus, ΔCTL contains a 4 aa linker. Each strain bears vanillate-induced ftsZ and xylose-induced CTL variant. Bar = 2 μm. Arrows highlight bulges. (b) Immunoblots of lysates from strains in (a) grown with vanillate and glucose or with xylose for 6.5 h using FtsZ (top) or SpmX antisera (bottom). Positions of molecular weight markers (kDa) are indicated (left). (c) Maximum widths of cells from strains from (a) grown with xylose for 4.5 h and for EG985 grown with vanillate and glucose (ΔCTL VG; expressing WT ftsZ). Data points represent the maximum width for each cell in that population. Data are combined from two independent imaging experiments for each strain. ΔCTL X data include two independent experiments with EG985 and one with EG852 (independent, isogenic strains for producing ΔCTL). n = 80, 354, 254, 274, 268, 242, 281, 151, 281, 212, 357, and 326 cells for samples as presented from left to right. Bar = average width, whiskers = s.d. (d) Growth and lysis of strains from (a). Cells were grown with xylose from time 0 and OD was monitored at 600 nm. Final plots were normalized for each data set and represent average data from three technical replicates. The time at which OD began to fall was interpreted as time of initiation of lysis. (e) ZapA-Venus fluorescence (yellow) overlaid on phase contrast (blue) images of cells producing the indicated FtsZ CTL variants and ZapA-Venus. Each variant was induced with xylose while depleting WT FtsZ for 5 h prior to imaging.
Similar to ΔCTL, we observed patchy, discontinuous ZapA-Venus localization in strains producing short-linker variants that induce early lysis (L5, L7, L9, and L11) (Fig. 4e). In contrast, the L14-producing strain that does not undergo bulging or lysis contained focused Z-rings resembling those found in WT cells. We conclude that a minimal linker of 13–14 amino acids is required to form focused Z-rings and to prevent FtsZ from inducing lethal changes to the cell envelope in C. crescentus.
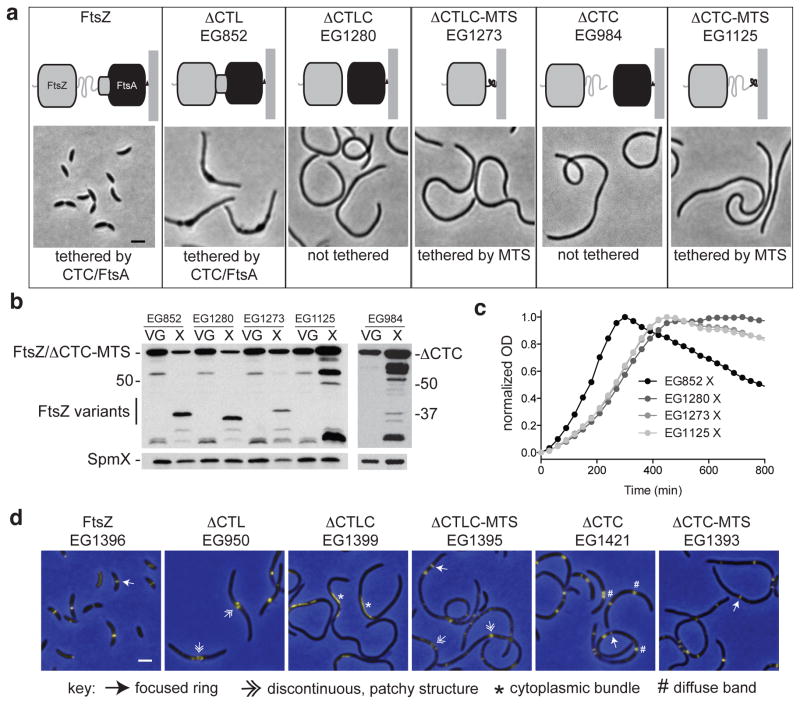
ΔCTL requires the CTC to induce bulging and lysis
We next asked if FtsZ lacking the CTL must be attached to the membrane to invoke its effects. Membrane attachment of FtsZ is mediated by interaction of the CTC with the membrane-tethering protein, FtsA27–29 (Fig. 5a). We therefore generated an FtsZ variant lacking both the CTL and CTC (ΔCTLC). Cells producing ΔCTLC and depleted of FtsZ grew as smooth filaments, similar to cells producing a variant lacking only the CTC (ΔCTC), but did not undergo bulging or rapid lysis (Fig. 5a–c), suggesting that membrane attachment is required for ΔCTL to induce its lethal effects. To determine if the mode of membrane attachment was important, we appended the membrane-targeting sequence (MTS) from E. coli MinD to ΔCTC or ΔCTLC to directly tether these variants to the membrane. Similar to ΔCTLC and ΔCTC, cells producing these constructs (ΔCTC-MTS or ΔCTLC-MTS) grew as smooth filaments that did not rapidly lyse (Fig. 5a–c). We conclude that ΔCTL requires the CTC to induce bulging and lysis. As the only known activity of the CTC in C. crescentus is binding to FtsA, our results indicate that membrane attachment by the native membrane anchor is required for the ΔCTL phenotype.
Figure 5. ΔCTL requires the membrane-anchoring CTC to invoke bulging and lysis.
(a) Schematics and representative phase contrast images of WT cells (FtsZ) and cells producing the indicated FtsZ variants and depleted of WT FtsZ for 5 h. ΔCTLC - FtsZ lacking the CTL and FtsA-binding CTC. MTS - membrane targeting sequence from E. coli MinD. ΔCTC - FtsZ lacking the CTC. Bar = 2 μm. (b) Immunoblots of lysates from strains in (a) grown with vanillate and glucose or with xylose for 5 h probed with FtsZ (top) or SpmX antisera. Positions of molecular weight markers (kDa) are indicated. (c) Growth and lysis of strains from (a). Cells were grown with xylose from time 0 and OD was monitored at 600 nm. Final plots were normalized for each data set and represent average data from three technical replicates. (d) ZapA-Venus fluorescence (yellow) overlaid on phase contrast (blue) images of cells producing the indicated FtsZ CTL variants and ZapA-Venus. Each variant was induced with xylose while depleting WT FtsZ for 5 h prior to imaging. Bar = 2 μm.
To determine if the membrane-targeting variants support Z-ring formation, we examined ZapA-Venus localization in each of the above strains. Although ΔCTC-MTS was nonfunctional, it recruited ZapA-Venus to Z-rings in a manner indistinguishable from WT (Fig. 5d). Apparently normal Z-rings were also observed in cells producing ΔCTLC-MTS, although patchy structures like those observed in ΔCTL-producing cells were more prevalent. In cells producing ΔCTLC, ZapA-Venus localized exclusively to cytoplasmic bundles. Eliminating the CTL in conjunction with the CTC was required for formation of these cytoplasmic bundles, as ZapA-Venus localized to diffuse, broad bands in ΔCTC-producing cells. Thus, although targeting to membranes via the CTC is not required for Z-ring formation, the CTC is required for ΔCTL to induce bulging and lysis. Deletion of the CTL may increase FtsZ polymer bundling, an effect that is most apparent in vivo when membrane attachment of ΔCTL is eliminated.
ΔCTL must polymerize to induce bulging and lysis
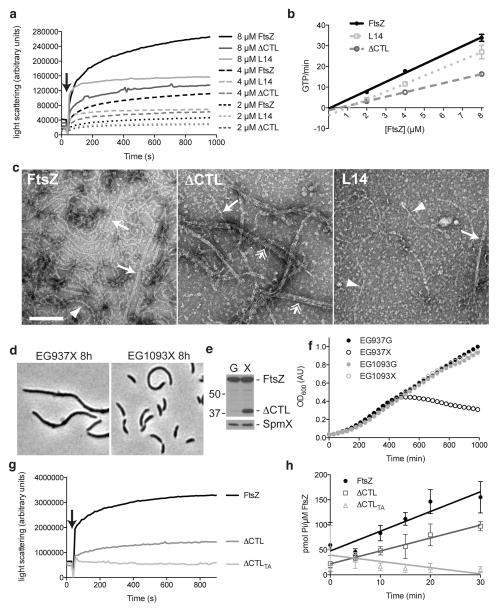
Prior work in B. subtilis demonstrated that FtsZ lacking the CTL polymerized poorly19 and we observed aberrant Z-ring structures in cells producing CTL variants. To address if C. crescentus ΔCTL has altered polymerization properties, we assessed its self-assembly in vitro. We purified WT FtsZ, ΔCTL, and, as a control for effects specific to the ΔCTL phenotype, the L14 short-linker variant that did not induce bulging and lysis but was non-functional for cytokinesis. Both ΔCTL and L14 exhibited reduced polymerization when compared to WT, as assessed by high-speed pelleting and right-angle light scattering (Fig. 6a, Supplementary Fig. 5a). Correspondingly, the GTPase rates of ΔCTL (1.86 ± 0.04 min−1 (mean ± standard error (s.e.), n=3) and L14 (2.89 ± 0.06 min−1, n=3) were 42 and 65% that of WT FtsZ (4.44 ± 0.10 min−1, n=3), respectively, at 4 μM (Fig. 6b). Whereas the estimated critical concentration for WT FtsZ was 0.09 μM, that of ΔCTL was increased to 0.57 μM and L14 to 0.97 μM, consistent with reduced longitudinal interactions (Fig. 6b).
Figure 6. ΔCTL must polymerize to cause bulging and lysis.
(a) Right-angle light scattering over time by solutions containing purified 2,4, or 8 μM WT FtsZ, ΔCTL, or L14. Arrow indicates addition of GTP to induce polymerization. Curves are averages of three experiments. (b) GTPase activity of FtsZ variants from (a) as a function of protein concentration. Averages ± s.d. from three replicates are plotted with linear curve fits. Extrapolated x-intercepts represent estimated critical concentration. (c) Negative stain electron micrographs of the indicated FtsZ variant at 8 μM in the presence of 2 mM GTP and 2.5 mM MgCl2. Arrows: paired protofilaments; double arrows: thick bundles; arrowheads: single protofilaments. Bar = 100 nm. (d) Phase contrast micrographs of EG937, bearing xylose-inducible ΔCTL, and EG1093, a spontaneous suppressor mutant of EG937 grown in xylose for 8 h. (e) Immunoblot of lysates from strain EG1093 grown in glucose or xylose for 24 h probed with FtsZ (top) or SpmX (bottom) antisera. Positions of molecular weight markers (kDa) are indicated (left). (f) Growth of EG937 and EG1093, with glucose (G) or xylose (X) added at time 0. (g) Right angle light scattering over time by solutions containing purified 8 μM WT FtsZ, ΔCTL, or ΔCTL T112A/A282T (ΔCTLTA). Arrow indicates addition of GTP to induce polymerization. Curves are averages of two experiments. (h) Inorganic phosphate liberated over time by 8 μM FtsZ variants from (f). Averages ± s.d. from three replicates are plotted with linear curve fits.
We next examined the polymers formed by these CTL variants by negative stain TEM. At 4 μM protein concentration, few polymers were observed for ΔCTL or L14, indicating that they may be unstable. At 8 μM, we observed distinct polymer structures for each variant (Fig. 6c). While WT FtsZ assembled into single or paired protofilaments, L14 produced short, curved protofilaments and ΔCTL formed long, highly bundled polymers. These data demonstrate that the CTL contributes to efficient polymerization of FtsZ and that deletion of the CTL leads to increased protofilament bundling.
To better understand the mechanism by which ΔCTL functions, we sought to identify suppressors of the bulging and lysis phenotype. To do this, we created a strain with WT ftsZ at the ftsZ locus and ΔCTL at the xylX locus and selected for clones that grew in the presence of xylose. One of the suppressors we isolated contained two point mutations in ΔCTL itself. This mutant grew equally well in the presence or absence of xylose and exhibited only mild filamentation in the presence of xylose (Fig. 6d–f). We confirmed that these are the only mutations required to suppress bulging and lysis by cloning ΔCTL with these point mutations into a vanillate-dependent ftsZ depletion background (Supplementary Fig. 5c–e). Interestingly, one of the mutations in the ΔCTL suppressor is in the tubulin signature motif required for GTP binding and hydrolysis (T112A)30, and the other is in the monomer-monomer interface (A283T)31 (Supplementary Fig. 1, 5b). Each of these mutations is expected to impact polymerization. We tested the activity of the ΔCTL double mutant (ΔCTLTA) and found that, as predicted, it does not polymerize and has no detectable GTPase activity (Fig. 6g, h). Thus, although ΔCTL has reduced polymerization activity as compared to WT FtsZ, our data suggest that it must polymerize to induce its toxic effects.
ΔCTL induces specific changes to peptidoglycan composition
We next turned our attention to the cell envelope to ask how it is being perturbed by ΔCTL. We postulated that local breaches in the PG weakened the cell wall at sites of ΔCTL assembly, permitting bulging and lysis due to turgor pressure. A similar bulging filament phenotype has been reported in E. coli cells treated with penicillin32.
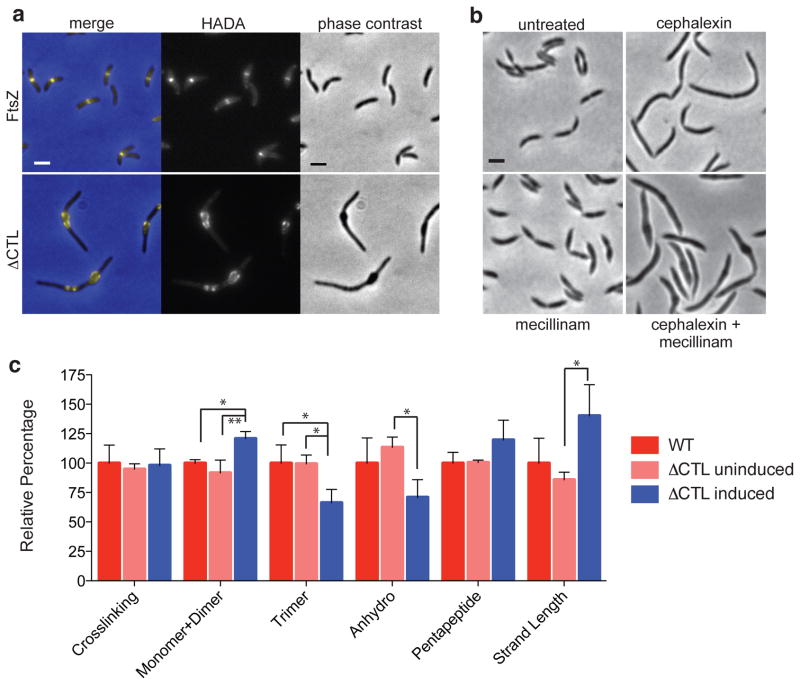
Penicillin inhibits the penicillin binding proteins (PBPs) that build the cell wall; therefore, we asked if PG synthesis is disrupted in cells producing ΔCTL. To answer this, we pulse-labeled cells with a fluorescent D-alanine derivative (HADA) that is incorporated into the lipid II PG precursor and can be used to image sites of new PG synthesis33,34. As expected, HADA labeling of cells producing WT FtsZ yielded midcell bands of fluorescence, consistent with the bulk of PG synthesis in C. crescentus occurring at midcell10 (Fig. 7a, top). Surprisingly, in cells producing ΔCTL we observed robust HADA labeling at bulges (Fig. 7a, bottom). However, the pattern of insertion over time was irregular for ΔCTL (Supplementary Fig. 6), perhaps reflecting altered Z-ring organization or the incipient effects of membrane bulging through holes in the cell wall. From this we conclude that PG synthesis occurs at sites of ΔCTL assembly at least up to the step of transglycosylation, based on the mechanism of HADA incorporation.
Figure 7. Peptidoglycan crosslinking and glycan strand length are altered in cells producing ΔCTL.
(a) Fluorescence (HADA) and phase contrast micrographs of strain EG852 grown with vanillate and glucose (top, FtsZ) or xylose (bottom, ΔCTL) for 5 h, then pulse-labeled with HADA for 5 min prior to imaging. Bar = 2 μm. (b) Phase contrast images of C. crescentus lacking β-lactamase (LS107) that were untreated, or treated with cephalexin, mecillinam, or both for 6 h. Bar = 2 μm. (c) Relative abundance of total muropeptide crosslinking, the indicated crosslinked species, anhydro glycan ends, pentapeptides, and glycan strand length of sacculi isolated from WT C. crescentus, or from EG852 grown in vanillate and glucose (ΔCTL uninduced) or in xylose (ΔCTL induced) for 5 h prior to sacculus preparation. Muropeptides were isolated and analyzed in triplicate for each sample. Values are expressed as a percentage of the average WT value for each feature. Average + s.d. are presented. n = 3 each. *: p<0.05, **: p<0.01 using 1-way ANOVA with Tukey’s Multiple Comparison post-test.
Penicillin targets numerous PBPs, including transpeptidases and bifunctional enzymes. Using more specific inhibitors, it was reported that to invoke the bulging filament phenotype in E. coli, the division-specific transpeptidase, PBP3/FtsI, must be inhibited together with either the elongation specific transpeptidase, PBP235, or the lytic transglycosylase, Slt7036,37, an enzyme that shortens glycan strands. We were able to recapitulate a bulging filament phenotype by treating C. crescentus cells lacking the β-lactamase gene with both cephalexin (a PBP3 inhibitor) and mecillinam (a PBP2 inhibitor) (Fig. 7b). Based on these data, we hypothesized that ΔCTL-mediated bulging and lysis results from reduced transpeptidation (crosslinking) and/or increased glycan strand length.
To directly examine the changes to the PG induced by ΔCTL, we performed ultra performance liquid chromatography (UPLC) analysis of muropeptides prepared from PG isolated from WT cells, or from the ΔCTL-expressing strain in the absence or presence of xylose (Supplementary Fig. 7). Total muropeptide crosslinking was similar in the three samples (Fig. 7c). However, we observed an ~30% decrease in the abundance of trimers (higher-order crosslinked species) in cells producing ΔCTL and a concomitant increase in the combined percentage of monomers and dimers. Our data and previous studies of C. crescentus PG38,39 show that WT cells have shorter glycan strands than organisms such as E. coli, manifesting larger anhydro peaks representing the ends of glycan strands that allow for detection of increases in glycan strand length with greater fidelity. Upon ΔCTL expression, we detected an ~50% increase in glycan strand length (Fig. 7c). These findings indicate that, consistent with the phenotypic similarity to chemical inhibition of PBP3 and PBP2 or Slt70, production of ΔCTL causes a decrease in higher-order transpeptidation and an increase in glycan strand length relative to cells producing WT FtsZ. The changes we observed in our muropeptide analysis may underrepresent the extent of changes to the PG at the bulges, since the analysis aggregates PG composition from the entire cell and the ΔCTL-induced changes are likely to be highly localized.
Midcell proteins are recruited to ΔCTL-mediated bulges
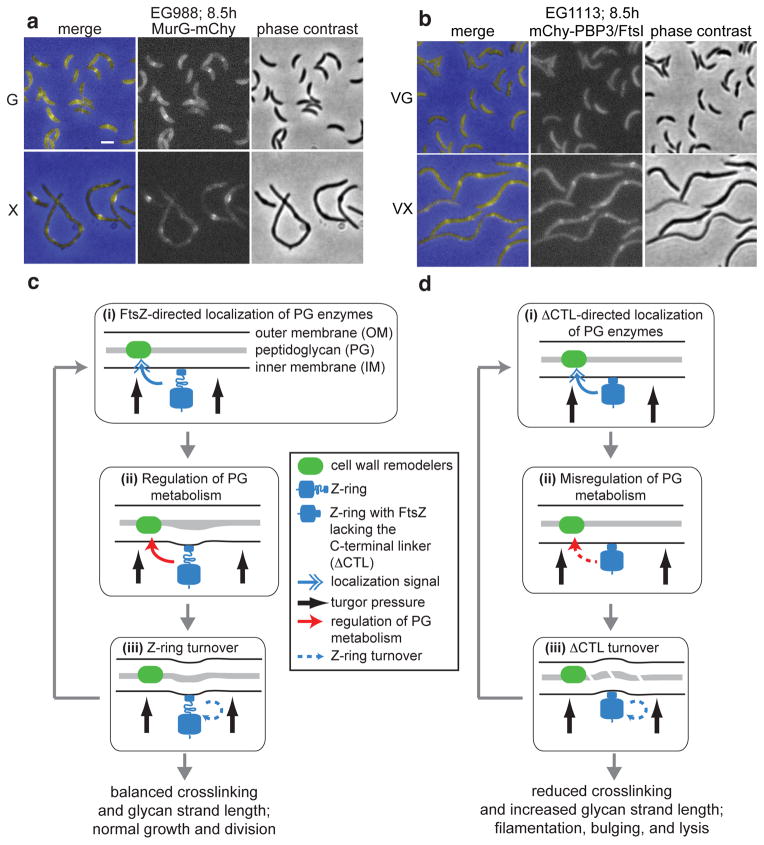
The mechanisms by which cytoskeletal proteins like FtsZ regulate cell wall metabolism are mostly unclear. One model posits that FtsZ serves only as a localization signal, bringing PG enzymes and their regulators to the appropriate location to carry out their functions. In C. crescentus, a number of proteins implicated in cell wall metabolism are sequentially enriched at midcell9. We hypothesized that in ΔCTL-producing cells, one or more of these factors might not be recruited to sites of FtsZ assembly, leading to alterations in PG metabolism. To test this, we examined the localization of FtsZ-binding proteins (ZapA, FzlA, and FzlC), early recruits to midcell implicated in PG metabolism (MurG and DipM), and late divisome proteins involved in PG metabolism (PBP3/FtsI, FtsW, FtsN) in cells producing ΔCTL. Surprisingly, for each fluorescent protein fusion, we observed enrichment at ΔCTL-mediated bulges (Fig. 8a, b, Supplementary Fig. 8a–f). In addition, we performed in vitro co-sedimentation assays for MipZ and FzlA, two essential binding partners of FtsZ in C. crescentus. We observed similar levels of co-sedimentation of each with FtsZ and ΔCTL (Supplementary Fig. 8g). We conclude that the CTL is dispensable for binding of FtsZ to these essential partners and that midcell proteins are enriched at sites of Z-ring assembly in the presence of ΔCTL. This points to a mode of regulation of PG metabolism by FtsZ that is in addition to its ability to recruit proteins to midcell.
Figure 8. PG enzymes are recruited to sites of ΔCTL assembly.
(a, b) Fluorescence, phase contrast, and merged images demonstrating localization of fluorescent fusions to divisome proteins in cells producing ΔCTL. MurG-mCherry and mCherry-FtsI were produced upon vanillate induction in the EG937 background. Cells were grown in the presence of glucose or xylose for 8.5 h prior to imaging. Vanillate was added 2 h prior to imaging to induce expression of the fluorescent fusion. (c) Graphical representation of the role of FtsZ in promoting PG remodeling in C. crescentus. (i) Once established at the incipient division site, the Z-ring promotes local assembly of the divisome, including PG enzymes and their regulators. (ii) We propose that, in addition to recruiting PG enzymes to midcell, the Z-ring initiates a signal that regulates their activity, allowing appropriate re-shaping and invagination of the cell envelope. (iii) The Z-ring turns over, making constriction an iterative process. (d) When ΔCTL is produced, midcell proteins are still recruited (i), but their activities are misregulated (ii), leading to reduced PG crosslinking, increased glycan strand length, envelope bulging, and cell lysis (iii).
The bacterial actin MreB also plays an important role in regulating PG metabolism and is enriched at midcell for much of the cell cycle in C. crescentus40,41. We therefore asked if MreB midcell localization is required for the ΔCTL-induced PG misregulation we observe. To do this, we took advantage of previously described MreB mutants that are not recruited to midcell, but that support normal growth and division: MreB(D16G)42 and MreB(Q26P)7. In both of these backgrounds, ΔCTL production induced filamentation, bulging, and lysis in a manner similar to WT (Supplementary Fig. 9). These data indicate that MreB is likely dispensable for the CTL-dependent mechanism by which FtsZ regulates PG metabolism.
DISCUSSION
Cytoskeletal proteins in bacteria are required to specify and maintain cell shape, yet the mechanisms by which they regulate PG remodeling to achieve cell shape changes are largely undefined. Here, we have provided evidence that the disordered CTL of FtsZ is required for the Z-ring to direct robust cell wall construction. We demonstrated that deletion of the CTL causes lethal defects in PG metabolism through a mechanism requiring ΔCTL polymerization and the membrane-anchoring CTC. Additionally, we determined that, while the ability of ΔCTL to recruit proteins to midcell is intact, ΔCTL production leads to altered Z-ring structure and specific changes in PG composition. Our characterization of the ΔCTL variant has therefore allowed us to propose a PG regulatory function for FtsZ that extends beyond its ability to simply recruit PG metabolic machinery to its site of assembly.
In this study, we set out to characterize the role of the long CTLs found in α-proteobacterial FtsZs. Functional analysis of divergent CTLs and CcCTL deletion variants indicated a sequence-dependent function for the CTL in C. crescentus (Figs. 1, 2, Supplementary Fig. 2). In addition to having notably long CTLs, α-proteobacteria also encode several FtsZ-binding proteins (ZBPs) that are conserved only in this group43–45. We initially hypothesized that the CTL might act as a binding platform for one or more essential, α-proteobacterial-specific ZBPs. This does not appear to be the case, however, at least for known ZBPs: MipZ and FzlA interact equally well with purified WT and ΔCTL FtsZ (Supplementary Fig. 8g).
An alternative sequence-dependent function may be to regulate FtsZ protein stability23,46 or otherwise affect FtsZ levels post-transcriptionally. FtsZ levels vary over the cell cycle in C. crescentus, being subject to both transcriptional and proteolytic regulation23,47. We observed reduced steady state levels of some CTL variants when expressed from equivalent constructs, and found that Nt34 and Ct36 were degraded more rapidly than WT FtsZ (Supplementary Fig. 2f). These effects on protein stability may be encoded in the CTL primary sequence as ClpXP protease recognition sites, as was recently demonstrated for E. coli FtsZ46. However, we have not identified any such sequences within the CcCTL. Using E. coli proteins in vitro, polymeric FtsZ was shown to be degraded more rapidly by ClpXP than monomeric FtsZ48. CTL-dependent effects on FtsZ turnover may therefore be mediated indirectly, by altering the monomer-polymer ratio of FtsZ. Effects of the CTL on FtsZ levels is likely only part of the story, however, as the RpCTL variant was present at nearly WT levels, yet failed to support division.
The sequence-dependent function of the C. crescentus CTL lies in contrast to observations from E. coli and B. subtilis, where nearly any disordered sequence similar in length to the native CTL was sufficient to support normal division19,20. In B. subtilis, however, the presence of a CTL was required to prevent cell lysis, and in E. coli, one CTL variant (Add64blk) caused a “bulging and lysis” defect and an increase in filament bundling similar to what we observed with ΔCTL19,20. The morphological changes associated with ΔCTL production in C. crescentus drew our attention to this phenomenon and highlighted a likely role for the CTL in regulating PG metabolism (Fig. 3). We posit that in C. crescentus the CTL is at least bifunctional: it fulfills a sequence-dependent function required for efficient division and a minimal-length-dependent role in preventing misregulation of PG remodeling. A length-dependent, sequence-independent role for the CTL in executing cytokinesis may also exist in C. crescentus: many of the CcCTL deletion variants we tested, while viable, performed cytokinesis inefficiently (Fig. 2). We hypothesize that the role of a minimal CTL in preventing cell lysis is conserved and, if so, explains the presence of a CTL of at least 9 residues in all FtsZ homologs bearing a CTC18.
The surprising consequences of ΔCTL production indicate misregulation of a subset of cell wall remodeling activities. This differs from complete loss of FtsZ function, wherein a Z-ring fails to form, PG metabolism becomes dispersed10, and cells grow as smooth filaments until lysis occurs many hours later49. Our muropeptide analysis revealed distinct alteration of the sacculus in ΔCTL-producing cells: they had reduced higher-order crosslinking and increased glycan strand length relative to cells producing WT FtsZ (Fig. 7c). The phenotypic similarity of ΔCTL production to inhibition of PBP3 and PBP2 or Slt70 along with the known activities of these enzymes is consistent with the possibility that they are inhibited by ΔCTL. Interestingly, a previous study reported similar changes to the cell wall when they inhibited MreB for several hours, then allowed cells to recover MreB function39. The authors postulated an increase in transglycosylation activity to explain the observed changes. Although the CTL-dependent effects we describe appear MreB-independent (Supplementary Fig. 9), a local increase in transglycosylation, without a coordinated increase in transpeptidation, could similarly explain the altered PG chemistry we observe with ΔCTL production.
The next challenge is to explain ΔCTL-induced changes to the cell wall mechanistically. One can rationalize loss of localized cell wall metabolism when Z-rings completely fail to form if FtsZ acts simply as a localization signal. However, in ΔCTL-producing cells, insertion of new PG material is still localized to sites of FtsZ assembly (Fig. 7a) and all proteins we examined that normally localize to midcell were enriched at ΔCTL-mediated bulges (Fig. 8a, b, Supplementary Fig. 8). These data disfavor a model for FtsZ function involving simple protein recruitment, although we cannot rule out the possibility that changes in protein localization in ΔCTL-producing cells occur that are too subtle to resolve by conventional light microscopy and/or are complicated by the geometry of ΔCTL expressing cells. It is possible that FtsZ regulates PG metabolism through a scaffolding function that is more complex than simply positioning PG enzymes and their regulators at midcell. For example, the Z-ring may regulate the precise molecular architecture, stoichiometry, sequential order and/or dynamics of assembly and disassembly of protein complexes required for PG metabolism at the division site.
There are unsolved mysteries regarding assembly of proteins downstream of FtsZ that hint at biophysical regulation of cell wall metabolism. In C. crescentus, MurG, which catalyzes the final step in PG precursor synthesis, and DipM, a protein involved in PG hydrolysis, localize to midcell immediately after assembly of the Z-ring in an FtsZ-dependent manner9,10,50–52. However, neither interacts directly with FtsZ, nor do they require any other early recruit to midcell for their localization9,43. In fact, DipM resides in the periplasm and requires its PG-binding domains for localization50,52, suggesting special characteristics of PG at sites of FtsZ assembly that mediate DipM recruitment. Additionally, a number of inner membrane proteins are excluded from sites of FtsZ assembly through an unknown mechanism43,53. These observations indicate that the Z-ring alters the biophysical characteristics of the membrane54,55 and/or PG locally to drive protein recruitment or exclusion. A similar biophysical mechanism might be invoked to regulate the activity of cell wall enzymes: we hypothesize that FtsZ initiates a physical signal that leads to local activation of PG metabolism (Fig. 8c). Such a signal is likely transduced through the CTC to FtsA and other effectors before ultimately being received by PG enzymes.
Membrane-associated FtsZ is capable of generating constrictive force on membranes in vitro4. However, whether this force is generated in vivo and, if so, how such force is used during growth and division are unknown. One possibility is that FtsZ-mediated force is the physical signal described above: force generated by the Z-ring would be transduced to the cell envelope and/or divisome proteins and used to mechanically regulate the activity of PG remodeling enzymes (Fig. 8c). The CTL has been proposed to behave as an entropic spring20 that is expected to affect the mechanical properties of FtsZ. Moreover, we observed altered FtsZ architecture for ΔCTL in cells and in vitro. We hypothesize that reduction of the CTL below a minimal length alters the physical properties of the Z-ring, including its ability to generate or transduce force, in a manner that leads to misregulation of cell wall remodeling (Fig. 8d). The unique phenotype associated with ΔCTL production provides a valuable genetic tool for further delineation of the mechanisms underlying the regulation of cell shape by the bacterial cytoskeleton.
Methods
Bacterial strains, plasmids, and growth conditions
Caulobacter crescentus NA1000 strains were grown in peptone yeast extract (PYE) medium or in minimal M2 medium at 30°C. Additives and antibiotics were used at the following concentrations in liquid (solid) media for C. crescentus: xylose 0.3 (0.2)%, glucose 0.2%, vanillate 0.5 (0.5) mM, myo-inositol 0.3%, gentamycin 1 (5) μg mL−1, kanamycin 5 (25) μg mL−1, spectinomycin 25 (100) μg mL−1, streptomycin (5 μg mL−1), cephalexin 1 μg mL−1, and mecillinam 5 μg mL−1. Prior to changes in induction conditions, cells were washed 2–3 times in plain media. Growth rate analyses were performed in 96-well plates with shaking at 30°C using a Tecan Infinite 200 Pro plate reader. Strains and plasmids used in this study are listed in Supplementary Data 1 and 2.
Light microscopy and image analysis
Cells were imaged during log phase of growth after immobilization on 1% agarose pads. Light microscopy was performed on a Nikon Eclipse Ti inverted microscope equipped with a Nikon Plan Fluor 100X (NA 1.30) oil Ph3 objective and Photometrics CoolSNAP HQ2 cooled CCD camera. Chroma filter cubes were used as follows: ET-EYFP for YFP and Venus, ET-ECFP for CFP, ET-EGFP for GFP, ET-dsRED for mCherry, and ET-DAPI for HADA. Images were processed in Adobe Photoshop. Automated cell width analysis was performed using MicrobeTracker56. Algorithm 4 was used for determining cell outlines, with the following parameter changes: areaMin = 150, areaMax = 5000, and wspringconst = 0.1. Maximum cell width along the entire length of the cell for each cell in the population was determined from this analysis. In cells with bulges, the maximum cell width corresponds to the width of the largest bulge.
Whole cell transmission electron microscopy
5 μL of cells were spotted onto carbon-coated mesh copper grids (Electron Microscopy Sciences, Fort Washington, PA) and allowed to adhere for 15 min. Grids were rinsed with ddH2O and stained with 7.5% uranyl magnesium acetate for 5 min. The grids were rinsed in ddH2O five times and allowed to dry briefly before placing in a grid box. Grids were imaged the next day at 80 kV using a JEOL JEM 1010 transmission electron microscope equipped with a 4k × 4k Gatan CCD Ultrascan 4000 SP model 890 camera.
Antibodies and immunoblotting
Purified full-length C. crescentus FtsZ was used to immunize a rabbit for antibody production (Josman, LLC). Specificity for FtsZ was demonstrated by immunoblotting lysates from strains depleted of FtsZ and observing a reduction in the signal at the expected molecular weight of full-length C. crescentus FtsZ (predicted 54 kDa, runs at ~65 kDa) (e.g. Supplementary Fig. 2c) and by producing FtsZ variants and observing new bands at the expected molecular weight (e.g. Supplementary Fig. 3). For immunoblotting against whole cell lysates, cells were harvested in log phase of growth and lysed in SDS-PAGE loading buffer by boiling for 5 min. SDS-PAGE and transfer of protein to nitrocellulose membrane were performed using standard procedures. FtsZ antiserum was used at 1:20,000 dilution. SpmX antiserum was used at 1:50,000 dilution57. Uncropped immunoblots are presented in Supplementary Figure 3.
Protein stability assay
Translation was inhibited by adding chloramphenicol (30 μg ml−1) to strains expressing WT FtsZ or linker variants as the only copy of FtsZ. Cells were harvested every 20 min for 2 h and lysates from each time point (equivalent total OD units loaded for each) were subjected to immunoblotting with FtsZ antiserum. Horseradish peroxidase-conjugated secondary antibody and ECL substrate were used for detection. Signal was detected using an ImageQuant LAS4000 mini chemiluminescence imaging system (GE Healthcare Life Sciences, Piscataway, NJ) and quantified with ImageQuant TL software. Protein half-life was calculated by plotting the relative concentration (normalized to amount of FtsZ or FtsZ variant at time 0) against time and fitting a one-phase decay curve using Graphpad Prism software (Graphpad Software, Inc., La Jolla, CA).
Protein purification
FtsZ, FtsZ variants, His6-FzlA and His6-MipZ were overproduced and purified as described previously43 with slight modifications. To purifiy FtsZ, Rosetta (DE3) pLysS E. coli cells bearing plasmid pMT219 were grown at 37°C to OD600 of 0.8–1.0 prior to induction with 0.5 mM IPTG for 3–4 h. Cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C, resuspended in 30 mL Buffer QA (50 mM Tris-HCl pH 8.0, 50 mM KCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol) per L of culture, snap frozen in liquid nitrogen and stored at −80°C until purification. Cell suspensions were thawed, one Mini Complete Protease Inhibitor tablet was added per 30 mL (Roche), lysozyme was added to 1 mg mL−1, PMSF was added to 2 mM, and DNAse I (New England Biolabs) was added to 2 units mL−1 final concentrations. Cell suspensions were incubated for 30–60 min at room temperature, sonicated, and centrifuged at 15,000 × g for 30 min at 4°C. Supernatant was filtered and loaded onto two HiTrap Q HP 5 mL columns (GE Life Sciences) joined in tandem and equilibrated in Buffer QA. Protein was eluted using a linear KCl gradient from 50–500 mM over 20 column volumes. Peak fractions containing FtsZ were combined and ammonium sulfate was added to 20% saturation. Ammonium sulfate precipitate was recovered by centrifugation at 10,000 × g for 10 min at 4°C. After resting overnight on ice, precipitates were resuspended in FtsZ Storage Buffer (50 mM HEPES-KOH pH 7.2, 50 mM KCl, 0.1 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol) and consecutively applied to a Superdex 200 10/300 GL column (GE Life Sciences) in FtsZ Storage Buffer. Peak fractions were combined, concentrated if necessary, snap frozen in liquid nitrogen, and stored at −80°C. ΔCTL, L14, and ΔCTLTA FtsZ variants were overproduced from plasmids pEG681, pEG723, pEG886, respectively, in Rosetta (DE3) pLysS E. coli as for FtsZ and purified following the same procedure used for FtsZ. The only exceptions were that ΔCTL precipitated at 30% ammonium sulfate and ΔCTLTA precipitated at 40% ammonium sulfate.
To purifiy His6-MipZ, Rosetta (DE3) pLysS E. coli cells bearing plasmid pMT183 were grown at 37°C to OD600 of 0.6 prior to induction with 0.5 mM IPTG for 3.25 h. Cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C, washed in phosphate-buffered saline, snap frozen in liquid nitrogen, and stored at −80°C until purification. Cells were thawed and resuspended in Ni lysis buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 10 mM imidazole, 1 mM EDTA, 10% glycerol). One Mini Complete Protease Inhibitor tablet was added per 30 mL (Roche), PMSF was added to 2 mM, and DNAse I (New England Biolabs) was added to 2 units mL−1 final concentrations. Cells were lysed by passage through a French Press twice at 15000 psi and lysates were centrifuged at 15,000 × g for 30 min at 4°C. His6-MipZ was isolated by binding to Ni-NTA agarose (Qiagen), washed in Ni wash buffer (Ni lysis buffer with 20 mM imidazole), and eluted in Ni elution buffer (Ni lysis buffer with 300 mM imidazole). Peak fractions were concentrated and applied to a Superdex 200 10/300 GL column (GE Life Sciences) equilibrated in MipZ Storage Buffer (50 mM HEPES-NaOH pH 7.2, 50 mM NaCl, 0.1 mM EDTA, 1 mM β-mercaptoethanol, 10% glycerol). Peak fractions were pooled, concentrated, snap frozen in liquid nitrogen, and stored at −80°C.
To purifiy His6-FzlA, Rosetta (DE3) pLysS E. coli cells bearing plasmid pEG327 were grown at 30°C to OD600 of 0.5 prior to induction with 0.5 mM IPTG for 4 h. Cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C, resuspended in FzlA lysis buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 20 mM imidazole, 10% glycerol), snap frozen in liquid nitrogen, and stored at −80°C until purification. Cells were thawed and lysozyme was added to 1 mg mL−1, one Mini Complete Protease Inhibitor tablet was added per 30 mL (Roche), MgCl2 was added to 2.5 mM and DNAse I (New England Biolabs) was added to 2 units mL−1 final concentrations. Cells were incubated at room temperature for 45 min, sonicated, and lysates were centrifuged at 15,000 × g for 30 min at 4°C. Lysate was filtered and applied to two 1 mL HisTrap FF columns joined in tandem (GE Life Sciences) and equilibrated in FzlA lysis buffer. Columns were washed in FzlA lysis buffer and His6-FzlA was eluted in FzlA elution buffer (FzlA lysis buffer with 300 mM imidazole). Peak fractions were combined, dialyzed into FzlA Storage Buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 5% glycerol), snap frozen in liquid nitrogen, and stored at −80°C.
FtsZ activity assays
FtsZ activity assays were performed in HEK50 buffer (50 mM HEPES-KOH pH 7.2, 50 mM KCl, 0.1 mM EDTA). MgCl2 was used at 2.5 mM and GTP was used at 2 mM. Light scattering was monitored using a Fluoromax-3 spectrofluorometer (Jobin Yvon, Inc.) with 350 nm excitation and emission wavelengths and 2 nm slits. GTPase activity was assayed using the SensoLyte MG Phosphate Assay Kit (AnaSpec) following the manufacturer’s protocol. GraphPad Prism software was used to fit curves and determine GTPase rates. For pelleting and co-sedimentation assays, FtsZ was polymerized for 15 min at room temperature, with additional proteins when indicated. Reactions were subjected to centrifugation at 250,000 × g for 15 min at 25°C, after which supernatant and pellet fractions were collected. Critical concentration was estimated as the x-intercept in GTPase activity vs. protein concentration plots.
For visualization of protofilaments by TEM, 8 μM of purified protein was polymerized in HEK50 buffer with 2.5 mM MgCl2 and 2 mM GTP for ~15 min, spotted onto glow-discharged carbon-coated mesh copper grids (Electron Microscopy Sciences, Hatfield, PA), dried after 2 min, and stained with 0.75% uranyl formate for 2 min. The grids were dried once again and the staining was repeated. The grids were completely dried and imaged using a Philips/FEI BioTwin CM120 transmission electron microscope equipped with an AMT XR80 8 megapixel CCD camera (AMT Imaging, USA).
HADA labeling
Strain EG852 was washed twice in PYE and resuspended in PYE with either vanillate and glucose (to induce WT ftsZ) or xylose (to induce ΔCTL). Cells were grown for 5 h at 30°C with shaking, HADA34 was added to a final concentration of 0.41 mM and cells were returned to the shaker for 5 min. Cells were removed, washed twice in phosphate buffered saline (PBS), resuspended in ice-cold 70% ethanol, and incubated on ice for 20 min. Cells were pelleted, resuspended in PBS, and imaged. For pulse-chase labeling, following 5 min incubation with HADA, cells were washed twice in PYE, resuspended in PYE with supplements, and returned to the incubator. Cells were withdrawn at time zero or indicated times and processed for imaging as above.
Muropeptide analysis
UPLC samples were prepared as described previously58, with modifications. Log-phase cultures of C. crescentus EG852 and EG864 were washed in PYE and diluted 1:10 in 500 mL plain PYE (EG864), PYE with vanillate plus glucose (EG852 ΔCTL uninduced), or PYE with xylose (EG852 ΔCTL induced). Cultures were grown for 5 h at 30°C to an OD600 of 0.3, then centrifuged at 6,000 × g for 10 min at 4°C. The resulting pellets were resuspended in 3 mL ice-cold water. Cell suspensions were then lysed by boiling in 6% SDS for 30 min and allowed to cool to room temperature. Lysed cell suspensions were ultracentrifuged at 400,000 × g and pelleted sacculi were digested with muramidase into muropeptides. Samples were pH adjusted and injected onto a Waters H Class UPLC system equipped with a BEH C18 1.7-μm column (Waters, MA, USA) and eluted with sodium phosphate buffers. Peaks were quantified and identified as particular muropeptide species from their elution times based on Takacs et al.39, from which the crosslinking density and strand length were calculated59.
Supplementary Material
Acknowledgments
We are grateful to Patrick Viollier for SpmX antiserum; to Yves Brun, Erkin Kuru, and Michael van Nieuwenzhe for HADA and guidance in its use; to Matt Welch and Rebecca Lamason for R. parkeri genomic DNA; to Arash Komeili for M. magneticum AMB-1 genomic DNA; to Yves Brun and Ellen Quardokus for assistance with TEM and for hosting ELM; to Peter Chien for advice on protein turnover assays; and to Lucy Shapiro and Martin Thanbichler for strains. We also thank Jie Xiao and the Xiao laboratory for helpful discussions. This work was funded by the JHUSOM (to EDG), an NIH Director’s New Innovator Award DP2OD006466 (to KCH), and an NSF Graduate Research Fellowship (to AM).
Footnotes
Author contributions
KS, SMD, KCH, and EDG designed the research. KS, AM, SMD, ELM, and EDG performed the research. KS, AM, SMD, KCH, and EDG analyzed the data. KS, AM, ELM, KCH, and EDG wrote the paper.
Competing Financial interests
The authors declare no competing financial interests.
References
- 1.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S, Shaevitz JW. The mechanics of shape in prokaryotes. Front Biosci (Schol Ed) 2013;5:564–574. doi: 10.2741/s390. [DOI] [PubMed] [Google Scholar]
- 3.Cabeen MT, et al. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osawa M, Anderson DE, Erickson HP. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc Natl Acad Sci USA. 2010;107:9182–9185. doi: 10.1073/pnas.0911517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken) 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aarsman MEG, et al. Maturation of the Escherichia coli divisome occurs in two steps. Molecular Microbiology. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- 8.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goley ED, et al. Assembly of the Caulobacter cell division machine. Molecular Microbiology. 2011;80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aaron M, et al. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Molecular Microbiology. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
- 11.Potluri LP, Kannan S, Young KD. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J Bacteriol. 2012;194:5334–5342. doi: 10.1128/JB.00859-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osawa M, Erickson HP. Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA. 2013;110:11000–11004. doi: 10.1073/pnas.1222254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meier EL, Goley ED. Form and function of the bacterial cytokinetic ring. Curr Opin Cell Biol. 2014;26:19–27. doi: 10.1016/j.ceb.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Löwe J, Amos LA. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 15.Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 16.Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaughan S, Wickstead B, Gull K, Addinall SG. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J Mol Evol. 2004;58:19–29. doi: 10.1007/s00239-003-2523-5. [DOI] [PubMed] [Google Scholar]
- 19.Buske P, Levin PA. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Molecular Microbiology. 2013;89:249–263. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner KAJA, Moore DA, Erickson HP. The C-terminal Linker of Escherichia coli FtsZ Functions as an Intrinsically Disordered Peptide. Molecular Microbiology. 2013;89:264–275. doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolin W, Corbo JC, Long SR. Cloning and characterization of a Rhizobium meliloti homolog of the Escherichia coli cell division gene ftsZ. J Bacteriol. 1991;173:5822–5830. doi: 10.1128/jb.173.18.5822-5830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quardokus E, Din N, Brun YV. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc Natl Acad Sci USA. 1996;93:6314–6319. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams B, Bhat N, Chien P, Shapiro L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Molecular Microbiology. 2014;93:853–866. doi: 10.1111/mmi.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boutte CC, et al. Genetic and computational identification of a conserved bacterial metabolic module. PLoS Genet. 2008;4:e1000310. doi: 10.1371/journal.pgen.1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strobel W, Möll A, Kiekebusch D, Klein KE, Thanbichler M. Function and localization dynamics of bifunctional penicillin-binding proteins in Caulobacter crescentus. J Bacteriol. 2014;196:1627–1639. doi: 10.1128/JB.01194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Din N, Quardokus EM, Sackett MJ, Brun YV. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Molecular Microbiology. 1998;27:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- 28.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Molecular Microbiology. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 29.Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science. 2013;341:392–395. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz U, Asmus A, Frank H. Autolytic enzymes and cell division of Escherichia coli. J Mol Biol. 1969;41:419–429. doi: 10.1016/0022-2836(69)90285-x. [DOI] [PubMed] [Google Scholar]
- 33.Cava F, de Pedro MA, Lam H, Davis BM, Waldor MK. Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 2011;30:3442–3453. doi: 10.1038/emboj.2011.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuru E, et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew Chem Int Ed Engl. 2012;51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imada A, Kintaka K, Nakao M, Shinagawa S. Bulgecin, a bacterial metabolite which in concert with beta-lactam antibiotics causes bulge formation. J Antibiot. 1982;35:1400–1403. doi: 10.7164/antibiotics.35.1400. [DOI] [PubMed] [Google Scholar]
- 37.Templin MF, Edwards DH, Höltje JV. A murein hydrolase is the specific target of bulgecin in Escherichia coli. J Biol Chem. 1992;267:20039–20043. [PubMed] [Google Scholar]
- 38.Desmarais SM, de Pedro MA, Cava F, Huang KC. Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. Molecular Microbiology. 2013;89:1–13. doi: 10.1111/mmi.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takacs CN, et al. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J Bacteriol. 2010;192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Figge RM, Divakaruni AV, Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Molecular Microbiology. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- 42.Dye NA, Pincus Z, Fisher IC, Shapiro L, Theriot JA. Mutations in the nucleotide binding pocket of MreB can alter cell curvature and polar morphology in Caulobacter. Molecular Microbiology. 2011;81:368–394. doi: 10.1111/j.1365-2958.2011.07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goley ED, Dye NA, Werner JN, Gitai Z, Shapiro L. Imaging-based identification of a critical regulator of FtsZ protofilament curvature in Caulobacter. Mol Cell. 2010;39:975–987. doi: 10.1016/j.molcel.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radhakrishnan SK, Pritchard S, Viollier PH. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev Cell. 2010;18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 45.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 46.Camberg JL, Viola MG, Rea L, Hoskins JR, Wickner S. Location of Dual Sites in E. coli FtsZ Important for Degradation by ClpXP; One at the C-Terminus and One in the Disordered Linker. PLoS ONE. 2014;9:e94964. doi: 10.1371/journal.pone.0094964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camberg JL, Hoskins JR, Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci USA. 2009;106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Din N, Quardokus EM, Sackett MJ, Brun YV. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Molecular Microbiology. 2001;40:1051–1063. doi: 10.1046/j.1365-2958.1998.00752.x. [DOI] [PubMed] [Google Scholar]
- 50.Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter. Molecular Microbiology. 2010;77:56–73. doi: 10.1111/j.1365-2958.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poggio S, Takacs CN, Vollmer W, Jacobs-Wagner C. A protein critical for cell constriction in the Gram-negative bacterium Caulobacter crescentus localizes at the division site through its peptidoglycan-binding LysM domains. Molecular Microbiology. 2010;77:74–89. doi: 10.1111/j.1365-2958.2010.07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Möll A, Schlimpert S, Briegel A, Jensen GJ, Thanbichler M. DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Molecular Microbiology. 2010;77:90–107. doi: 10.1111/j.1365-2958.2010.07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meredith DH, Plank M, Lewis PJ. Different patterns of integral membrane protein localization during cell division in Bacillus subtilis. Microbiology (Reading, Engl ) 2008;154:64–71. doi: 10.1099/mic.0.2007/013268-0. [DOI] [PubMed] [Google Scholar]
- 54.Strahl H, Bürmann FBU, Hamoen LW. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun. 2014;5:3442. doi: 10.1038/ncomms4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arumugam S, Petrov EP, Schwille P. Cytoskeletal pinning controls phase separation in multicomponent lipid membranes. Biophys J. 2015;108:1104–1113. doi: 10.1016/j.bpj.2014.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Molecular Microbiology. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desmarais SM, Cava F, de Pedro MA, Huang KC. Isolation and preparation of bacterial cell walls for compositional analysis by ultra performance liquid chromatography. J Vis Exp. 2014:e51183. doi: 10.3791/51183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glauner B. Separation and Quantification of Muropeptides with High-Performance Liquid Chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.