Abstract
The dose-dependent bioactivity of small molecules on cells is a crucial factor in drug discovery and personalized medicine. Although small-molecule microarrays are a promising platform for miniaturized screening, it has been a challenge to use them to obtain quantitative dose-response curves in vitro, especially for lipophilic compounds. Here we establish a small-molecule microarray assay capable of controlling the dosage of small lipophilic molecules delivered to cells by varying the sub-cellular volumes of surface supported lipid micro- and nanostructure arrays fabricated with nanointaglio. Features with sub-cellular lateral dimensions were found necessary to obtain normal cell adhesion with HeLa cells. The volumes of the lipophilic drug-containing nanostructures were determined using a fluorescence microscope calibrated by atomic-force microscopy. We used the surface supported lipid volume information to obtain EC-50 values for the response of HeLa cells to three FDA-approved lipophilic anticancer drugs, docetaxel, imiquimod and triethylenemelamine, which were found to be significantly different from neat lipid controls. No significant toxicity was observed on the control cells surrounding the drug/lipid patterns, indicating lack of interference or leakage from the arrays. Comparison of the microarray data to dose-response curves for the same drugs delivered liposomally from solution revealed quantitative differences in the efficacy values, which we explain in terms of cell-adhesion playing a more important role in the surface-based assay. The assay should be scalable to a density of at least 10,000 dose response curves on the area of a standard microtiter plate.
Introduction
The dosage of drugs is a critical factor in determining their efficacy at scales ranging from cellular to the whole organism. For instance, it has been demonstrated that although microtubule stabilizers such as docetaxel affect microtubule mass at high concentration, at 100 fold lower dosage they work in a completely different manner to kinetically stabilize the microtubules without affecting the mass1, 2. Currently, most high throughput screens in drug discovery involve testing 105 - 106 candidate molecules on cells at single concentrations, and only performing dosage studies on the positive hits3, 4. This presents the problem of potentially missing biologically relevant information obtainable at different dosages4-6. Success in using microarrays for rapid analysis of large biomolecules such as nucleic acids, proteins, and carbohydrates have led to recent efforts to produce small-molecule microarrays (SMMs) for in vitro cell culture assays7-12. Since they were first developed13, SMMs have gone from being used as probes for ligand interaction, to applications in proteomics, bioactivity screens and whole cell interactions14-16. Although SMMs show potential in the miniaturization of high-throughput screening, a challenge lies in the variation of dosage and cellular uptake for cell-based assays. Many SMMs covalently link different molecules to the surface and measure cell adhesion as a readout15, 17, 18, which is suitable for drugs that target cell surface receptors, yet incompatible with drugs that must be internalized by the cell.
One promising approach to address the dosage issue is the sandwiched system, which is capable of obtaining dose resonse curves at a surface density of >2000 assays on the area of a standard glass slide19. The system involves microarraying drug solutions onto posts capable of addressing indidvidual microwells where cells can be cultured. By depositing different amounts of small molecules onto the posts, different dosages can be screened. Another approach to address the dosage issue is to make use of the concentration gradient that arises from the diffusion of drugs out of polymer complexes into which they have been embedded10. A challenge still exists in obtaining microarray-based dose-response curves for lipophilic compounds that do not readily dissolve in aqueous solutions, which is important as the majority of compounds in small molecule high-throughput screening libraries are lipophilic20. We previously demonstrated a lipid multilayer microarray in which lipophilic small molecules are encapsulated in a lipid volume on the surface until cells adhere and internalize the lipids and small molecules.21 In that assay, we were able to elicit maximum cellular responses from the surface equivalent to solution delivered dosages of up to ~40 μM.
In order to scale up lipid multilayer microarrays for medium and eventually high throughput cell culture screening on a chip, a scalable fabrication and quality control process is needed. We previously used dip-pen nanolithography (DPN) as it can control lipid multilayer volumes and integrate different materials onto a single surface in a direct-write process. However, DPN is still limited in the number of different materials that can be integrated and the uniformity of deposited lipid multilayer volumes over large surface areas (>0.01 mm2)22, 23. To overcome this limit, we developed a new method of lipid multialyer nanofabrication which we call nanointaglio24, 25. Intaglio is a mode of printing that deposits an ink from the recesses of the polymeric stamp rather than from the surface relief, the latter being the common mode of microcontact printing24, 25. Importantly, nanointaglio enables the reproducible control over the volumes of materials for cell culture assays as depicted in Figure 1, while opening the way for scaling up the number of materials screened. The volume of material deposited from the recesses of a nanointaglio stamp is determined by the size of the recesses in the stamp and the number of prints between inking steps. Combining pin-spotting technology with nanointaglio printing has the capability to drastically scale up the throughput of nanointaglio as a screening platform24.
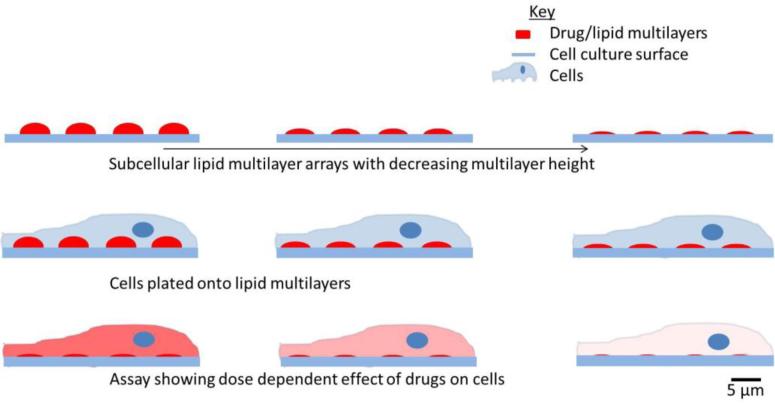
Figure 1.
Lipid multilayer volume dependent dose control. Arrays of varying volume are fabricated by nanointaglio and volume is determined by quantitative fluorescence microscopy. Cells are then cultured on the patterned surfaces and an assay is performed to determine the dose dependent effect of the multilayers on the cells.
Quality control is an essential step in micro- and nano-fabrication. Measuring the heights of printed features by atomic force microscopy (AFM) is the most accurate way to quantify the multilayer volumes but the need for high throughput and a sterile environment prior to cell culture make this process impractical for cell based screening. To address this issue, we previously developed a method of high-throughput optical calibration of lipid multilayer heights by fluorescence microscopy26. This calibration is done by measuring the fluorescence intensities of fluorescently labeled lipid multilayers at different exposure times and comparing these values to AFM-measured heights. From the calculated heights and lateral dimensions of the lipid multilayer spots we can then calculate the volume of the lipid dots and hence the dosage of the encapsulated drugs.
To quantify the dosage delivered to cells from lipid multilayer microarrays, we need to determine: 1) whether the lipid arrays affect the initial adhesion of cells to the pattern enough to interfere with the assay, 2) how much drug is present per unit area for areas with different multilayer heights, and 3) how the surface delivery compares to solution delivery. Here we achieve these goals by investigating cell adhesion to lipid spots of various dimensions, quantifying the volume of the lipid multilayer arrays with AFM-calibrated fluorescence microscopy, and finally obtaining dose-response curves from surface and solution delivery for three different drugs.
Experimental
Materials and Method
Preparation of liposomal drugs
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (sodium salt) (DOPS), cholesterol and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine-B-sulfonyl)(ammonium salt) (rhodamine-PE) each dissolved in chloroform were aliquoted into a glass vial in the molar ratio 55:35:9.5:0.527.
We chose this mixture because of the expected low toxicity due to the biocompatibility of this lipid formulation and because of the use of the lipids in transfection28. Drugs were added to the aliquoted lipids at a proportion of 10% by mass. The drug was omitted from the negative control. The chloroform was evaporated from the mixture using a gentle stream of nitrogen gas. The vial was then placed in a vacuum for 20 minutes to remove residual chloroform. For nanointaglio delivery, ethanol was added and the dried lipids resolubilized, except for imiquimod where a chloroform/methanol solution (1:1) was used for resolubilization. For the solution delivery, liposomes were formed by adding Hank's Balanced Salt Solution (HBSS) to the vial of dried lipids to form liposomes and encapsulate the drugs. The suspension was then sonicated for 10 minutes. Docetaxel was kindly provided by Diego Zorio of the Department of Chemistry at Florida State University. Imiquimod and triethylenemelamine were obtained from the Approved Oncology Drug Set II (National Cancer Institute (NCI)/ National Institutes of Health Developmental Therapeutics Program). Lipids used in supplementary live-cell experiments were a mixture of DOPC and 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP) in a 7:3 ratio with 1 mol% fluorescein-PE added. All lipids were purchased from Avanti Polar Lipids Inc., Alabaster, AL, USA.
Inking and stamping
The lipid/drug mixtures were arrayed on the palettes by pipetting a 0.5 μL aliquot for each of the lipid/drug mixtures in ethanol (inks) individually onto a glass slide. The inked palettes were then dried in a vacuum for 24 hrs. A polydimethylsiloxane (PDMS) stamp with well features of 5 μm diameter and 2.5 μm depth, covering 19% of the stamp surface, was inked by pressing the patterned surface onto the ink palette24. In order to vary the dosage, the inks were stamped multiple times onto poly-D-lysine-coated optical dishes procured from MatTek Corporation, Ashland, MA USA. We used poly-D-lysine due to poly-L-lysine's susceptibility to degradation by extracellular enzymes released by cells. Alignment of stamp wells with the inks on the pallet is unnecessary here as the surface of the stamp is completely covered with the 5 μm diameter microwell features.
Cell culture
Nanointaglio delivery
The HeLa cells used for the experiments were purchased from American Type Culture Collection (ATCC), Manassas, VA, USA. The cells were grown to 70% confluence before use. The printed lipid multilayer array was kept in a glove box for 2 hrs to ensure the stability of the lipids during immersion in aqueous media29. The HeLa cells were gently seeded over the multilayer microarrays by adding 2 mL of the cell suspension at a density of 200,000 cells/mL. The cells were incubated 72 hrs for the toxicity study and 12 hrs for the adhesion study. After incubation, the cells were washed with HBSS buffer, stained for the nuclei, imaged, and counted to determine the final viability. DAPI staining was by incubation the cells with the DAPI dye for 10 minutes and washing the cells twice. Syto 9 and propidium iodide staining were done by incubating each dye with the appropriate cells for 15 minutes and washing the cells twice.
Solution delivery
Cells were seeded in 24-well plates 24 hrs prior to the experiment. Each well was seeded with 500 μL of cells suspended in media at 100,000 cell/mL. The cells were seeded so that they would have a confluence of 70% at the time of the experiment. Cells were incubated with liposomally-encapsulated drugs for 72 hrs, were then rinsed three times before staining with DAPI (Life Technologies, Eugene, OR, USA) and imaging for viability determination. The images for the live-cell experiment were taken at desired time points over the 72 hr period. Cell culture medium was not changed over the duration of the experiments.
Adhesion assays
Cells were incubated over the nanointaglio pattern and the lipid blobs for 12 hrs. The cells were then washed with PBS buffer, fixed in 4% formaldehyde, and immunostained for the adhesion molecule vinculin, for cytoskeletal actin, and for the nuclei. Vinculin was immunostained with anti-vinculin-FITC, actin was stained with TRITC-conjugated Phalloidin, and nuclei were stained with DAPI. All the fluorescent stains were purchased from EMD Millipore, Billerica, Massachusetts, USA.
Fluorescence microscopy
Epifluorescence microscopy was done using a Ti-E inverted microscope (Nikon Instruments, Melville, NY, USA) fitted with a Retiga SRV (QImaging, Surrey, BC, Canada) CCD camera (1.4 MP, Peltier cooled to −45 °C). Rhodamine-PE doped lipid structures were imaged using the G-2E/C filter, DAPI was imaged using the UV-2E/C fluorescence filter, and Syto 9 (Invitrogen, Eugene, OR, USA) and fluorescein (Avanti polar Lipids) were both imaged using the B-2E/C. Immunofluorescent confocal microscopy was performed using a DeltaVision pDV (GE Healthcare, Pittsburgh, PA) to collect images, which were then deconvolved with SoftWorX 3.7.0 (Applied Precision, Issaquah, WA) and compiled with Adobe Phososhop CS6 (Adobe Systems)30. Live-cell imaging was done using an Olympus Viva View fluorescent incubator microscope.
Atomic Force Microscopy (AFM) imaging
AFM heights of the neat lipid and lipid-encapsulated drug prints were measured in tapping mode with a Dimension Icon AFM (Bruker, Billerica, MA, USA) and tapping mode AFM cantilevers (FESPA, 8 nm nominal tip radius, 10-15 μm tip height, 2.8 N m−1 spring constant, Bruker, Billerica, MA, USA).
Calibration and dosage quantification
Optical calibration was done as previously described19. Briefly, nanointaglio printing used fabricated rhodamine-PE doped lipid multilayers with varying heights by printing multiple times from a PDMS stamp with wells 5 μm in diameter and 2.5 μm in depth. The fluorescence intensity in grey values (g.v.) of individual dots was then measured at different exposure times (0.02s, 0.04s, 0.08s, 0.2s, 0.4s, 0.8s, 2s) and the optical response of the camera for each dot was determined in units of g.v./s whch we call “sensitivity”. The AFM heights of the same fluorescently-measured lipid multilayers were also measured. The camera sensitivity was plotted against the AFM-measured heights and a linear regression of the resulting graph was used to determine the calibration factor. With this factor, lipid dot heights can be calculated from the optical fluorescence microscopy data using the formula, . We measured the number and maximum fluorescence intensities of constituent dots of each spot using the ImageJ software and converted the intensities to heights using the calibration formula. We determined the volume of materials per dot by multiplying the area of each dot by its calculated height, assuming the dot shape of a cylinder. The total volume of the lipid spots was then determined by summing up the volumes of the individual constituent dots. The mass of the printed lipids was then calculated assuming a density of 1g/L. Since we added the drugs to the lipids in a 1:9 ratio we were able to calculate the total mass of drugs per unit area and use that value as our dosage. The EC-50 of each drug was determined by plotting the dose-response curve with the Origin® program using the growth sigmoidal dose response function, . The EC-50 values were generated from the graph.
Statistics
Comparisons of adhesion between the surface-supported lipids and the controls were done using the student t-test (p = 0.05). The adhesion experiments were done in duplicates with four prints per sample. For the dose-response experiments we used four replicates with two prints per sample for each dosage, for a total of 40 samples per dose-response curve.
Results and discussion
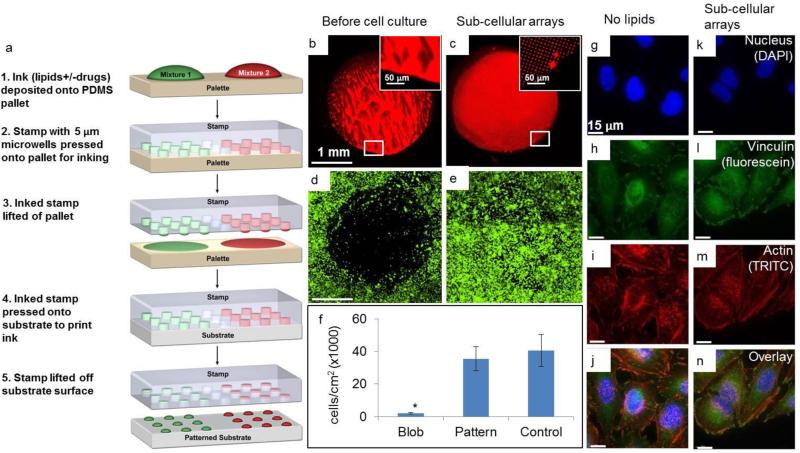
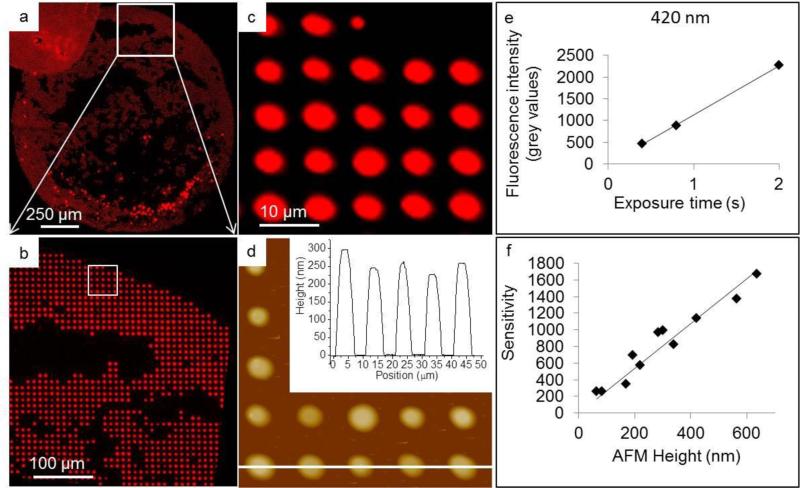
In order to establish the cell culture assay and determine the dosage limits of the lipid-based delivery system, we first investigated the adhesion of HeLa cells to lipid multilayer patterns with sub-cellular and super-cellular lateral feature sizes (Figure 2). Figure 2a shows a schematic of the nanointaglio process used to fabricate the lipid multilayer arrays24. An inked stamp can be used to print multiple times, and as ink is depleted from the stamp the multilayer heights decrease. In this experiment we printed eight times per inking. In the first prints from a freshly inked nanointaglio stamp, excess ink on the stamp resulted in contiguous lipid multilayers, which we refer to here as blobs. We compared the adhesion of HeLa cells to these blobs and adhesion to patterns with distinct sub-cellular features (5 μm diameter dots arranged in a grid with a pitch of 10 μm). The cells were cultured for 12 hours and the substrate area in the same dish, without any lipid patterns, was used as a control. Typically the HeLa cells used here ranged between 20-50 μm in diameter. On average each cell covered ~2-5 dots. Figure 2 shows fluorescence micrographs of the blobs and sub-cellular patterns, before and after cell culture. From the fluorescence data, it can be seen that cells adhere to the area patterned with sub-cellular features, but not to the blob area. Since the cells do not grow on blobs, the maximum deliverable dosage to the cells will be limited to the maximum nanointaglio heights. Importantly, quantitative analysis of the data shows no significant difference (p>.05 after 12 replicates) between the adhesion of cells to the sub-cellular patterns and adhesion to the control. It is not surprising that the cells do not adhere to the blob areas, as lipid bilayers have been shown to prevent cellular adhesion31-33. However, it is striking that the sub-cellular patterns have such a negligible affect on the adhesion of HeLa cells. In order to determine whether morphological differences could be detected between the controls and the sub-cellular patterns, cells adhered to the control and to the sub-cellular arrays were stained for the focal adhesion-associated protein vinculin, for the cytoskeletal protein actin, and for nuclei. Figure 2(g-n) are representative fluorescence micrographs that indicate no observable morphological differences induced by the sub-cellular arrays after 12 hours. To determine whether cell viability can be assayed by counting the number of cells remaining on the surface after culture on the sub-cellular arrays, we carried out live-cell imaging on sub-cellular lipid multilayer arrays with and without docetaxel (Supplementary Figure 1 and Supplementary Videos 1-3 available as ESI). These data show the cells adhering and proliferating over the patterned areas over a timescale of over 72 hours. From the videos, it can be seen that the cells take a few hours longer to spread on the lipid controls (Supplementary video 2) than on the poly-D-lysine-coated glass coverslip (Supplementary video 1), but after 12 hours they both adhere and appear morphologically the same. However, cells cultured on the drug-containing spots (Supplementary video 3) begin to die before fully spreading on the surface, as indicated by propidium iodide staining. These data suggest that the cells likely take up the surface-supported liposomal formulations as they are adhering. Next we determined how much drug was deposited in each nanointaglio multilayer spot for dosage calculations. This was was done using AFM of fluorescently-labeled lipid multilayer arrays to calibrate a fluorescence microscope for quantitative determination of lipid multilayer volumes (Figure 3)19. Figures 3(a-c) shows a fluorescence image of one of the sample areas used for calibration, and Figure 3d shows an AFM image of the same area shown in Figure 3c. Figures 3(e-f) show typical calibration curves that allow us to quantify lipid dot volumes and dosages in units of ng/mm2 from fluorescence micrographs, as described in detail in the experimental section. The dosages obtained from 5 prints ranged from 0.005 ng/mm2 to 1.5 ng/mm2. The dosage for each lipid control was the amount of lipids that equalled the amount of the drug/lipid mixture. As lipid multilayers on certain surfaces tend to spread in humid air or under water29, we found the reliability of the calibration process to depend on using a surface that sufficiently arrested spreading of the lipid multilayers before and during AFM imaging. The surface also had to be compatible with cell culture. Poly-D-lysine coated optical glass-bottom dishes were found suitable for this purpose. We tried multiple surfaces for this experiment (data not shown), including tissue culture polystyrene dishes, untreated glass slides, and plasma treated glass surfaces, and found the poly-D-lysine coated surface worked best in preventing lipid spreading in air. The fidelity of the calibration was tested by applying it to new samples not used in the calibration and comparing the calculated heights to the AFM-measured heights. The average deviation of the calibrated heights from the AFM-measured heights was 15%. We attribute this variation to some minimum, unavoidable spreading during the process of AFM imaging where humidity could not be controlled.
Figure 2.
Multilayers with subcellular lateral dimensions are necessary for surface supported lipid multilayer delivery to HeLa cells. (a) Schematic showing the process of nanointaglio printing. (b) and (c) Lipid multilayer blob and a spot with subcellular dots, respectively. (d) and (e) Cell survival populations when cultured on (b) and (c), respectively. (f) The graph shows a significant difference between the cell survival on blobs and the negative control with no lipids (p = 0.05, students t-test). After 12 replicate experiments, no significant difference was observed between the cell survival on the nanointaglio pattern and cell survival on the negative control with no lipids. Images were taken after 12 hrs of cell incubation in culture medium. (g)-(j) Fluorescent confocal images of DAPI stained nuclei, FITC conjugated anti-vinculin (focal adhesions) and TRITC-conjugated phalloidin (actin) for the cells cultured over a control area without lipids further indicating that the cells adhered normally to the nanointaglio patterned surfaces. (k)-(n) Fluorescent confocal images of DAPI stained nuclei, FITC conjugated anti-vinculin (focal adhesions) and TRITC-conjugated phalloidin (actin) for the cells cultured over the nanointaglio printed subcellular lipid multilayers.
Figure 3.
AFM fluorescent calibration of lipid multilayers fabricated using nanointaglio. (a) Fluorescent micrograph of a spot printed using the nanointaglio method. (b) Magnified section of (a) indicated by white square in (a).(c) Magnified fluuorescence image of sample lipid multilayer area used for AFM-optical calibration, indicated by white square in (b). (d) AFM profile of sample drug/lipid multilayer print.Insert is a section analysis of indicated area showing height range of 225-300 nm. (e) Plot of fluorescence intensity by exposure time, to determine sensitivity for a sample height of 420 nm. The graph in (e) was obtained from different image than shown in (a), (b), (c) and (d). (f) Calibration curve obtained by plotting the sensitivities or slopes of a line fitted to plots of fluorescence intensity versus exposure time for a variety of heights against their AFM measured heights.
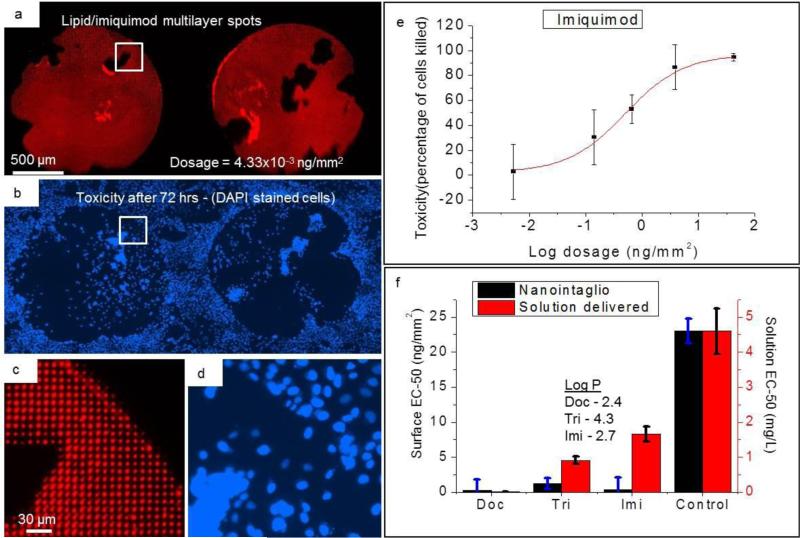
To test the dose-dependent delivery capability of nanointaglio, we chose three hydrophobic drugs, docetaxel, imiquimod, and triethylenemelamine with octanol water partition coefficients (LogP) of 2.4, 4.3, and 2.734, respectively. Non uniform ink deposition onto the stamp, for instance by coffee ring effects associated with drying liquids, is dealt with in two ways. First, sacrificial printing steps remove excess ink from the stamp. Second, areas of the stamp that don't contain enough ink to print are not counted in the dosage calculation, as described in more detail in the methods section. Lipid-multilayer-encapsulated drugs and neat lipid controls were printed by nanointaglio to obtain several different dosages. Figure 4(a) shows the fluorescently doped lipid-multialyer-encapsulated imiquimod. After printing, cells were then cultured over the multilayers for 72 hrs and subsequently stained with DAPI, then counted to determine the toxicity of the drugs to the cells. Figure 4(b) shows the toxicity after 72 hrs. Figures 4(c) and 4(d) show zoomed in images of the respective areas indicated in 4(a) and 4(a). Dose-response curves such as that shown in Figure 4(e) were generated from the patterns, and from these the EC-50 values were obtained from fits to a signmoidal dose-response function. Panel (f) of figure 4 shows the EC-50 values generated from both nanointaglio and solution normalized to the nanointaglio negative control with no lipids.
Figure 4.
Comparison between nanointaglio and solution dose response. (a) Fluorescence image of lipid multilayer spots with encapsulated imiquimod. (b) Toxicity of lipid/imiquimod multilayers to cells. Cells die over area patterned with imiquimod after 72 hrs. Cell nuclei stained with DAPI blue. (c) Magnified fluorescence image of area indicated by white box in (a) showing the subcellular lateral dimensions of the lipid/imiquimod multilayers (d) Magnified fluorescence image of area indicated by white box in (b) showing DAPI-stained nuclei of cells that remain adhered to the surface after multiple rinses. (e) Dose-response curve of imiquimod obtained from the nanointaglio pattern. Error bars are standard deviation from 8 replicates. (f) Comparison between nanointaglio and solution-delivered EC-50 values for docetaxel (Doc), triethylenemelamine (Tri), and imiquimod (Imi). The values are normalized to the nanointaglio negative control (with no drugs). Experiments were done with 8 replicates.
Solution delivery data is shown in Supplementary Figure 2 available as ESI. To establish the quantiative dose-respone microarray assay, the inks were deposited onto the palette array by hand with a spot sizes of approximately 1.5 mm in diameter. Five dosages were used, and 8 replicate spots were made for each dosage, so that the total area used for each dose-response curve for a single drug was 70 mm2. Once the assay is established, it should be scalable using a standard microarrayer to print smaller spots of 200 μm diameter onto a palette24 to allow us to obtain more than 10,000 EC-50 values from the area of a microtiter plate.
The EC-50 values obtained for the drug microarrays differed significantly from their controls without drugs, an expected result as they are all FDA approved anti-cancer drugs. The EC-50 values of docetaxel and imiquimod were each 2 orders of magnitude lower than the lipid control whereas triethylenemelamine was one order of magnitude lower than the lipid control. The control used in the microarray was the amount of lipid that equalled the mass of lipid/drug mixture for each dosage. Docetaxel produced the highest toxicity of the three microarrayed drugs, a result which we found to be consistent with other toxicity studies involving the drugs we used here35.
Quantitative comparison between the toxicities of the microarray and solution based delivery methods showed some differences. While docetaxel appeared less toxic when delivered from the microarray than from solution, triethylenemelamine and imiquimod were more toxic from the microarrays. We speculate that a difference in the adhesion of the cells may be responsible for this variation in cytotoxicity. Docetaxel kills cells by preventing mitotic cell division, yet this compound could also be expected to interfere with cytoskeletal remodelling during adhesion. Since adhesion typically occurs before cell division, docetaxel-induced delays in adhesion on the microarray might lead to delays in cell division and hence a higher EC-50 in the microarray assay than for solution delivery to already adhered cells.
At the patient level, there is a need to identify drug combinations and dosages that maximize efficacy on an individual basis. One example of this is the Feedback System Control II (FSC.II system) that provides a reproducible way to phenotypically identify a safe therapeutic window maximum for drugs, together with the combinations and dosages that perform more efficaciously than other randomly-sampled mixtures36. The quantitative, small-molecule microarray-based assay shown here for the first time has the unique potential for ex vivo testing of primary patient cells. Although here we tested it with a model system, we have taken the first steps towards a portable small-molecule assay capable of screening multiple drug combinations. Nanointaglio presents the opportunity of making dose-dependent high-throughput screening a benchtop process achievable in many research and clinical laboratories. The combination of nanointaglio and microarray technology to miniaturize HTS means that minimal quantities of cells will be required for assays, and the arrays can be portable. This makes the technology promising for applications in personalized medicine where limited amounts of patients’ primary cells are available for single or combinatorial drug testing, and where patient-specific efficacy is highly desired.
Conclusions
We have developed a method of generating quantitative dose-response curves from microarrays of liposomal small molecules. This will allow for the incorporation of a dose-response aspect in the microarray process, while also creating an avenue for optically monitoring the effects of other dosages not previously accessible. The combination of pinspotting microarray technology with nanointaglio will provide a platform to perform this screen for a larger number of drugs simultaneously. In addition, this method opens the door for potential applications in personalized medicine, where a minimal quantity of pimary cells is available for ex vivo assays to determine optimal treatment.
Supplementary Material
Acknowledgements
The authors thank the National Cancer Institute (NCI)/National Institutes of Health Developmental Therapeutics Program for the Approved Oncology Drug Set II for the panel of drugs, Diego Zorio for providing the docetaxel, Jingjiao Guan providing the polymer stamps, and Jen Kennedy for proofreading. This work was supported by NIH R01 GM107172.
Footnotes
† Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI:10.1039/x0xx00000x
Notes and references
- 1.Jordan MA. Current Medicinal Chemistry - Anti-Cancer Agents. 2002;2:1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 2.Jimenez-Barbero J, Amat-Guerri F, Snyder JP. Current Medicinal Chemistry - Anti-Cancer Agents. 2002;2:91–122. doi: 10.2174/1568011023354416. [DOI] [PubMed] [Google Scholar]
- 3.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Nat. Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 4.Bibette J. Proc. Natl. Acad. Sci. U. S. A. 2012;109:649–650. doi: 10.1073/pnas.1119350109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ru Zang DL, Tang I-Ching. Jufang Wang and Shang-Tian Yang, International Journal of Biotechnology for Wellness Industries. 2012;1:31–51. [Google Scholar]
- 6.Goktug AN, Chai SC, Chen T. Data Analysis Approaches in High Throughput Screening. 2013 [Google Scholar]
- 7.Angres B. Expert Rev. Mol. Diagn. 2005;5:769–779. doi: 10.1586/14737159.5.5.769. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Mochon JJ, Tourniaire G, Bradley M. Chem. Soc. Rev. 2007;36:449–457. doi: 10.1039/b511848b. [DOI] [PubMed] [Google Scholar]
- 9.Uttamchandani M, Walsh DP, Yao SQ, Chang YT. Curr Opin Chem Biol. 2005;9:4–13. doi: 10.1016/j.cbpa.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Bailey SN, Sabatini DM, Stockwell BR. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16144–16149. doi: 10.1073/pnas.0404425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arrabito G, Reisewitz S, Dehmelt L, Bastiaens PI, Pignataro B, Schroeder H, Niemeyer CM. Small. 2013;9:4243–4249. doi: 10.1002/smll.201300941. [DOI] [PubMed] [Google Scholar]
- 12.Arrabito G, Schroeder H, Schroder K, Filips C, Marggraf U, Dopp C, Venkatachalapathy M, Dehmelt L, Bastiaens PIH, Neyer A, Niemeyer CM. Small. 2014;10:2870–2876. doi: 10.1002/smll.201303390. [DOI] [PubMed] [Google Scholar]
- 13.MacBeath G, Koehler AN, Schreiber SL. J. Am. Chem. Soc. 1999;121:7967–7968. [Google Scholar]
- 14.Tavana H, Jovic A, Mosadegh B, Lee QY, Liu X, Luker KE, Luker GD, Weiss SJ, Takayama S. Nat. Mater. 2009;8:736–741. doi: 10.1038/nmat2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong JA, Neel DV, Wassaf D, Caballero F, Koehler AN. Curr. Opin. Chem. Biol. 2014;18:21–28. doi: 10.1016/j.cbpa.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arrabito G, Galati C, Castellano S, Pignataro B. Lab Chip. 2013;13:68–72. doi: 10.1039/c2lc40948h. [DOI] [PubMed] [Google Scholar]
- 17.Falsey JR, Renil M, Park S, Li SJ, Lam KS. Bioconjugate Chem. 2001;12:346–353. doi: 10.1021/bc000141q. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Uttamehandani M, Sun HY, Yao SQ. QSAR Comb. Sci. 2006;25:1009–1019. [Google Scholar]
- 19.Wu JH, Wheeldon I, Guo YQ, Lu TL, Du YN, Wang B, He JK, Hu YQ, Khademhosseini A. Biomaterials. 2011;32:841–848. doi: 10.1016/j.biomaterials.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuegg J, Cooper MA. Curr. Top. Med. Chem. 2012;12:1500–1513. doi: 10.2174/156802612802652466. [DOI] [PubMed] [Google Scholar]
- 21.Kusi-Appiah AE, Vafai N, Cranfill PJ, Davidson MW, Lenhert S. Biomaterials. 2012;33:4187–4194. doi: 10.1016/j.biomaterials.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Wang YH, Giam LR, Park M, Lenhert S, Fuchs H, Mirkin CA. Small. 2008;4:1666–1670. doi: 10.1002/smll.200800770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenhert S, Sun P, Wang YH, Fuchs H, Mirkin CA. Small. 2007;3:71–75. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- 24.Lowry TW, Kusi-Appiah A, Guan JJ, Van Winkle DH, Davidson MW, Lenhert S. Adv. Mater. Interfaces. 2014:1. doi: 10.1002/admi.201300127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nafday OA, Lowry TW, Lenhert S. Small. 2012;8:1021–1028. doi: 10.1002/smll.201102096. [DOI] [PubMed] [Google Scholar]
- 26.Nafday OA, Lenhert S. Nanotechnology. 2011:22. doi: 10.1088/0957-4484/22/22/225301. [DOI] [PubMed] [Google Scholar]
- 27.Hariri H, Bhattacharya N, Johnson K, Noble AJ, Stagg SM. J. Mol. Biol. 2014;426:3811–3826. doi: 10.1016/j.jmb.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandyopadhyay P, Kren BT, Ma XM, Steer CJ. Biotechniques. 1998;25:282. doi: 10.2144/98252gt03. [DOI] [PubMed] [Google Scholar]
- 29.Lenhert S, Brinkmann F, Laue T, Walheim S, Vannahme C, Klinkhammer S, Xu M, Sekula S, Mappes T, Schimmel T, Fuchs H. Nat. Nanotechnol. 2010;5:275–279. doi: 10.1038/nnano.2010.17. [DOI] [PubMed] [Google Scholar]
- 30.Culver-Cochran AE, Chadwick BP. Plos One. 2012:7. doi: 10.1371/journal.pone.0050023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersson AS, Glasmastar K, Sutherland D, Lidberg U, Kasemo B. Journal of Biomedical Materials Research Part A. 2003;64A:622–629. doi: 10.1002/jbm.a.10442. [DOI] [PubMed] [Google Scholar]
- 32.Groves JT, Dustin ML. J. Immunol. Methods. 2003;278:19–32. doi: 10.1016/s0022-1759(03)00193-5. [DOI] [PubMed] [Google Scholar]
- 33.Kam L, Boxer SG. J. Biomed. Mater. Res. 2001;55:487–495. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 34.DrugBank [04-24-2015]; http://www.drugbank.ca/drugs/DB00724 (2015)
- 35.Hoeksema KA, Jayanthan A, Cooper T, Gore L, Trippett T, Boklan J, Arceci RJ, Narendran A. OncoTargets Ther. 2011;4:149–168. doi: 10.2147/OTT.S21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Lee D-K, Chen K-Y, Chen J-Y, Zhang K, Silva A, Ho C-M, Ho D. ACS nano. 2015;9:3332–3344. doi: 10.1021/acsnano.5b00638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.