Abstract
Previous studies have demonstrated the metabolism of ritodrine through sulfation. The current study was designed to identify the human SULTs that are capable of sulfating ritodrine and to investigate how genetic polymorphism of the major ritodrine-sulfating SULT, SULT1A3, may affect its sulfating activity. A systematic analysis revealed that of the 13 known human SULTs, SULT1A1, SULT1A3, and SULT1C4, were capable of mediating the sulfation of ritodrine, with SULT1A3 displaying the strongest sulfating activity. Effects of genetic polymorphism on the sulfating activity of SULT1A3 were examined. By employing site-directed mutagenesis, 4 SULT1A3 allozymes were generated, expressed, and purified. Purified SULT1A3 allozymes were shown to exhibit differential sulfating activity toward ritodrine. Kinetic studies further demonstrated differential substrate affinity and catalytic efficiency among the SULT1A3 allozymes. Collectively, these results provided useful information concerning the differential metabolism of ritodrine through sulfation in different individuals.
Keywords: Sulfation, ritodrine, cytosolic sulfotransferase, SULT, SULT1A3, genetic polymorphism
1. Introduction
Preterm labor is known to be a major cause of perinatal mortality and morbidity (Neilson et al., 2014). Ritodrine is a tocolytic agent used for treating preterm labor (Neilson et al., 2014). Ritodrine stimulates the β2-adrenergic receptor in the body, causing an attenuation of uterine contractility (Neilson et al., 2014). Due to the lack of uterine selectivity, however, it may lead to a number of adverse effects for the mother and the fetus (Yaju and Nakayama, 2006; Kimura et al., 2013; Driul et al., 2014). Cardiac side effects, including increased heart rate and systolic blood pressure, myocardial ischemia, and pulmonary edema, are most common for the mother (Yaju and Nakayama, 2006; Kimura et al., 2013; Driul et al., 2014). Since ritodrine can cross the placental barrier, it may produce similar side effects in the fetus. The therapy with ritodrine is thus associated with these adverse effects which may vary among patients. To better understand its therapeutic effects as well as the adverse effects in different individuals, it is important to clarify the mechanism underlying the metabolism of ritodrine. Previous studies indicated that ritodrine was eliminated primarily through sulfation and glucuronidation (Pacifici et al., 1993; Pacifici, 2005), and both the mother treated with ritodrine and neonate delivered by treated mother were found to excrete sulfate and glucuronide conjugates of ritodrine in urine (Brashear et al., 1988).
Sulfate conjugation is a major pathway operated in humans and other vertebrates for the biotransformation and excretion of a diverse array of xenobiotics including drugs (Mulder and Jakoby, 1990; Falany and Roth, 1993; Weinshilboum and Otterness, 1994). The responsible enzymes, called the cytosolic sulfotransferases (SULTs), catalyze the transfer of a sulfonate group from the active sulfate, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to an acceptor substrate compound containing a hydroxyl or an amino group (Lipmann, 1958). Sulfate conjugation by these enzymes may result in the inactivation of the substrate compounds and/or increase their water-solubility, thereby facilitating their removal from the body (Mulder and Jakoby, 1990; Falany and Roth, 1993; Weinshilboum and Otterness, 1994). Previous studies have demonstrated the sulfation of ritodrine by human liver and duodenum cytosols (Pacifici et al., 1998), and several human SULTs have been shown to display ritodrine-sulfating activity (Nishimuta et al., 2005). To clarify further the involvement of SULT-mediated sulfation in the metabolism of ritodrine, it is prudent to identify all human SULTs that are capable of mediating the sulfation of ritodrine. Moreover, in view of the individual differences in susceptibility to the adverse effects of ritodrine, it is an intriguing issue whether SULT genetic polymorphism may affect the metabolism of ritodrine through sulfation. Like with many other genes, single nucleotide polymorphisms (SNPs) of SULT genes have been reported (Glatt et al., 2000; Lindsay et al., 2008; Daniels and Kadlubar, 2013). For example, four non-synonymous coding SNPs (cSNPs) for the SULT1A3 gene were detected by sequencing DNA samples from African-American and Caucasian-American subjects (Thomae et al., 2003; Hildebrandt et al., 2004). Since SULT1A3 has been shown to be capable of sulfating ritodrine (Nishimuta et al., 2005), it is important to find out whether its genetic polymorphism may have a significant impact on the metabolism of ritodrine through sulfation, thereby influencing the efficacy and side effects of the drug in different individuals.
In this communication, we report a systematic analysis of the sulfating activity of all known human SULTs toward ritodrine. Different allozymes of SULT1A3, a major ritodrine-sulfating SULT, were generated, expressed, purified, and characterized with respect to their kinetic parameters in mediating ritodrine sulfation.
2. Materials and Methods
2.1. Materials
Ritodrine was a product of Santa Cruz Biotechnology Inc. (Dallas, TX). Adenosine 5′-triphosphate (ATP), 3′-phosphoadenosine-5′-phosphosulfate (PAPS), N-2-hydroxylpiperazine-N′-2-ethanesulfonic acid (HEPES), dimethyl sulfoxide (DMSO), Trizma base, dithiothreitol (DTT), and silica gel thin-layer chromatography (TLC) plates were from Sigma Chemical Company (St. Louis, MO, USA). Ultrafree-MC 5000 NMWL filter units were products of Millipore (Bedford, MA, USA). Carrier-free sodium [35S]sulfate was a product of Perkin-Elmer (Waltham, MA, USA). Ecolume scintillation cocktail was purchased from MP Biomedicals, LLC. (Irvine, CA, USA). Recombinant human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase was prepared as previously described (Yanagisawa et al., 1998). EX Taq DNA polymerase was a product of Takara Bio (Mountain View, CA, USA). Protein molecular weight markers were from New England Biolabs, Inc. (Ipswich, MA, USA). Oligonucleotide primers were synthesized by MWG Biotech (Huntsville, AL, USA). X-ray films were purchased from BioExpress (Kaysville, UT, USA). All other chemicals were of the highest grade commercially available.
2.2. Preparation of the human SULTs
Recombinant human P-form (SULT1A1 and SULT1A2) and M-form (SULT1A3) phenol SULTs, thyroid hormone SULT (SULT1B1), two SULT1Cs (SULT1C2, SULT1C3, and SULT1C4), estrogen SULT (SULT1E1), dehydroepiandrosterone (DHEA) SULT (SULT2A1), two SULT2B1s (SULT2B1a and SULT2B1b), a neuronal SULT (SULT4A1) and SULT6B1, expressed using pGEX-2TK or pET23c prokaryotic expression system, were prepared as described previously (Sakakibara et al., 1998a; Sakakibara et al., 1998b; Pai et al., 2002; Sakakibara et al., 2002; Suiko et al., 2002).
2.3. Generation, expression, and purification of SULT1A3 allozymes
The QuikChange site-directed mutagenesis kit from Stratagene was used for the generation of cDNAs encoding SULT1A3 allozymes. Briefly, wild-type SULT1A3 cDNA packaged in pGEX-2TK prokaryotic expression vector was used as the template in conjunction with specific mutagenic primers (see Table 1 for the mutagenic primers used). The amplification conditions were 12 cycles of 30 s at 95°C, 1 min at 55°C, and 6 min at 68°C. The “mutated” SULT1A3 sequences were verified by nucleotide sequencing (Sanger et al., 1977). pGEX-2TK vector harboring individual mutated SULT1A3 cDNA was transformed into competent XL1-Blue E. coli cells. The transformed cells, grown to A600 nm = ~0.5 in 1 liter of LB medium supplemented with 100 μg/ml ampicillin and induced with 0.1 mM IPTG overnight at room temperature, were collected by centrifugation and homogenized in 20 ml of an ice-cold lysis buffer (10 mM Tris-HCl, pH 8.0, 150 mm NaCl, and 1 mM EDTA) using an Aminco French press. The crude homogenate thus prepared was subjected to centrifugation at 10,000 × g for 30 min at 4°C. The supernatant collected was fractionated using 0.5 ml of glutathione-Sepharose, and the bound fusion protein was treated with 2 ml of a thrombin digestion buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2.5 mM CaCl2) containing 5 units/ml bovine thrombin. Following a 1-h incubation at room temperature with constant agitation, the preparation was subjected to centrifugation. The recombinant enzyme present in the supernatant collected was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to enzymatic characterization as described below.
Table 1.
Primer sets used for the site-directed mutagenesis of human SULT1A3
| SULT1A3 Allozyme | Amino Acid Substitution | Mutagenic Primer Set |
|---|---|---|
| SULT1A3*2 | Lys234Asn | 5′-ttcaaggagatgaagaacaaccctatgaccaactac-3′ 5′-gtagttggtcatagggttgttcttcatctccttgaa-3′ |
| SULT1A3*3 | Pro101Leu | 5′-actctgaaagacacaccgctcccacggctcatcaagtca-3′ 5′-tgacttgatgagccgtgggagcggtgtgtctttcagagt-3′ |
| SULT1A3*4 | Pro101His | 5′-actctgaaagacacaccgcacccacggctcatcaagtca-3′ 5′-tgacttgatgagccgtgggtgcggtgtgtctttcagagt-3′ |
| SULT1A3*5 | Arg144Cys | 5′-tcctactaccatttccactgtatggaaaaggcgcaccct-3′ 5′-agggtgcgccttttccatacagtggaaatggtagtagga-3′ |
2.4. Sulfotransferase assay
The sulfating activity of the recombinant human SULTs was determined using PAP[35S] as the sulfonate donor. The reaction mixture for the standard enzymatic assay, prepared in a final volume of 20 μl, contained, 50 mM MOPS at pH 7.0, 14 μM of PAP[35S], 1 mM DTT, and 50 μM substrate. Stock solutions of the substrates, prepared in DMSO, were used in the enzymatic assay. Controls with water or DMSO replacing substrate were also included. The reaction was started by the addition of the enzyme, allowed to continue at 37°C for 10 min (5 min in case of the kinetic assays), and terminated by placing the tube containing the reaction mixture on a heating block at 100°C for 3 min. The precipitates were cleared by centrifugation at 15,000×g for 3 min and the supernatant was subjected to the analysis of [35S]sulfated product. Afterwards, 1 μl of the reaction mixture was spotted on a silica TLC plate and the spotted TLC plate was subjected to TLC analysis using a solvent system containing n-butanol: acetonitrile in a ratio of 3:2 (by volume). Upon completion of TLC, the TLC plate was air-dried and autoradiographed by using an X-ray film. The radioactive spot corresponding to the sulfated product was located and cut out and eluted in 0.5 ml water in a glass vial. 4.5 ml of Ecolume scintillation liquid was added to each vial, mixed thoroughly, and the radioactivity therein was counted by using a liquid scintillation counter. Each experiment was performed in quadruplicate, together with a control without substrate. The cpm count obtained was used to calculate the specific activity in the unit of nmol of sulfated product/min/mg enzyme. For the kinetic studies on the sulfation of ritodrine, varying concentrations (5 μM, 6.67 μM, 10 μM, 25 μM, 50 μM, 100 μM, 250 μM, and 500 μM) were used with 50 mM HEPES, pH 7.0, as the buffer according to the procedure described earlier. The specific activity data were analyzed based on the Michaelis-Menten kinetics to calculate the kinetic constants. Statistical analyses were performed using SPSS v17.0 statistics software. One-way ANOVA was used to estimate the ANOVA P-value.
2.5. Miscellaneous methods
The sulfonate donor, PAP[35S], was synthesized from ATP and carrier-free [35S]sulfate using the bifunctional human ATP sulfurylase/APS kinase (Yanagisawa et al., 1998). The synthesized PAP[35S] was adjusted to the desired concentration and specific activity by the addition of nonradioactive (cold) PAPS. SDS-PAGE was performed on a 12% polyacrylamide gel using the method of Laemmli (1970). Protein determination was based on the method of Bradford (1976) with bovine serum albumin as a standard.
3. Results
3.1. Identification of the human SULTS capable of sulfating ritodrine
To identify the enzymes that are responsible for the sulfation of ritodrine, 13 known human SULTs (SULT1A1, SULT1A2, SULT1A3, SULT1B1, SULT1C2, SULT1C3, SULT1C4, SULT1E1, SULT2A1, SULT2B1a, SULT2B1b, SULT4A1, SULT6B1), previously cloned, expressed, and purified, were examined for sulfating activity with different concentrations (5, 50 and 100 μM) of ritodrine as substrate. Results obtained showed that ten (SULT1A2, SULT1B1, SULT1C2, SULT1C3, SULT1E1, SULT2A1, SULT2B1a, SULT2B1b, SULT4A1 and SULT6B1) of the 13 SULTs displayed no detectable activities. Of the other three SULTs (SULT1A1, SULT1A3, SULT1C4), SULT1A3 exhibited considerably stronger sulfating activity than the other two towards ritodrine. Table 2 shows the specific activities of these three enzymes with 50 μM ritodrine as a substrate. The 50 μM substrate concentration was chosen based on two considerations, one being that serum levels of ritodrine in patients have been reported to be in the low μM range (Pacifici, 2005) and the other being the considerably higher Km values (in the hundred μM range) found for purified SULT enzymes (cf. Table 3).
Table 2.
Specific activities of human SULTs with ritodrine as a substrate*
| Substrate | Specific Activity (nmol/min/mg)* | ||
|---|---|---|---|
| SULT1A1 | SULT1A3 | SULT1C4 | |
| Ritodrine | 2.34 ± 0.02 | 47.74 ± 2.83 | 22.14 ± 3.70 |
| Standard** | 32.83 ± 0.21 | 33.53 ± 0.83 | 16.01 ± 1.23 |
Specific activity refers to nmol ritodrine sulfated/min/mg purified enzyme. Data represent means ± S.D. derived from three experiments. The concentration of ritodrine tested in the reaction mixture was 50 μM.
Standard refers to the prototype substrate commonly used to assay for the sulfating activity of the indicated SULT. p-Nitrophenol was used for SULT1A1 and SULT1C4; and dopamine was used for SULT1A3. The concentration of these substrates used in the reaction mixtures was 50 μM.
Table 3.
Kinetic constants of the human SULT1A3 allozymes in catalyzing the sulfation of ritodrine*
| SULT1A3 Allozyme | Km (μM) | Vmax (nmol/min/mg) | Vmax/Km |
|---|---|---|---|
| SULT1A3*1 | 121.8 ± 6.7 | 163.9 ± 8.4 | 1.35 |
| SULT1A3*2 (Lys234Asn) | 117.8 ± 6.9 | 95.2 ± 3.9 | 0.81a |
| SULT1A3*3 (Pro101Leu) | 192.5 ± 10.3 | 161.2 ± 9.0 | 0.84a |
| SULT1A3*4 (Pro101His) | 137.6 ± 5.2 | 55.2 ± 3.2 | 0.40a |
| SULT1A3*5 (Arg144Cys) | 248.5 ± 10.0 | 178.5 ± 10.2 | 0.72a |
Results represent means± S.D. derived from three determinations.
Statistical significance versus SULT1A3*1 (P-value < 0.001)
3.2. Effects of genetic polymorphism on the ritodrine-sulfating activity of SULT1A3 allozymes
To find out how different amino acid changes resulting from non-synonymous coding SNPs (Thomae et al., 2003; Hildebrandt et al., 2004) may affect the sulfating activity of the SULT1A3 protein products, our approach was to prepare recombinant SULT1A3 allozymes for enzymatic characterization with ritodrine as substrate.
3.2.1. Preparation of human SULT1A3 allozymes
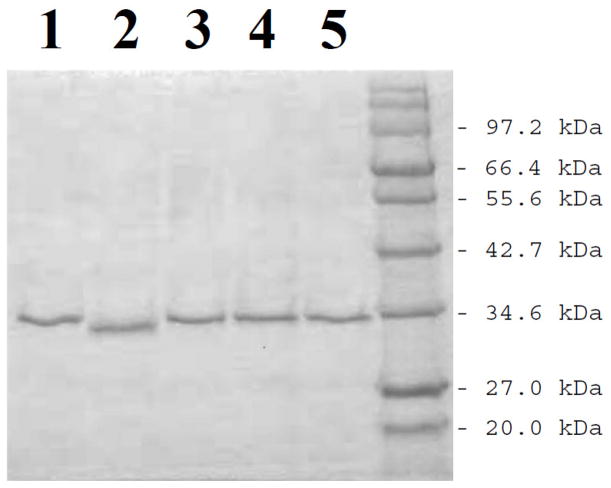
Based on the procedure described in the Materials and Methods section, cDNAs encoding different SULT1A3 allozymes packaged in pGEX-2TK prokaryotic expression vector were individually transformed into BL 21 E. coli host cells for expressing the recombinant enzymes. As shown in Fig. 1, the recombinant SULT1A3 allozymes (designated SULT1A3*2, SULT1A3*3, SULT1A3*4, and SULT1A3*5) fractionated from the homogenates of transformed E. coli cells using glutathione-Sepharose and cleaved off the bound fusion proteins by thrombin digestion appeared to be highly homogeneous upon SDS-polyacrylamide gel electrophoresis, with apparent molecular weights similar to predicted molecular weight (34,196) of wild-type SULT1A3 (designated SULT1A3*1).
Fig. 1.
SDS gel electrophoretic pattern of the purified human SULT1A3 allozymes. SDS-PAGE was performed on a 12% gel, followed by Coomassie blue staining. Samples analyzed in lanes 1 through 5 correspond to SULT1A3*1, SULT1A3*2, SULT1A3*3, SULT1A3*4, and SULT1A3*5. Positions of protein molecular weight markers co-electrophoresed are indicated on the right.
3.2.2. Characterization of the ritodrine-sulfating activity of human SULT1A3 allozymes
Purified SULT1A3 allozymes were assayed for sulfating activity using ritodrine as substrate. As shown in Fig. 2, compared with wild-type SULT1A3 (SULT1A3*1), the the other four SULTA3 allozymes all exhibited significantly lower sulfating activity toward ritodrine. Of the four, SULT1A3 *3 (Pro101His) showed the lowest ritodrine sulfating activity (15.9 nmol/min/mg) which is nearly three times as low compared with SULT1A3*1 (cf. Table 2). To examine further the effects of genetic polymorphism on SULT1A3-mediated ritodrine sulfation, kinetic experiments were performed on wild-type and SULT1A3 allozymes. Assays were carried out using varying concentrations (range from 0 to 500 μM) of ritodrine at pH 7.0. Data obtained were used to generate Lineweaver-Burk double-reciprocal plots in order to calculate the Km, Vmax, and Vmax/Km for each of the SULT1A3 allozymes in catalyzing the sulfation of ritodrine. The kinetic parameters, Km, Vmax, and Vmax/Km thus obtained are compiled in Table 3. It was noted that both Pro101Leu and Arg144Cys variations led to a dramatic increase in the Km value, implying that these variation might have led to a decrease in the binding affinity for ritodrine. Lys234Asn and Pro101His variations, on the other hand, both led to a dramatic decrease in the Vmax value, implying their effect on the catalysis of ritodrine sulfation. Calculated Vmax/Km values, which reflects the catalytic efficiency of different SULT1A3 allozymes, varied in the range of 0.40 – 1.35, which in general correlate reasonably well with the specific activity data shown in Fig. 2. Collectively, these data implied a dramatic effect of genetic polymorphism on the sulfation of ritodrine in individuals with different SULT1A3 genotypes.
Fig. 2.
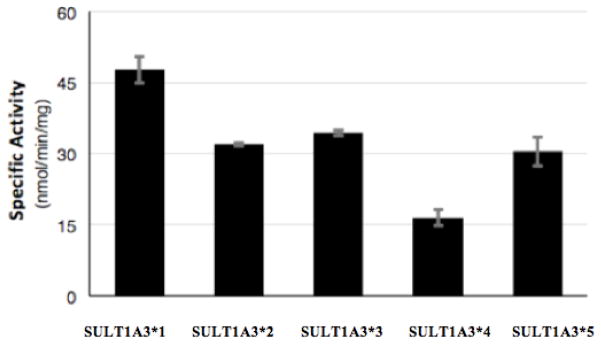
Specific activities of the sulfation of ritodrine by human SULT1A3 allozymes. Concentration of ritodrine used in the enzymatic assays was 50 μM. Data shown represent mean ± standard deviation derived from four determinations.
4. Discussion
Previous studies had demonstrated that sulfation represents an important pathway in the biotransformation of ritodrine (Pacifici et al., 1998; Nishimuta et al., 2005). The current study aimed to identify those human SULTs that are capable of sulfating ritodrine. A systematic analysis revealed that three of the thirteen human SULTs, SULT1A1, SULT1A3, and SULT1C4 were capable of sulfating ritodrine, with SULT1A3 displaying considerably stronger sulfating activity than the other two. These results indicated that human SULT1A3 is likely the major enzyme responsible for the sulfation of ritodrine in the body. While SULT1A3 is known to be expressed in many organs including brain, lung, liver, kidney and gastrointestinal tract (Dooley et al., 2000), SULT1C4, which displayed the second highest specific activity toward ritodrine, has been reported to be expressed at much higher levels in fetal tissues including fetal lung, liver, small intestine and kidney (Sakakibara et al., 1998b; Stanley et al., 2005). This latter finding may imply a role for SULT1C4 in mediating the sulfation of ritodrine in fetus and neonate. SULT1A1, which exhibited a low but significant ritodrine-sulfating activity, had previously been shown to be expressed in the liver, as well as brain, gastrointestinal tract, platelets, and placenta (Barker et al., 1994).
An intriguing question is whether and how genetic polymorphism of SULT1A3 may affect its sulfating activity. Evidence of genetic polymorphism of SULT1A3 was first observed in a study showing large variations of its enzymatic activity in platelet samples prepared from a cohort of 232 individuals (Price et al., 1998). By sequencing DNA samples derived from 60 African-American and 60 Caucasian–American subjects, four non-synonymous coding SNPs of the SULT1A3 gene were later detected (Thomae et al., 2003; Hildebrandt et al., 2004). Using the site-directed mutagenesis technique followed by recombinant protein expression, corresponding SULT1A3 allozymes (SULT1A3*2, SULT1A3*3 and SULT1A4*) were prepared and characterized in comparison with the wild-type enzyme (SULT1A3*1). Activity data shown in Fig. 2 revealed indeed significant variations in ritodrine-sulfating activity among the four SULT1A3 allozymes. It is noted that the activity data of SULT1A3 allozymes shown in Fig. 2 differ considerably from the activity data of the same allozymes previously reported using dopamine as a substrate (Thomae et al., 2003; Hildebrandt et al., 2004). It is possible that the discrepancy may be, in part at least, due to the different substrates utilized. Additionally, the previous studies used transfected COS-1 cell lysates as the source of the allozymes, whereas purified SULT1A3 allozymes were used in the present study. Subsequent kinetic experiments revealed that the amino acid variations in the four SULT1A3 allozymes resulted in increased Km value and/or decreased Vmax value, indicating their effects on binding affinity for and/or catalytic activity toward ritodrine sulfation (cf. Table 3). These results thus provided support for the effects of genetic polymorphism on the sulfation of ritodrine in individuals with different SULT1A3 genotypes.
In summary, the current study demonstrated that of the thirteen known human SULTs, SULT1A1, SULT1A3, and SULT1C2 were capable of mediating the sulfation of ritodrine. Genetic polymorphism of SULT1A3, a major ritodrine-sulfating SULT, appeared to exert a dramatic effect on the sulfating activity of SULT1A3 allozymes. These findings may in the future aid in the development of individualized regimens of ritodrine for more effectively and safely treating preterm labor during pregnancy.
Acknowledgments
This work was supported in part by a grant (#R03HD071146) from National Institutes of Health.
Abbreviations
- SULT
cytosolic sulfotransferase
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- PCR
polymerase chain reaction
- SNP
single nucleotide polymorphism
Footnotes
Statement of Conflict of Interest: None declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barker EV, Hume R, Hallas A, Coughtrie WH. Dehydroepiandrosterone sulfotransferase in the developing human fetus: quantitative biochemical and immunological characterization of the hepatic, renal, and adrenal enzymes. Endocrinol. 1994;134:982–989. doi: 10.1210/endo.134.2.8299591. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brashear WT, Kuhnert BR, Wei R. Maternal and neonatal urinary excretion of sulfate and glucuronide ritodrine conjugates. Clin Pharmacol Ther. 1988;44:634–641. doi: 10.1038/clpt.1988.205. [DOI] [PubMed] [Google Scholar]
- Daniels J, Kadlubar S. Sulfotransferase genetic variation: from cancer risk to treatment response. Drug Metab Rev. 2013;45:415–422. doi: 10.3109/03602532.2013.835621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley TP, Haldeman-Cahill R, Joiner J, Wilborn TW. Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem Biophys Res Commun. 2000;277:236–245. doi: 10.1006/bbrc.2000.3643. [DOI] [PubMed] [Google Scholar]
- Driul L, Londero AP, Adorati-Menegato A, Vogrig E, Bertozzi S, Fachechi G, et al. Therapy side-effects and predictive factors for preterm delivery in patients undergoing tocolysis with atosiban or ritodrine for threatened preterm labour. J Obstet Gynaecol. 2014;24:1–6. doi: 10.3109/01443615.2014.930094. [DOI] [PubMed] [Google Scholar]
- Falany C, Roth JA. In: Human Drug Metabolism; From Molecular Biology to Man. Jeffery EH, editor. Boca Raton: CRC Press; 1993. pp. 101–115. [Google Scholar]
- Glatt H, Engelke CE, Pabel U, Teubner W, Jones AL, Coughtrie MW, Andrae U, Falany CN, Meinl W. Sulfotransferases: genetics and role in toxicology. Toxicol Lett. 2000;112–113:341–348. doi: 10.1016/s0378-4274(99)00214-3. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Carrington DP, Thomae BA, Eckloff BW, Schaid DJ, Yee VC, Weinshilboum RM, Wieben ED. Genetic diversity and function in the human cytosolic sulfotransferases. Pharmacogenomics J. 2007;7:133–143. doi: 10.1038/sj.tpj.6500404. [DOI] [PubMed] [Google Scholar]
- Hildebrandt MA, Salavaggione OE, Martin YN, Flynn HC, Jalal S, Wieben ED, Weinshilboum RM. Human SULT1A3 pharmacogenetics: gene duplication and functional genomic studies. Biochem Biophys Res Commun. 2004;321:870–878. doi: 10.1016/j.bbrc.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Jaju PB, Dhabadi VB. Nifedipine versus ritodrine for suppression of preterm labor and analysis of side effects. J Obstet Gynaecol India. 2011;61:534–537. doi: 10.1007/s13224-011-0083-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kato T, Nohara R. Pulmonary edema complicating ritodrine infusion in a patient with premature labor. Intern Med. 2013;52:155. doi: 10.2169/internalmedicine.52.8986. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lindsay J, Wang LL, Li Y, Zhou SF. Structure, function and polymorphism of human cytosolic sulfotransferases. Curr Drug Metab. 2008;9:99–105. doi: 10.2174/138920008783571819. [DOI] [PubMed] [Google Scholar]
- Lipmann F. Biological sulfate activation and transfer. Science. 1958;128:575–580. doi: 10.1126/science.128.3324.575. [DOI] [PubMed] [Google Scholar]
- Mulder GJ, Jakoby WB. In: Drug Metabolism. Mulder GJ, Jakoby WB, editors. London: Taylor and Francis; 1990. pp. 107–61. [Google Scholar]
- Neilson JP, West HM, Dowswell T. Betamimetics for inhibiting preterm labour. Cochrane Database Syst Rev. 2014;2:CD004352. doi: 10.1002/14651858.CD004352.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimuta H, Tsujimoto M, Ogura K, Hiratsuka A, Ohtani H, Sawada Y. Inhibitory effects of various beverages on ritodrine sulfation by recombinant human sulfotransferase isoforms SULT1A1 and SULT1A3. Pharm Res. 2005;22:1406–1410. doi: 10.1007/s11095-005-5263-y. [DOI] [PubMed] [Google Scholar]
- Pacifici GM. Sulfation of drugs and hormones in mid-gestation human fetus. Early Hum Dev. 2005;81:573–581. doi: 10.1016/j.earlhumdev.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Pacifici GM, Kubrich M, Giuliani L, de Vries M, Rane A. Sulphation and glucuronidation of ritodrine in human foetal and adult tissues. Eur J Clin Pharmacol. 1993;44:259–264. doi: 10.1007/BF00271368. [DOI] [PubMed] [Google Scholar]
- Pacifici GM, Quilici MC, Giulianetti B, Spisni R, Nervi M, Giuliani L, Gomeni R. Ritodrine sulphation in the human liver and duodenal mucosa: Interindividual variability. Eur J Drug Metab Pharmacokinet. 1998;23:67–74. doi: 10.1007/BF03189829. [DOI] [PubMed] [Google Scholar]
- Pai TG, Sugahara T, Suiko M, Sakakibara Y, Xu F, Liu MC. Differential xenoestrogen-sulfating activities of the human cytosolic sulfotransferases: molecular cloning, expression, and purification of human SULT2B1a and SULT2B1b sulfotransferases. Biochim Biophys Acta. 2002;1573:165–170. doi: 10.1016/s0304-4165(02)00416-6. [DOI] [PubMed] [Google Scholar]
- Price RA, Cox NJ, Spielman RS, Van Loon JA, Maidak BL, Weinshilboum RM. Inheritance of human platelet thermolabile phenol sulfotransferase (TL PST) activity. Genet Epidemiol. 1988;5:1–15. doi: 10.1002/gepi.1370050102. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y, Suiko M, Pai TG, Nakayama T, Takami Y, Katafuchi J, Liu MC. Highly conserved mouse and human brain sulfotransferases: molecular cloning, expression, and functional characterization. Gene. 2002;285:39–47. doi: 10.1016/s0378-1119(02)00431-6. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y, Takami Y, Nakayama T, Suiko M, Liu MC. Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form phenol sulfotransferases. J Biol Chem. 1998a;273:6242–6247. doi: 10.1074/jbc.273.11.6242. [DOI] [PubMed] [Google Scholar]
- Sakakibara Y, Yanagisawa K, Katafuchi J, Ringer DP, Takami Y, Nakayama T, Suiko M, Liu MC. Molecular cloning, expression, and characterization of novel human SULT1C sulfotransferases that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J Biol Chem. 1998b;273:33929–33935. doi: 10.1074/jbc.273.51.33929. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley EL, Hume R, Coughtrie MW. Expression profiling of human fetal cytosolic sulfotransferase involved in steroid and thyroid hormone metabolism and in detoxification. J Mol Cell Endocrinol. 2005;240:32–42. doi: 10.1016/j.mce.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Suiko M, Sakakibara Y, Liu MC. Sulfation of environmental estrogen-like chemicals by human cytosolic sulfotransferases. Biochem Biophys Res Commun. 2002;267:80–84. doi: 10.1006/bbrc.1999.1935. [DOI] [PubMed] [Google Scholar]
- Thomae BA, Rifki OF, Theobald MA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catecholamine sulfotransferase (SULT1A3) pharmacogenetics: functional genetic polymorphism. J Neurochem. 2003;87:809–819. doi: 10.1046/j.1471-4159.2003.02027.x. [DOI] [PubMed] [Google Scholar]
- Weinshilboum R, Otterness D. In: Conjugation-Deconjugation Reactions in Drug Metabolism and Toxicity. Kaufmann FC, editor. Berlin: Springer-Verlag; 1994. pp. 45–78. [Google Scholar]
- Yaju Y, Nakayama T. Effectiveness and safety of ritodrine hydrochloride for the treatment of preterm labour: a systematic review. Pharmacoepidemiol Drug Saf. 2006;15:813–822. doi: 10.1002/pds.1317. [DOI] [PubMed] [Google Scholar]
- Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, Takayanagi K, Natori Y, Liu MC. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. Biosci Biotechnol Biochem. 1998;62:1037–1040. doi: 10.1271/bbb.62.1037. [DOI] [PubMed] [Google Scholar]