Abstract
The emergence of New Delhi metallo-β-lactamase 1 (NDM-1) has become established as a major public health threat and represents a new challenge in the treatment of infectious diseases. In this study, we report a high incidence and endemic spread of NDM-1-producing carbapenem-resistant Enterobacter cloacae isolates in Henan province, China. Eight (72.7%) out of eleven non-duplicated carbapenem-resistant E. cloacae isolates collected between June 2011 and May 2013 were identified as NDM-1 positive. The bla NDM-1 gene surrounded by an entire ISAba125 element and a bleomycin resistance gene ble MBL in these isolates were carried by diverse conjugatable plasmids (IncA/C, IncN, IncHI2 and untypeable) ranging from ~55 to ~360 kb. Molecular epidemiology analysis revealed that three NDM-1-producing E. cloacae belonged to the same multilocus sequence type (ST), ST120, two of which were classified as extensively drug-resistant (XDR) isolates susceptible only to tigecycline and colistin. The two XDR ST120 E. cloacae isolates co-harbored bla NDM-1, armA and fosA3 genes and could transfer resistance to carbapenems, fosfomycin and aminoglycosides simultaneously via a conjugation experiment. Our study demonstrated NDM-1 was the most prevalent metallo-β-lactamase (MBL) among carbapenem-resistant E.cloacae isolates and identified a potential endemic clone of ST120 in Henan province. These findings highlight the need for enhanced efforts to monitor the further spread of NDM-1 and XDR ST120 E. cloacae in this region.
Introduction
Enterobacter cloacae (E. cloacae) is an important nosocomial pathogen causing various infections including urinary tract, skin and soft tissue, lower respiratory tract, wounds, biliary tract, intravenous catheters and central nervous system and intrinsically resistant to ampicillin and narrow-spectrum cephalosporins owing to chromosomal cephalosporinase[1]. Recently, a new antibiotic named Teixobactin was reported to have excellent activity against Gram-positive pathogens without detectable resistance. However, this agent was ineffective against most Gram-negative bacteria containing Enterobacteriaceae (Escherichia coli: Teixobactin MIC = 25μg/ml; Klebsiella pneumoniae: Teixobactin MIC > 25μg/ml)[2]. Due to the increase in multiple drug-resistant Gram-negative bacteria, carbapenems have become the last resort antibiotics in treatment of infections caused by these pathogens including E. cloacae. The emergence of resistance to carbapenems, mediated by carbapenemases in clinical Enterobacteriaceae such as E. cloacae isolates represents a serious public health concern worldwide. To date, both metallo-(IMP-8, NDM-1, VIM-1) and non-metallo-(KPC-2) β-lactamases have been reported in carbapenem-resistant E. cloacae[3–6].
New Delhi metallo-β-lactamase 1 (NDM-1), a metallo-β-lactamase (MBL) capable of hydrolyzing all β-lactams but monobactams, was first identified in a carbapenem-resistant Klebsiella pneumoniae strain recovered from a Swedish patient who was hospitalized in India in 2008[7], and mainly detected in carbapenem-resistant Acinetobacter spp. in mainland China[8–10]. Only sporadic reports of NDM-1-producing E. cloacae until the high prevalence and endemic spread of NDM-1-positive Enterobacteriaceae was observed in Henan province, China[11]. Thus, the aim of this study is to investigate the current prevalence and molecular characteristics of the NDM-1-producing E. cloacae in Henan province.
Materials and Methods
Bacterial strains and antibiotic susceptibility testing
A total of 112 non-duplicate E. cloacae clinical isolates were obtained from three hospitals located in the middle [the First Affiliated Hospital of Zhengzhou University (ZZ), n = 69], western [the central hospital of Sanmenxia city (SMX), n = 12], and southern [the central hospital of Zhumadian city (ZMD), n = 31] regions of Henan Province, north-central China from June 2011 to May 2013. Of the 112 isolates tested, 11 isolates (9.8%) (ZZ: n = 7; SMX: n = 1; ZMD: n = 3) were categorized as carbapenem-resistant (Ertapenem, MIC ≥ 2 μg/ml or Imipenem, MIC ≥ 4 μg/ml). All isolates were identified by VITEK2 compact (bioMerieux, France) and 16S rRNA gene sequencing. Antimicrobial susceptibilities for the NDM-1 producing isolates and transconjugants were initially tested using the VITEK2 system and then were followed by measuring the MIC using the broth microdilution method (for imipenem, ertapenem, ciprofloxacin, levofloxacin, gentamicin, amikacin, aztreonam, chloramphenicol and tetracycline), the VITEK2 system (for trimethoprim/sulfamethoxazole, piperacillin/ tazobactam, ceftazidime and cefepime), and the agar dilution method (for fosfomycin), respectively, according to the Clinical Laboratory Standards Institute (CLSI) guidelines(2013). Mueller-Hinton broth (MHB) was used as the test medium in the broth microdilution method, and Mueller-Hinton agar (MHA) containing 25 μg/ml glucose 6-phosphate was used for fosfomycin testing in the agar dilution method. Bacterial suspensions of 0.5 McFarland turbidity for antimicrobial susceptibility testing were prepared by using fresh bacterial colonies taken directly from MHA plates that were incubated at 37°C for 16 to 20 h. Colistin and tigecycline MICs were determined by E-test (AB bioMérieux, France), and results were interpreted as recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2013). E. coli ATCC 25922 was used as quality control strain.
Detection of resistance determinants
All of the carbapenem-resistant E. cloacae isolates were screened for carbapenemase production by using the modified Hodge test and imipenem-EDTA double-disk synergy test according to the CLSI guidelines. PCR and nucleotide sequencing were employed to screen for the presence of carbapenemases encoding genes[12], extended-spectrum-β-lactamase (ESBL) genes, plasmid-mediated AmpC genes, 16S rRNA methyltransferase genes, and fosfomycin resistance determinants [13–17](Table 1).
Table 1. Detection of resistance determinants in the 11 carbapenem-resistant E. cloacae isolates.
| Antimicrobial category | Associated resistance determinants |
|---|---|
| β-lactams | AmpC genes: bla MOX, bla CMY, bla LAT, bla BIL, bla DHA, bla ACC, bla MIR, bla ACT, bla FOX |
| ESBLs genes: bla TEM, bla SHV, bla CTX-M groups 1, 2, 8, 9 and 26 | |
| Carbapenemase genes: bla IMP, bla VIM, bla KPC, bla NDM, bla OXA-1-like | |
| Aminoglycosides | 16S methylase genes: armA, rmtA-E, and npmA |
| Phosphonic acids (Fosfomycin) | fosA, fosB, fosC and fosX, |
Pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST)
PFGE of XbaI-digested (TaKaRa, Japan) genomic DNA of bla NDM-1-positive E. cloacae and reference marker Salmonella serotype Braenderup strain (H9812) was performed using a contour-clamped homogeneous electric field (CHEF)-Mapper XA PFGE system (Bio-Rad, USA) for 22 h at 6 V/cm and 14°C, with a pulse angle of 120° and pulse times from 5 to 35 s. Comparison of the PFGE patterns was performed with InfoQuest FP software version 4.5 (Bio-Rad Laboratories, USA) using the Dice similarity coefficient. Clusters were defined as DNA patterns sharing >85% similarity. MLST was carried out as described previously[18], the database available at http://pubmlst.org/ecloacae was used for assigning STs.
Conjugation Experiments
The transfer of carbapenem resistance was tested using a conjugation test (broth mating method), E. coli J53(sodium azide resistant) was used as the recipient strain. Transconjugants were selected on Mueller-Hinton agar containing sodium azide (100 μg/ml) and imipenem (1μg/ml). The presence of the bla NDM-1 gene and other resistance determinants according to phenotype in transconjugants were determined by using PCR and sequencing.
Plasmid analysis and genetic environment of the bla NDM-1 gene
Plasmid analysis was performed as described previously[19]. Briefly, Genomic DNA was digested with S1 nuclease (TaKaRa, Japan) and separated by PFGE as above, but with a switch time from 2.16 to 63.8 s for 18 h run time. Then, the DNA fragments were transferred to nylon membranes (Millipore, USA), hybridized with digoxigenin-labelled bla NDM-1-specific probe and detected using a nitroblue tetrazolium-5-bromo-4-chloro -3-indolylphosphate (NBT/BCIP) colour detection kit (Roche Applied Sciences, Germany). The genetic context of the bla NDM-1 gene was investigated by PCR mapping and subsequent sequencing, the primers were used as described previously[11].
Results
Detection of bla NDM-1 positive isolates
Eight out of the eleven (72.7%) non-duplicate carbapenem-resistant E. cloacae isolates, were identified as bla NDM-1 positive, which were obtained from blood (n = 3), urine (n = 2), sputum (n = 2) and wound (n = 1) specimens. Additionally, 2 isolates were IMP-4 positive, and 1 isolate did not contain the carbapenemase genes (bla NDM, bla KPC, bla VIM, bla IMP, and bla OXA-48-like) screened in this study. The 8 bla NDM-1-positive E. cloacae were obtained from two hospitals in two different cities in Henan Province, including the First Affiliated Hospital of Zhengzhou University (n = 6) and the central hospital of Zhumadian city (n = 2). The clinical data of the 8 isolates were summarized in Table 2. These isolates were collected from 8 individual patients, consisting of 5 male (52.5%) and 3 female (37.5%) with a mean age of 29.7 years, including 2 infants(ECL-2, ECL-36). Of note, 3 patients (37.5%) died of infections.
Table 2. Characteristics of bla NDM-1-positive E. cloacae.
| Isolate | Clinical features | STs a | Associated resistance determinants b | Plasmid type carrying bla NDM-1/ Plasmid size (kb) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Age/Sex | Specimen | Ward | Outcome | β-lactamases | 16SrRNA methylase | Others | |||
| ECL-2 | 7m/female | sputum | Cardial Surgery | discharge | ST177 | TEM-1、EBC、CMY-2、CTX-M-1 | RmtB | - | Untypeable/70 |
| ECL-4 | 48y/male | blood | ICU | discharge | ST88 | TEM-1、ACT-20、CTX-M-3 | - | - | N/65 |
| ECL-27 | 57y/male | blood | ICU | discharge | ST90 | ACT-20、CTX-M-G9 | - | - | Untypeable/55 |
| ECL-36 | 15d/male | sputum | NICU | death | ST41 | MIR-2 | - | - | A/C/160 |
| ECL-37 | 37y/male | urine | Urology | discharge | ST120 | ACT-20、CTX-M-3 | - | - | Untypeable/55 |
| ECL- 62 | 25y/female | urine | Neurosurgery | death | ST120 | ACT-20、CTX-M-15 | ArmA | fosA3 | HI2/340 |
| ECL- ZMD10 | 49y/male | wound | Burn unit | discharge | ST120 | - | ArmA | fosA3 | Untypeable/360 |
| ECL- ZMD12 | 21y/female | blood | Hematology | death | ST93 | - | ArmA | - | A/C/55 |
a ST: Sequence type determined by multilocus sequence typing (MLST)
b Resistance markers that are co-transferred with bla NDM-1by conjugation are underlined. Minus signs indicate negative results.
Antimicrobial susceptibility testing and detection of resistance genes
All of the bla NDM-1 carrying isolates were resistant to carbapenems, cephalosporins, monobactams (aztreonam), β-lactam/β-lactamase inhibitor combinations, trimethoprim /sulfamethoxazole, but susceptible to colistin (MICs ≤ 2 μg/ml), and 5 out of 8 isolates (62.5%) exhibited resistance against tigecycline according to the EUCAST breakpoint, with MICs of ≥ 2 μg/ml (Table 3). The modified Hodge test and imipenem-EDTA double-disk synergy test yielded positive results for all isolates. PCR and sequencing results showed most of the bla NDM-1-carrying E. cloacae isolates (6/8, 75%) harbored ESBL genes (bla TEM-1, bla CTX-M-3, bla CTX-M-9, bla CTX-M-15), AmpC genes (bla ACT-20, bla CMY-2, bla MIR-2), or both. Other carbapenemase-encoding genes, including bla KPC, bla VIM, bla IMP, and bla OXA-48-like, were not detected in any of the bla NDM-1-positive isolates. Moreover, 4 isolates (50%) harbored 16S methylase genes (armA or rmtB), exhibited high-level resistance to amikacin (MIC > 256 μg/ml), and 2 isolates (25%) carried a plasmid-mediated fosfomycin resistance gene, fosA3 (Table 2).
Table 3. Antibiotic susceptibilities of bla NDM-1-positive E. cloacae and transconjugants (μg/mL).
| Isolate no. a | Antibiotics b | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TZP | CAZ | FEP | IPM | ETP | CIP | LEV | GEN | AMK | SXT | ATM | CHL | TET | FOS | TGC | CST | |
| ECL-2 | >256 | >256 | >256 | 32 | >32 | 1 | 1 | >256 | >256 | >320 | >256 | 64 | >256 | 32 | 2 | 0.5 |
| ECL-4 | >256 | >256 | >256 | 64 | >32 | 16 | >32 | 8 | 16 | >320 | >256 | 32 | 128 | 64 | 16 | 0.5 |
| ECL-27 | >256 | >256 | >256 | >64 | >32 | >32 | 16 | 64 | 2 | >320 | >256 | 64 | >256 | 16 | 3 | 1 |
| ECL-36 | >256 | >256 | >256 | 32 | >32 | <0.25 | <0.25 | 32 | <2 | >320 | 256 | 8 | 4 | 8 | 2 | 1 |
| ECL-37 | >256 | >256 | >256 | 16 | 32 | 16 | >32 | >256 | >256 | >320 | >256 | 32 | 128 | 64 | 3 | 1 |
| ECL-62 | >256 | >256 | >256 | 64 | 32 | 16 | 32 | >256 | >256 | >320 | >256 | 256 | >256 | >512 | 4 | 1 |
| ECL-ZMD10 | 64 | >256 | >256 | 8 | 32 | >32 | >32 | >256 | >256 | >320 | 256 | 256 | 256 | 128 | 1 | 1 |
| ECL-ZMD12 | >256 | >256 | >256 | 8 | 32 | >32 | >32 | >256 | >256 | >320 | >256 | 256 | 256 | 32 | 3 | 2 |
| E. coli Transconjugant Strains | ||||||||||||||||
| ECL-2-J53 | >256 | >256 | >256 | 32 | 32 | 1 | 0.5 | 64 | 64 | >320 | 128 | 8 | 64 | 16 | 1 | 0.5 |
| ECL-4-J53 | >256 | >256 | >256 | 32 | 32 | 16 | 32 | 2 | 8 | >320 | 128 | 8 | 32 | 16 | 4 | 0.5 |
| ECL-27-J53 | >256 | >256 | >256 | 32 | >32 | 16 | 8 | 16 | 2 | >320 | >256 | 32 | 64 | 8 | 1.5 | 1 |
| ECL-36-J53 | 64 | >256 | >256 | 32 | >32 | <0.25 | <0.25 | 32 | <2 | >320 | <1 | 8 | 8 | 8 | 1.5 | 1 |
| ECL-37-J53 | >256 | >256 | >256 | 16 | 32 | 16 | 32 | 16 | >64 | >320 | >256 | 16 | 128 | 32 | 1.5 | 1 |
| ECL-62-J53 | >256 | >256 | >256 | 32 | 16 | 16 | 32 | 64 | >64 | >320 | >256 | 32 | 128 | >512 | 2 | 1 |
| ECL-ZMD10-J53 | 64 | >256 | >256 | 4 | 16 | 32 | 16 | >256 | >256 | >320 | 128 | 256 | 256 | 128 | 1 | 0.5 |
| ECL-ZMD12-J53 | >256 | >256 | >256 | 8 | 32 | 32 | 32 | >256 | 16 | <20 | >256 | 256 | 256 | 32 | 3 | 1 |
| EC J53 | <4 | <1 | <1 | <1 | <0.5 | <0.25 | <0.25 | <1 | <2 | <20 | <1 | 8 | 2 | 2 | 0.25 | 0.5 |
a ECL, E. cloacae strains; For the transconjugants, all were E. coli J53 harboring plasmids from the respective clinical isolates. All of the bla NDM-1-positive isolates were multidrug-resistant (MDR) strains, the XDR isolates are highlighted in bold type.
b Abbreviations used:TZP, piperacillin/tazobactam (0.5/4-256/4); CAZ, ceftazidime (0.03–256); FEP, cefepime (0.015–256); IPM, imipenem(0.06–64); ETP, ertapenem(0.004–32); CIP, ciprofloxacin (0.004–32); LEV, levofloxacin (0.008–32); GEN, gentamicin (0.25–256); AMK, amikacin (0.5–256); ATM, aztreonam (0.06–256); CHL, chloroamphenicol (0.016–256); TET, tetracycline (0.016–256); FOS, fosfomycin (0.25–512); TGC, tigecycline (0.016–256); CST, colistin (0.016–256). The numbers in parentheses indicate the test range (μg/mL) for each agent.
Molecular epidemiology
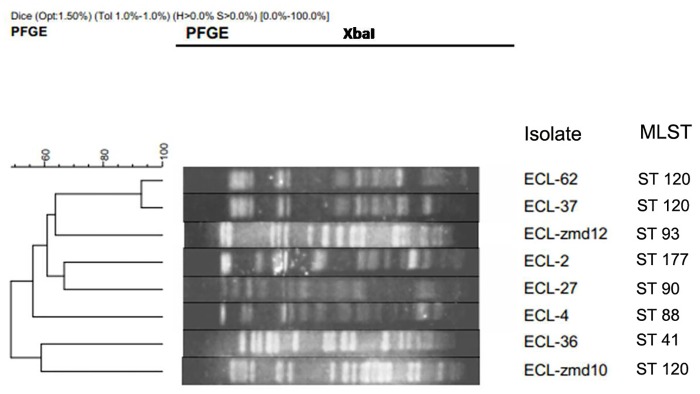
Based on a cutoff of 90% genetic similarity, seven PFGE subtypes were identified among the eight isolates. The linkage between PFGE subtype and MLST type was shown in Fig 1. Two isolates obtained from two different wards of the same hospital shared the same PFGE pattern, suggesting they were clonally related, the remaining strains were characterized by unique genotypes. MLST typing revealed 6 STs (ST120[n = 3], ST93[n = 1], ST177[n = 1], ST90[n = 1], ST88[n = 1], and ST41[n = 1]), and 3 isolates belonged to ST120, which were obtained from two different hospitals located in geographically separated areas (the First Affiliated Hospital of Zhengzhou University and the central hospital of Zhumadian city).
Fig 1. Dendrogram showing pulsed-field gel electrophoresis (PFGE) analysis and multilocus sequence typing (MLST) results for 8 bla NDM-1-positive E. cloacae isolates.
Plasmid analysis and flanking regions of the bla NDM-1 gene
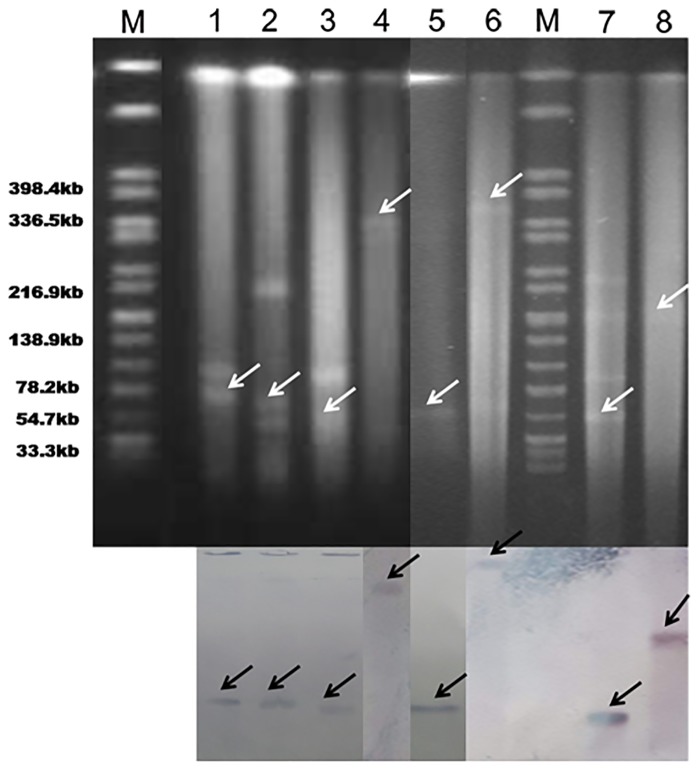
Conjugation experiments revealed that all of the NDM-1 plasmids were successfully transferred to E. coli J53, conferring resistance to carbapenems and cephalosporins in transconjugants. In addition, co-transfer of bla NDM-1 and other resistance determinants (bla TEM-1, bla CTX-M-3,15/G9, bla ACT-20, and fosA3) was observed in several isolates (Table 3). The analysis of PFGE profiles of S1 nuclease-digested genomic DNA and Southern blot hybridization showed that bla NDM-1 was located on diverse plasmids with sizes from~ 55 to~ 360 kb (Fig 2). The NDM-1-encoding plasmids belonged to different plasmid replicon types including IncA/C (n = 2), IncHI2 (n = 1), IncN (n = 1), and untypeable (n = 4) (Table 2 and Fig 2). PCR mapping and sequencing revealed that the entire ISAba125 element was located upstream of bla NDM-1 and that the bleomycin resistance gene ble MBL and truncated trpF gene encoding the phosphoribosylanthranilate isomerase were located immediately downstream of the bla NDM-1 gene in all of the 8 isolates (S1 Fig).
Fig 2. Detection of bla NDM-1 carrying plasmids by S1 nuclease PFGE and Southern hybridization.
Lanes M, marker (Salmonella H9812); Lane 1, ECL-2; Lane 2, ECL-4; Lane 3, ECL-37; Lane 4, ECL-62; Lane 5, ECL-27; Lane 6, ECL-ZMD10; Lane 7, ECL-ZMD12; Lane 8, ECL-36.
Discussion
In China, NDM-1 was commonly identified in Acinetobacter spp. isolated from clinical, environmental and farm animal samples but only reported sporadically in Enterobacteriaceae[8,10,20]. Our recent study demonstrated the prevalence of NDM-1 among carbapenem-resistant Enterobacteriaceae (CRE) in Henan province with an incidence of 33.3% and revealed new molecular epidemiological characteristics of CRE in China[11]. As a continued investigation, a pretty high proportion (8/11, 72.7%) of bla NDM-1 positive strains was identified among carbapenem-resistant E. cloacae isolates in this study, indicating NDM-1 was the dominant MBL as a mechanism of resistance to carbapenems in E. cloacae isolates in this region. By contrast, reports from Spain and other southern Europe countries revealed that VIM-1 was the most prevalent MBL among the carbapenem-resistant E. cloacae[21]. The prevalence rate of carbapenem-resistant E. cloacae in each hospital (ZZ: 10.1%, 7/69; SMX: 8.3%, 1/12; ZMD: 9.7%, 3/31) in our study was higher than that reported in Spain (5.1%). In addition, a conjugative IncHI2 plasmid of 300 kb plays an important role in dissemination of bla VIM-1 among different E. cloacae clones[21], however, NDM-1 plasmids identified in carbapenem-resistant E. cloacae isolates in this study belonged to multiple replicon types and with various sizes. Observations above demonstrate the importance of the local epidemiological factors in the emergence of specific types of carbapenemases in different regions.
In our study, ISAba125 was located upstream of the bla NDM-1, while ble MBL and a truncated trpF gene were located downstream of the bla NDM-1 in each E. cloacae isolate, Analysis of the genetic environment of bla NDM-1 revealed that the region flanking bla NDM-1 is very similar to some Acinetobacter spp. isolated in China. Recent studies highlighted the potential of Acinetobacter spp. as a reservoir for the dissemination of NDM-1 towards Enterobacteriaceae[22,23]. Given that bla NDM-1 was mostly detected in Acinetobacter spp. in China, we proposed that the acquisition of bla NDM-1 in E. cloacae may be originally from Acinetobacter spp. under antibiotics selective pressure, and insertion elements may contribute to the spread of bla NDM-1 among E. cloacae isolates.
Besides mobile genetic elements mediated bla NDM-1 transfer, clonal spread is another factor involved in the prevalence of NDM-producing Enterobacteriaceae at local and regional level. Outbreaks of NDM-1-producing Klebsiella pneumoniae ST147 and ST231 have been reported in Xi’an, China and Ontario, Canada, respectively[24,25]. Our study identified a potential prevalent clone of ST120 among the 8 carbapenem-resistant E. cloacae isolates in Henan province. However, this ST was different from some widespread E. cloacae STs (ST66, ST78, ST108 and ST114) that reported in Europe countries, exhibiting expanded-spectrum cephalosporins resistant phenotype [26]. Since limited numbers were obtained, the spread of the ST120 isolates in this region still need to be further monitored. It is noteworthy that two out of the three ST120 isolates (ECL-62 and ECL-ZMD10) were identified as extensively drug-resistant (XDR) bacteria susceptible only to tigecycline and colistin. Moreover, The two XDR ST120 E. cloacae isolates co-harbored bla NDM-1, armA and fosA3 genes and could transfer resistance to carbapenems, fosfomycin and aminoglycosides simultaneously by conjugation. Aminoglycosides (gentamycin, amikacin, tobramycin) and fosfomycin were considered as the most common antibiotics for the treatment of infections due to carbapenemase production[27]. The dissemination of E. cloacae ST120 isolates will seriously limit the future therapeutic options.
In conclusion, our study demonstrated NDM-1 was the most prevalent MBL among carbapenem-resistant E. cloacae isolates in Henan province, and identified a potential endemic clone of ST120. The emergence of XDR E. cloacae ST120 isolates is worrying, early detection and surveillance of NDM-1 producing E. cloacae are urgently needed to prevent their further spread.
Supporting Information
The boxed arrows indicate the positions and directions of transcription of the genes. The gray-shaded areas represent regions sharing >99% DNA identity.
(TIFF)
Acknowledgments
The authors would like to acknowledge Dr. Tao He from the College of Veterinary Medicine, China Agricultural University for data analysis of PFGE.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funding provided by Medical Science and Technique Foundation of Henan Province (Grant no. 112102310165 and no.201303046).
References
- 1. Yang FC, Yan JJ, Hung KH, Wu JJ (2012) Characterization of Ertapenem-resistant Enterobacter cloacae in a Taiwanese University Hospital. J. Clin. Microbiol 50: 223–226. 10.1128/JCM.01263-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, et al. (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517: 455–459. 10.1038/nature14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennett JW, Herrera ML, Lewis JS, Wickes BW, Jorgensen JH (2009) KPC-2-producing Enterobacter cloacae and Pseudomonas putida coinfection in a liver transplant recipient. Antimicrob. Agents Chemother 53:292–294. 10.1128/AAC.00931-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yan JJ, Ko WC, Chuang CL, Wu JJ (2002) Metallo-β-lactamase-producing Enterobacteriaceae isolates in a university hospital in Taiwan: prevalence of IMP-8 in Enterobacter cloacae and first identification of VIM-2 in Citrobacter freundii . J. Antimicrob. Chemother 50:503–511. [DOI] [PubMed] [Google Scholar]
- 5. Castanheir M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE (2011) Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob. Agents Chemother 55:1274–1278. 10.1128/AAC.01497-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heller I, Grif K, Orth D (2012) Emergence of VIM-1-carbapenemase-producing Enterobacter cloacae in Tyrol, Austria. J. Med. Microbiol 61:567–571. 10.1099/jmm.0.038646-0 [DOI] [PubMed] [Google Scholar]
- 7. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. (2009) Characterization of a New Metallo-β-Lactamase Gene, bla NDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob Agents Chemother 53:5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Y, Zhou ZH, Jiang Y, Yu YS (2011) Emergence of NDM-1-producing Acinetobacter baumannii in China. J Antimicrob Chemother 66: 1255–1259. 10.1093/jac/dkr082 [DOI] [PubMed] [Google Scholar]
- 9. Sun Y, Li Q, Chen S, Song Y, Liu J, Guo X, et al. (2014) Characterization and Plasmid Elimination of NDM-1-Producing Acinetobacter calcoaceticus from China. PLoS One 9: e106555 10.1371/journal.pone.0106555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang C, Qiu S, Wang Y, Qi L, Hao R, Liu X, et al. (2013) Higher Isolation of NDM-1 Producing Acinetobacter baumannii from the Sewage of the Hospitals in Beijing. PLoS One 8: e64857 10.1371/journal.pone.0064857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qin S, Fu Y, Zhang Q, Qi H, Wen J, Wen J, et al. (2014) High Incidence and Endemic Spread of NDM-1-Positive Enterobacteriaceae in Henan Province, China. Antimicrob. Agents Chemother 58:4275–4282. 10.1128/AAC.02813-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clinical and Laboratory Standards Institute 2013. Performance standards for antimicrobial susceptibility testing; Twenty-Third Informational Supplement. M100–S23. [Google Scholar]
- 13. Doi Y, Arakawa Y (2007) 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis 45:88–94. [DOI] [PubMed] [Google Scholar]
- 14. Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD (2012) Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol 50: 3877–3880. 10.1128/JCM.02117-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leflon-Guibout V, Jurand C, Bonacorsi S, Espinasse F, Guelfi MC, Duportail F, et al. (2004) Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob. Agents Chemother 48:3736–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perez-Perez FJ, Hanson ND (2002) Detection of plasmid-mediated AmpC-β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol 40:2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woodford N, Fagan EJ, Ellington MJ (2006) Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum-β-lactamases. J. Antimicrob. Chemother 57:154–155. [DOI] [PubMed] [Google Scholar]
- 18. Tohru MA, Kayoko H, Norio O, Masahiro S, Teruo K (2013) Multilocus Sequence Typing (MLST) for Characterization of Enterobacter cloacae . PLoS One 8: e66358 10.1371/journal.pone.0066358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barton BM, Harding GP, Zuccarelli AJ (1995) A general method for detecting and sizing large plasmids. Anal. Biochem 226:235–240. [DOI] [PubMed] [Google Scholar]
- 20. Wang B and Sun DC (2014) Detection of NDM-1 carbapenemase-producing Acinetobacter calcoaceticus and Acinetobacter junii in environmental samples from livestock farms. J Antimicrob Chemother. 10.1093/jac/dku405 [DOI] [PubMed] [Google Scholar]
- 21. Villa J, Viedma E, Brañas P, Orellana MA, Otero JR, Chaves F (2014) Multiclonal spread of VIM-1-producing Enterobacter cloacae isolates associated with In624 and In488 integrons located in an IncHI2 plasmid. Int J Antimicrob Agents 43:451–455. 10.1016/j.ijantimicag.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 22. Bogaerts P, Huang TD, Rezende de Castro R, Bouchahrouf W, Glupczynski Y (2013). Could Acinetobacter pittii act as an NDM-1 reservoir for Enterobacteriaceae? J Antimicrob Chemother 68:2414–2415. 10.1093/jac/dkt201 [DOI] [PubMed] [Google Scholar]
- 23. Partridge SR, Iredell JR(2012) Genetic contexts of bla NDM-1 . Antimicrob Agents Chemother 56:6065–6067. 10.1128/AAC.00117-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X, Xu X, Li Z, Chen H, Wang Q, Yang P, et al. (2014) An outbreak of a nosocomial NDM-1-producing Klebsiella pneumoniae ST147 at a teaching hospital in mainland China. Microb Drug Resist 20:144–149. 10.1089/mdr.2013.0100 [DOI] [PubMed] [Google Scholar]
- 25. Borgia S, Lastovetska O, Richardson D, Eshaghi A, Xiong J, Chung C, et al. (2012) Outbreak of carbapenem-resistant enterobacteriaceae containing bla NDM-1, Ontario, Canada. Clin Infect Dis 55:e109–17. 10.1093/cid/cis737 [DOI] [PubMed] [Google Scholar]
- 26. Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJ, Carmeli Y, et al. (2015) MLST reveals potentially high-risk international clones of Enterobacter cloacae . J Antimicrob Chemother.70(1):48–56. 10.1093/jac/dku359 [DOI] [PubMed] [Google Scholar]
- 27. Rafailidis PI, Falagas ME (2014) Options for treating carbapenem-resistant Enterobacteriaceae . Curr Opin Infect Dis 27:479–483. 10.1097/QCO.0000000000000109 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The boxed arrows indicate the positions and directions of transcription of the genes. The gray-shaded areas represent regions sharing >99% DNA identity.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.