Abstract
We evaluated the limits of detection (LoD) for an 11-plex PCR-Luminex assay performed on Whatman FTA Elute cards smeared with stool containing pathogens associated with travelers’ diarrhea. LoDs ranged between 102-105 CFU, PFU or cysts/g for most pathogens except Cryptosporidium. Campylobacter and norovirus LoD increased with prolonged storage of cards.
Keywords: PCR, travelers’ diarrhea, stool card
1. INTRODUCTION
Travelers’ diarrhea (TD) is frequently experienced by civilians and deployed military personnel traveling to developing countries. Although the identification of enteropathogens in epidemiologic studies has been greatly facilitated by the use of polymerase chain reaction (PCR) assays (Al Amri, Senok, Ismaeel, Al-Mahmeed, & Botta, 2007; Aranda, Fagundes-Neto, & Scaletsky, 2004; Couturier, Lee, Zelyas, & Chui, 2011), testing remains limited due to requirements for collection, storage and transportation of diarrheal specimens. As a result, epidemiological data is largely derived from cohorts with on-site testing and storage facilities.
Self-collected stool smears obtained on a filter-paper matrix is an appealing alternative for stool collection and storage in field conditions (Grimes, et al., 2008; Orlandi & Lampel, 2000). However, the utility of filter-paper and impact on PCR detection is unknown. We performed a pilot study to compare the limits of detection (LoD) of a multiplex PCR assay between stool samples and smeared Whatman™ FTA Elute cards, and determine the impact of prolonged storage and environmental conditions on detection from cards. Enteropathogens tested included norovirus [GI, GII], Salmonella enteritidis, S. typhimurium, enterotoxigenic Escherichia coli, enterohemorrhagic E. coli, enteroaggregative E. coli, Shigella sonnei, Campylobacter jejuni, Giardia lamblia and Cryptosporidium.
2. MATERIALS AND METHODS
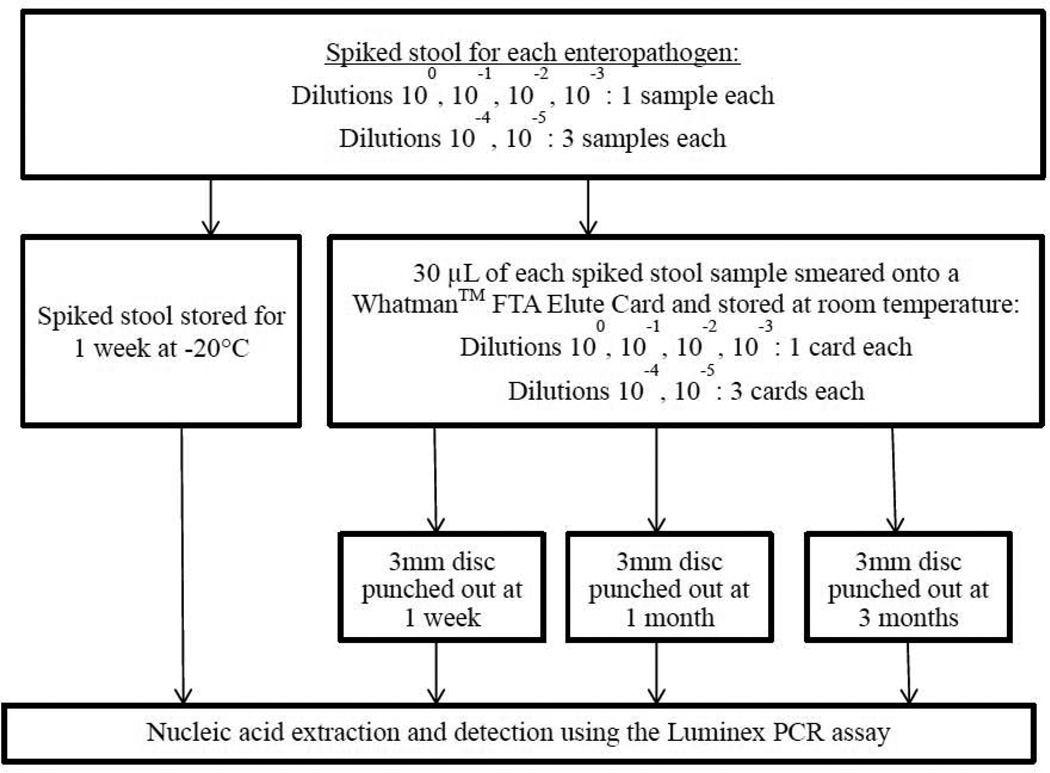
Reference strains from commercial entities (American Type Culture Collection, Waterborne Inc., Zeptometrix Corp.) or clinical isolates (enteroaggregative E. coli, Campylobacter) were used. Bacterial strains were inoculated into Mueller Hinton (MH) broth, incubated overnight, and the CFU/g concentration was determined by measuring the turbidity at 600 nm. Known concentrations of parasite oocysts and viruses were serially diluted in phosphate-buffer saline and de-ionized water, respectively. Five serial 1:10 dilutions were performed for each enteropathogen, and 50 µL of each dilution was spiked into 500 µL of a fecal suspension (15g healthy pathogen-negative stool [Bioreclamation, LLC] and 45 mL MH broth). 30 µL of spiked stool samples at 100, 10−1, 10−2, 10−3 dilutions was smeared onto Whatman™ FTA Elute cards (Figure 1). Spiked stool and smears were done in triplicate for the 4th and 5th dilutions. Stool samples were stored at −20°C and tested at 1 week. Stool cards were stored at room temperature and a 3mm disc was punched out from each card at 1 week, 1 month and 3 months. One stool card for each undiluted enteropathogen sample was stored at 4°C and in a humid incubator at 31°C for 1 week to determine the impact of environmental conditions on detection. Spiked stool samples were extracted using the Qiagen QiaAmp Stool Extraction kit (Qiagen, Les Ulis, France) and punched discs were extracted according to the manufacturer’s protocol (GEHealthCareLifeSciences, 2011).
Figure 1.
Preparation, storage and detection of spiked stool and smeared Whatman™ FTA Elute Card.
xTAG analyte-specific reagents (Luminex Molecular Diagnostics, Toronto, Canada) for Salmonella, ETEC, EHEC, Shigella, Campylobacter, Giardia, Cryptosporidium, norovirus GI and GII were used in the mastermix. Primers targeting the EAEC aggR gene (Forward: 5’-CGAAAAAGAGATTATAAAAATTAAC-3’, Reverse: 5’-GCTTCCTTCTTTTGTGTAT-3’) (Barletta, Ochoa, & Cleary, 2013) were first tested on DNA, then modified for the xTAG panel, and testing repeated to confirm detection. 10 µL of the extraction product was mixed with 15 µL of the mastermix (1.5 µL of xTAG DNase/RNase free water, 7.5 µL of Qiagen OneStep RT-PCR Buffer 5X, 0.5 µL of BSA, 1.45 µL of Qiagen 10mM dNTPs, 0.08 µL of 6M tetramethylammonium choride, 2.0 µL of Qiagen OneStep RT-PCR Enzyme Mix and 0.167 µL of each primer). A negative control (nuclease-free water) and an internal control (MS2) were used. PCR and hybridization was performed according to a published protocol (Navidad, Griswold, Gradus, & Bhattacharyya, 2013). Samples were analyzed using the xTAG Data Analysis Software (TDAS) and results reported as mean fluorescent intensity (MFI) units. Samples with an MFI ≥ 300 were considered positive, and the fourth and fifth dilutions were positive if the MFI was ≥ 300 for all 3 samples (Navidad, et al., 2013).
3. RESULTS
Overall, the LoDs ranged between 102 and 105 CFU, PFU, or cysts/g for most enteropathogens. LoDs were comparable (within 1-2 logs) between stool samples and stool cards at 1 week (Table 1). Cryptosporidium was not detected in spiked stool and had a high LoD in the stool card, probably due to the lack of oocyst disruption (e.g. bead-beating) during sample processing. No sustained increase in the LoD at 3 months was noted for most pathogens except Campylobacter which increased at 1 month and could not be detected at 3 months, and norovirus which increased by 1-2 logs. Cryptosporidium could not be detected at 1 month, and we elected not to test the card at 3 months after identifying the issue with extraction. No difference in detection with varying environmental conditions was noted except for Cryptosporidium and Campylobacter, which were either poorly detected or not detected when stored at either 4°C or 31°C.
Table 1.
Results of testing for limit of detection for pathogensa
| Pathogen(s) | Spiked stoolb 1 week |
Whatman™ FTA Elute Card with spiked stool smearb | ||
|---|---|---|---|---|
| 1 week | 1 month | 3 months | ||
| Enterotoxigenic E. coli (LT/ST) | 105/105 | 103/104 | 102/103 | 103/103 |
| Enterohemorrhagic E. coli (stx1/stx2) | 105/104 | 103/103 | 106/105 | 102/102 |
| Enteroaggregative E. coli | 105 | 104 | 105 | 104 |
| Salmonella typhimurium | 104 | 101 | 104 | 102 |
| Salmonella enteritidis | 102 | 104 | 103 | 103 |
| Shigella sonnei | 103 | 103 | 102 | 102 |
| Campylobacter jejuni | 104 | 105 | 106 | Not detected |
| Giardia lamblia | 103 | 100 | 101 | 101 |
| Cryptosporidium parvum | Not detected | 106 | Not detected | Not performed |
| Norovirus GIb | 104 | 103 | 105 | 104 |
| Norovirus GIIb | 102 | 103 | 105 | 105 |
Concentration at which pathogen was detected by multiplex PCR (mean fluorescence intensity value for detection was ≥ 300); one sample tested for each pathogen for dilutions 100-10−3; for dilutions 10−4 and 10−5 samples were tested in triplicate and mean MFI was used to determine limit of detection
Bacteria LoD reported as CFU/g, parasites reported as cysts/g, norovirus reported as PFU/g
4. DISCUSSION
Our results indicate that the FTA Elute card may be an effective method of storing genomic material from most diarrheal pathogens. Comparable LoDs were observed between stool samples and stool cards, indicating effective storage of genomic material and sequestration of factors inhibiting PCR. The LoDs observed were comparable to those reported in the literature (Liu, et al., 2012; Navidad, et al., 2013) and within the range associated with symptomatic infection (Granato, et al., 2010; Lampel, 2005). Orlandi et al. successfully detected Cryptosporidium oocyts from FTA stool cards without cyst disruption (Orlandi & Lampel, 2000), but we were unable to replicate these findings, and thus suggest including bead-beating for extraction. We also observed difficulty in detecting Campylobacter and an increase in norovirus GI/GII LoD with prolonged storage. Prior reports have documented successful storage and detection of Campylobacter for 7 months and norovirus for 11 weeks on the FTA Card (Delacour, Dubrous, & Koeck, 2010; Owens & Szalanski, 2005). Further testing is needed to evaluate the stability and detection of these pathogens with long-term storage.
As a pilot study, the small sample size limited our assessment of assay precision and reproducibility. In addition, issues related to quality of self-collected stool smears during travel, and its impact on detection we not evaluated. The use of filter-paper cards and a qualitative assay does not address quantification of clinically relevant pathogen load, pathogen phenotype and evaluation of the host immune response (e.g. by measuring fecal cytokines), which are important in ascribing etiology and correlating with disease attribution. We plan to further examine the utility of the FTA Elute Card paired with a quantitative PCR (TaqMan Array Card), using diarrheal specimens that have undergoing prior microbiologic workup, and self-collected stool smears obtained during travel.
HIGHLIGHTS.
We evaluated the detection limits for a PCR assay targeting travelers’ diarrhea pathogens.
WhatmanTM FTA Elute cards were smeared with spiked stool for testing
The limit of detection ranged between 102-105 CFU, PFU or cysts/g for most pathogens
The limit of detection for Campylobacter and norovirus increased with prolonged storage
Cryptosporidium was poorly detected from spiked stool and smeared stool cards
ACKNOWLEDGEMENTS
The authors thank their collaborators at Naval Medical Research Center, Bethesda MD and Naval Medical Research Unit-6, Peru for providing clinical isolates needed for assay validation.
Supported by the Infectious Disease Clinical Research Program (IDCRP), a Department of Defense (DoD) program executed through the Uniformed Services University of the Health Sciences, the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), under the Inter-Agency Agreement Y1-AI-5072.
The content of this publication is the sole responsibility of the authors and does not necessarily reflect the views or policies of the NIH or the Department of Health and Human Services, Uniformed Services University of the Health Sciences, the DoD or the Departments of the Army, Navy or Air Force. Mention of trade name, commercial products, or organizations does not imply endorsement by the US Government.
Some authors are employees of the U.S. Government. This work was prepared as part of their official duties. Title 17 U.S.C. 105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. 101 defines a United States Government work as a work prepared by a military service member or employee of the United States Government as part of that person’s official duties.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Al Amri A, Senok AC, Ismaeel AY, Al-Mahmeed AE, Botta GA. Multiplex PCR for direct identification of Campylobacter spp. in human and chicken stools. J Med Microbiol, 2007;56:1350–1355. doi: 10.1099/jmm.0.47220-0. [DOI] [PubMed] [Google Scholar]
- Aranda KR, Fagundes-Neto U, Scaletsky IC. Evaluation of multiplex PCRs for diagnosis of infection with diarrheagenic Escherichia coli and Shigella spp. J Clin Microbiol. 2004;42:5849–5853. doi: 10.1128/JCM.42.12.5849-5853.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta F, Ochoa TJ, Cleary TG. Multiplex real-time PCR (MRT-PCR) for diarrheagenic. Methods Mol Biol. 2013;943:307–314. doi: 10.1007/978-1-60327-353-4_21. [DOI] [PubMed] [Google Scholar]
- Couturier MR, Lee B, Zelyas N, Chui L. Shiga-toxigenic Escherichia coli detection in stool samples screened for viral gastroenteritis in Alberta, Canada. J Clin Microbiol. 2011;49:574–578. doi: 10.1128/JCM.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacour H, Dubrous P, Koeck JL. Noroviruses: a challenge for military forces. J R Army Med Corps. 2010;156:251–254. doi: 10.1136/jramc-156-04-10. [DOI] [PubMed] [Google Scholar]
- GEHealthCareLifeSciences. 2011 [Google Scholar]
- Granato PA, Chen L, Holiday I, Rawling RA, Novak-Weekley SM, Quinlan T, Musser KA. Comparison of premier CAMPY enzyme immunoassay (EIA), ProSpecT Campylobacter EIA, and ImmunoCard STAT! CAMPY tests with culture for laboratory diagnosis of Campylobacter enteric infections. J Clin Microbiol. 2010;48:4022–4027. doi: 10.1128/JCM.00486-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes KA, Mohamed JA, Dupont HL, Padda RS, Jiang ZD, Flores J, Belkind-Gerson J, Martinez-Sandoval FG, Okhuysen PC. PCR-based assay using occult blood detection cards for detection of diarrheagenic Escherichia coli in specimens from U. S. travelers to Mexico with acute diarrhea. J Clin Microbiol. 2008;46:2227–2230. doi: 10.1128/JCM.00073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampel KA. Foodborne pathogens: microbiology and molecular biology. Norfolk, United Kingdom: Caister Academic Press; 2005. [Google Scholar]
- Liu J, Gratz J, Maro A, Kumburu H, Kibiki G, Taniuchi M, Howlader AM, Sobuz SU, Haque R, Talukder KA, Qureshi S, Zaidi A, Haverstick DM, Houpt ER. Simultaneous detection of six diarrhea-causing bacterial pathogens with an in-house PCR-luminex assay. J Clin Microbiol. 2012;50:98–103. doi: 10.1128/JCM.05416-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navidad JF, Griswold DJ, Gradus MS, Bhattacharyya S. Evaluation of Luminex xTAG gastrointestinal pathogen analyte-specific reagents for high-throughput, simultaneous detection of bacteria, viruses, and parasites of clinical and public health importance. J Clin Microbiol, 2013;51:3018–3024. doi: 10.1128/JCM.00896-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi PA, Lampel KA. Extraction-free, filter-based template preparation for rapid and sensitive PCR detection of pathogenic parasitic protozoa. J Clin Microbiol. 2000;38:2271–2277. doi: 10.1128/jcm.38.6.2271-2277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens CB, Szalanski AL. Filter paper for preservation, storage, and distribution of insect and pathogen DNA samples. Journal of medical entomology. 2005;42:709–711. doi: 10.1093/jmedent/42.4.709. [DOI] [PubMed] [Google Scholar]