Abstract
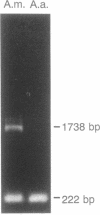
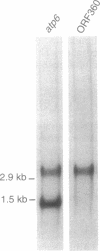
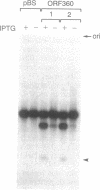
In eukaryotes, horizontal gene transfer is a rare event. Here we show that the mitochondrial genome of a lower fungus, Allomyces macrogynus, has an extra DNA segment not present in a close relative, Allomyces arbusculus. This insert consists of the C terminus of a foreign gene encoding a subunit of the ATP synthetase complex (atp6) plus an open reading frame encoding an endonuclease. The inserted atp6 portion is fused in phase to the resident gene, resulting in expression of a hybrid atp6 gene and the displacement of the original C-terminal atp6 region. We present evidence that this insertion may have been acquired by interspecific transfer and we discuss the possible role of the endonuclease in this process.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K. E., Hoess R. H. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 1992 Jan;5(1):87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Quirk S., Clyman J., Belfort M. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 1990 Jul 11;18(13):3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Werner S. The mitochondrial URF1 gene in Neurospora crassa has an intron that contains a novel type of URF. J Mol Biol. 1985 Nov 20;186(2):231–242. doi: 10.1016/0022-2836(85)90100-7. [DOI] [PubMed] [Google Scholar]
- Campbell A., Schneider S. J., Song B. Lambdoid phages as elements of bacterial genomes (integrase/phage21/Escherichia coli K-12/icd gene). Genetica. 1992;86(1-3):259–267. doi: 10.1007/BF00133724. [DOI] [PubMed] [Google Scholar]
- Chu F. K., Maley G. F., West D. K., Belfort M., Maley F. Characterization of the intron in the phage T4 thymidylate synthase gene and evidence for its self-excision from the primary transcript. Cell. 1986 Apr 25;45(2):157–166. doi: 10.1016/0092-8674(86)90379-x. [DOI] [PubMed] [Google Scholar]
- Collins R. A., Saville B. J. Independent transfer of mitochondrial chromosomes and plasmids during unstable vegetative fusion in Neurospora. Nature. 1990 May 10;345(6271):177–179. doi: 10.1038/345177a0. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988 Nov 25;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Nash H. A. The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell. 1983 Dec;35(3 Pt 2):795–803. doi: 10.1016/0092-8674(83)90112-5. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Domenico J. M. Sequence analysis of mitochondrial DNA from Podospora anserina. Pervasiveness of a class I intron in three separate genes. J Mol Biol. 1988 Dec 20;204(4):815–839. doi: 10.1016/0022-2836(88)90044-7. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Michel F., McNally K. L. DNA sequence analysis of the apocytochrome b gene of Podospora anserina: a new family of intronic open reading frame. Curr Genet. 1989 Dec;16(5-6):407–418. doi: 10.1007/BF00340720. [DOI] [PubMed] [Google Scholar]
- Dalgaard J. Z., Garrett R. A., Belfort M. A site-specific endonuclease encoded by a typical archaeal intron. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5414–5417. doi: 10.1073/pnas.90.12.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeley R. G., Gordon J. I., Burns A. T., Mullinix K. P., Binastein M., Goldberg R. F. Primary activation of the vitellogenin gene in the rooster. J Biol Chem. 1977 Nov 25;252(22):8310–8319. [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Anderson K. L., Alberro M. R. A naturally occurring horizontal gene transfer from a eukaryote to a prokaryote. J Mol Evol. 1990 Nov;31(5):383–388. doi: 10.1007/BF02106053. [DOI] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations--a review. Gene. 1989 Oct 15;82(1):91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Dürrenberger F., Rochaix J. D. Characterization of the cleavage site and the recognition sequence of the I-CreI DNA endonuclease encoded by the chloroplast ribosomal intron of Chlamydomonas reinhardtii. Mol Gen Genet. 1993 Jan;236(2-3):409–414. doi: 10.1007/BF00277141. [DOI] [PubMed] [Google Scholar]
- Eddy S. R., Gold L. Artificial mobile DNA element constructed from the EcoRI endonuclease gene. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1544–1547. doi: 10.1073/pnas.89.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble F. S., Thorner J. Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature. 1992 May 28;357(6376):301–306. doi: 10.1038/357301a0. [DOI] [PubMed] [Google Scholar]
- Hardy C. M., Clark-Walker G. D. Nucleotide sequence of the COX1 gene in Kluyveromyces lactis mitochondrial DNA: evidence for recent horizontal transfer of a group II intron. Curr Genet. 1991 Jul;20(1-2):99–114. doi: 10.1007/BF00312772. [DOI] [PubMed] [Google Scholar]
- Heinemann J. A. Genetics of gene transfer between species. Trends Genet. 1991 Jun;7(6):181–185. doi: 10.1016/0168-9525(91)90433-q. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Hill C. W., Gray J. A., Brody H. Use of the isocitrate dehydrogenase structural gene for attachment of e14 in Escherichia coli K-12. J Bacteriol. 1989 Jul;171(7):4083–4084. doi: 10.1128/jb.171.7.4083-4084.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell M. G. Horizontal transfer. Curr Opin Genet Dev. 1992 Dec;2(6):868–873. doi: 10.1016/s0959-437x(05)80109-1. [DOI] [PubMed] [Google Scholar]
- Kostriken R., Strathern J. N., Klar A. J., Hicks J. B., Heffron F. A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell. 1983 Nov;35(1):167–174. doi: 10.1016/0092-8674(83)90219-2. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M., Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- Lang B. F. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe: highly homologous introns are inserted at the same position of the otherwise less conserved cox1 genes in Schizosaccharomyces pombe and Aspergillus nidulans. EMBO J. 1984 Sep;3(9):2129–2136. doi: 10.1002/j.1460-2075.1984.tb02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May G., Taylor J. W. Independent transfer of mitochondrial plasmids in Neurospora crassa. Nature. 1989 May 25;339(6222):320–322. doi: 10.1038/339320a0. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Genetic exchanges between bacteriophage T4 and filamentous fungi? Cell. 1986 Aug 1;46(3):323–323. doi: 10.1016/0092-8674(86)90651-3. [DOI] [PubMed] [Google Scholar]
- Quirk S. M., Bell-Pedersen D., Belfort M. Intron mobility in the T-even phages: high frequency inheritance of group I introns promoted by intron open reading frames. Cell. 1989 Feb 10;56(3):455–465. doi: 10.1016/0092-8674(89)90248-1. [DOI] [PubMed] [Google Scholar]
- Sharma M., Ellis R. L., Hinton D. M. Identification of a family of bacteriophage T4 genes encoding proteins similar to those present in group I introns of fungi and phage. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6658–6662. doi: 10.1073/pnas.89.14.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shub D. A., Goodrich-Blair H. Protein introns: a new home for endonucleases. Cell. 1992 Oct 16;71(2):183–186. doi: 10.1016/0092-8674(92)90345-d. [DOI] [PubMed] [Google Scholar]
- Smith J. M., Dowson C. G., Spratt B. G. Localized sex in bacteria. Nature. 1991 Jan 3;349(6304):29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- Tian G. L., Michel F., Macadre C., Slonimski P. P., Lazowska J. Incipient mitochondrial evolution in yeasts. II. The complete sequence of the gene coding for cytochrome b in Saccharomyces douglasii reveals the presence of both new and conserved introns and discloses major differences in the fixation of mutations in evolution. J Mol Biol. 1991 Apr 20;218(4):747–760. doi: 10.1016/0022-2836(91)90263-6. [DOI] [PubMed] [Google Scholar]