Abstract
A molecular pathway analysis has been performed in order to complement previous genetic investigations on Schizophrenia. 4,486 Schizophrenic patients and 4,477 controls served as the investigation sample. 3,521 Bipolar patients and 3,195 controls served as replication sample. A molecular pathway associated with the neuronal pruning activity was found to be enriched in subjects with Schizophrenia compared to controls. HLA-C and HLA-DRA had more SNPs associated with both Schizophrenia and Bipolar Disorder than expected by chance.
Keywords: Schizophrenia, Pathway Analysis, Pruning
1. Introduction
Schizophrenia (SCZ) is a complex disorder ranking at the top ten causes of long-term disability worldwide (Beers, 2004). SCZ runs in families (Sullivan et al., 2003) and a number of independent investigations confirmed that genetic variations may be associated with SCZ (Hosak et al., 2012; Li et al., 2010; Rujescu et al., 2009; Shi et al., 2009; Stefansson et al., 2009; Walker et al., 2010; Williams et al., 2011). However, the complete picture of its genetic background remains elusive (O’Connell et al., 2011). Separating the impact of common genetic variants with a small effect towards SCZ from the false positive associations (Cantor et al., 2010) may prove to be difficult. A molecular pathway analysis may help by taking into account several variations harbored by different genes per time, clustered in consistent molecular groups. This perspective may successfully capture the signal of a number of genetic variations with a small single effect. This approach is consistent with the polygenetic nature of SCZ and it may reveal some aspects of the molecular basis of the disorder. We undertook a genome-wide molecular pathway analysis to test the hypothesis that the genetic variations associated with SCZ segregate in specific molecular pathways within the genome. The study was conducted in a large sample of subjects with SCZ and controls (n=8963) and then replicated in a large sample (n=6716) of subjects with Bipolar Disorder (BD). The shared genetic background of the two disorders (Cross-Disorder Group of the Psychiatric Genomics, 2013; Ivleva et al., 2010) is the rationale of this replication.
2. Methods
2.1. Pathway analysis: a tool for the study of polygenic disorders
A large pool of genes harboring variations with little impact on the risk for SCZ may prove difficult to be identified in common genetic association approaches. An entire biological process rather than single mutations in one or more random genes were then tested in the present study, in order to cluster variations with small impact on SCZ in consistent molecular pathways, so that the genetic effect on the risk for SCZ may be more easily recognized. The alteration of a biological process may be caused by a single mutation at the level of one of its nodal point or by multiple mutations with little effects on different sets of genes with a more marginal role in the pathway. This could explain the considerable amount of genes found associated with Schizophrenia and Bipolar Disorder, along with the inconsistent findings in literature. The pathway analysis stresses this hypothesis. Group of genes were then clustered in molecular networks. The resulting Network as a whole was then tested as a genetic risk factor for SCZ, and the finding replicated in BP subjects.
2.2. Flowchart of the study
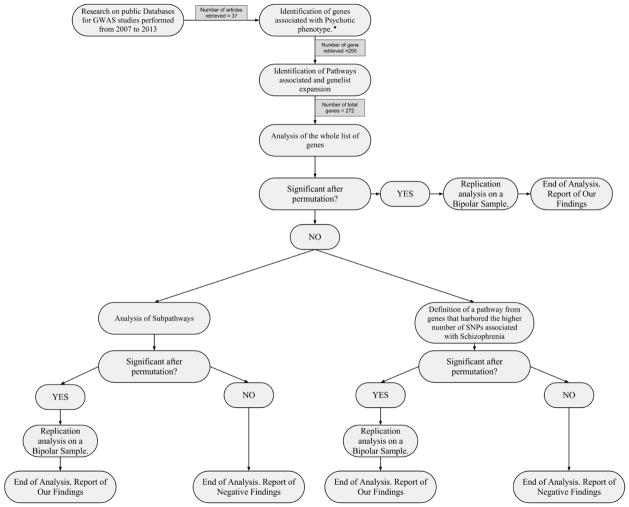
The study was developed as shown in the following diagram (Fig. 1).
Fig 1.
In the above figure is presented the flowchart of our analyses. Each node indicate a different stage of our work and the possible development of the analyses according to the data obtained at each stage.
*Psychotic phenotype: psychotic symptoms included any positive or negative schizophrenic symptoms in absence of organic disorder
The key points of the study are shown in step 3 and 4 - the genes extracted from literature were used to create a molecular network, that was then enriched and tested for association with SCZ; and point 5b - where the genes that held the highest number of variations associated with SCZ (at least ten times the expected number by chance) were clustered in highly significant clusters, enriched and re-submitted to the permutation analyses.
2.3. Dataset
Data of the SCZ and BD samples under investigation was retrieved from public NIHM databases. Table S1 in supplementary section reports the characteristics of the samples.
2.4. Genetic quality control
Genetic quality control included relatedness testing and principal components analyses. Identical subjects (pi-hat > 0.9) were identified and one was included after random choice. Relatives (pi-hat > 0.2) were excluded as well, by randomly choosing one representative. Principal component estimation was done with the non-related subset of individuals. The population structure and the study of origin were chosen as covariates along with the gender.
2.5. Imputation
Imputation was run for the genes that belong to the pathway under analysis. The data of HapMap 1000 genomes database on CEU population served for the imputation. IMPUTE2 (Howie et al., 2009) was used to impute the missing genetic information.
2.6. Pathway enrichment analysis
The genomic sections that contain the genes included in the selected networks were identified through PLINK annotation. These sections were imputed, checked for quality (info>0.9) and pruned (r2>0.2) and their association with the phenotype (schizophrenic patients vs controls or bipolar patients vs controls) tested (see supplementary materials). The prevalence of variations significantly (p<0.01) associated with the outcome in the pathway under analysis and randomly generated pathways were tested for a significant different distribution (Fisher exact test). Fisher test identifies the significant clustering of SNPs associated with the phenotype in the original pathway by comparing the distribution of SNPs in two pathways, the original and a random generated one. This operation was repeated 106 times (permutation) selecting different groups of random SNPs as a control group. 106 Fisher Tests where then conducted and the permuted p was extracted according to Phipson and Smyth (2010).
2.7. Toolset for the analysis
Cytoscape (Saito et al., 2012) and the related add-on GeneMania (Warde-Farley et al., 2010; Zuberi et al., 2013) were used to analyze and enrich the set of genes initially chosen from literature. The add-on MCode served to identify a number of 8 sub-clusters. Executables (BASH language) were written for operationalize the analyses (available upon request). R (R Core Team, 2014), PLINK (International Schizophrenia et al., 2009; Purcell et al., 2007), SNPTESTv2.3.0 (Marchini et al., 2007) and IMPUTE2 were part of the executables and served for the analyses.
2.8. Testing the hypothesis
This analysis was conducted in a sample of GWAS that partially overlapped those used for the selection of the candidate pathways. Apparently, this is like searching a confirmation of a finding starting from the same specimen that provided that result. In this case though, we used a method for analyzing the data that differs from the methods used in the GWAS that provided the results from which we started. Thus, the replication of a set of findings by using this new method can be used as a validation of the method itself. In particular, we expected to validate the results from the larger GWAS published so far that used a sample partially overlapping the one investigated here. If confirmed, this finding would pave the way to the use of this method in new independent samples. Moreover, our analysis differs from previous publications in that we analyzed a mega-sample that derived from the merging of individual samples.
3. Results
Lambda values did not show evidence of major population stratification factors neither in SCZ - Lambda of 1.04 (Standard Error[SE] = 4.15e-05; Lambda1000 = 1.02; SE = 1.50e-05); nor in the BD - Lambda = 1.23 (SE = 4.42e-05; Lambda1000 = 1.02; SE = 4.19e-06). The observed inflation factor in the total sample was interpreted as indicative of a large number of weakly associated SNPs consistent with the disease’s polygenic inheritance (Schizophrenia Psychiatric Genome-Wide Association Study, 2011).
The network obtained in step 3 of the flowchart (Fig. 1) was significantly associated with SCZ (permutated(106) p value = 0.037).
From the clusters elaborated in point 5b of the flowchart (Fig. 1), the one associated with the activity of HLA-DRA and HLA-C were found to have more variations associated with the phenotype than expected by chance. These systems are involved in pruning in the CNS (Faissner et al., 2010; Stephan et al., 2012). HLA-DRA was found to harbor more variations than expected by chance in the samples: 19 out of 108 in the SCZ sample (17% versus the expected 1%), and 2 out of 89 in the BD sample (2% versus the expected 1%). HLA-C was also found to harbor more significant variations than expected by chance: 18 out of 73 in the SCZ sample (24% versus the expected 1%), and 2 out of 62 in the BD sample (3% versus the expected 1%). Rs9268853 and rs9268858 were significantly associated with the phenotypes in both samples. The minor allele was more represented in cases than in controls in both samples. The additional pathways investigated showed no significant associations.
4. Discussion
Consistently with previous findings in literature (Gilvarry et al., 1996; Muller et al., 1999a; Rothermundt et al., 2001; Strous and Shoenfeld, 2006), we found evidence of an immune/autoimmune component in SCZ.
The immune hypothesis of SCZ has gained credibility thanks to studies performed in recent years (Krause et al., 2012; Saetre et al., 2007; Sequeira et al., 2012) and especially after the detection of novel functions of the immune molecules in the brain and bidirectional cross-talk between the Immune System(IS) and the Central Nervous System (CNS) (Tian et al., 2012).
Several studies (Axelsson et al., 1982; Karanikas, 2011; Muller et al., 1999b; Schwarz et al., 2000) that have reported an impaired blood-brain barrier in SCZ subjects, suggest that psychotic phenotype may be caused by degenerative immune-processes, similarly to Multiple Sclerosis (MS) (Amital and Shoenfeld, 1993; Ruutiainen et al., 1981).
Moreover, i) immune-inflammatory genes are up-regulated in the CNS (Saetre et al., 2007); ii) immune-inflammatory events have a critical role in the remodeling of glutamatergic synapses (Fourgeaud and Boulanger, 2010) whose disruption increase the risk for SCZ; iii) Immune System modulates the synaptic function (Schmitt et al., 2011).
Finally, closer to the interest of this paper, rs9268853 and rs9268858, two of the SNPs we found to be significantly associated with both SCZ and BD, are also associated with MS (NCBI dataset).
Recent studies indicate an increased microgliosis-astrogliosis ratio in SCZ (Wierzba-Bobrowicz et al., 2005), pointing that microglia cells may play an active role in the molecular processes involved with this disease.
Activated Microglia elements can act as antigen-presenting cells in CNS. These cells normally express low levels of MHCII proteins but pathological or inflammatory conditions up-regulate their expression (Kreutzberg, 1996; O’Keefe et al., 2002), possibly altering Microglia functions.
Considering the role of Microglia in neurodevelopment (Alliot et al., 1999) and in neuronal remodeling mechanics, including apoptosis, axon remodeling and synaptic pruning (Pont-Lezica et al., 2011), this activation could cause serious consequences in the shape and functions of neuronal nets. This scenario is consistent with typical deficits of SCZ and is also consistent with the age at onset of the disease, early adulthood, which is characterized by an intense pruning activity (Paus et al., 2008). This mechanism is also consistent with a recent hypothesis that underline the relevance of neuronal pruning in the development both SCZ and BD (Strakowski et al., 2012).
4.1. BD replication
SCZ and BD may share a relevant part of their genetic background (Shao and Vawter, 2008). Most association studies linking the MHC region with BD involve the classical HLA genes. Modrego and Ferrández (2000) observed an increased frequency of HLA-DR2 and HLA-DR3 in patients with BD. In 2009, the International Schizophrenia Consortium detected a hotspot on chromosome 6p22, in which are located HLA genes, with a highly significant association with both SCZ and BD (International Schizophrenia et al., 2009). Consistently with this set of evidence, we found that the same genes (HLA-C and HLA-DRA) harbored more variations associated with the disorder than expected by chance in both the SCZ and BD samples.
4.2. Limitations
The analysis is based on the knowledge of the variants associated with the disease, and the enrichment was conducted starting from the genes known to be associated with SCZ. This means that this approach will explore only pathways linked with the first selection of genes. As a result, this method cannot provide new unexpected genes associated with SCZ, even though it is designed to enlarge the list of possible candidates in consistent metabolic pathways. Further, it has to be noted that our positive findings in HLA region may be influenced by the presence of LD pairs. HLA region is known for having a very complex LD structure that is not removed by normal pruning. We tried to limit this bias using a very stringent cutoff, however some LD pairs may have survived. Our method has proven to provide results consistent with the previous investigations and more classic GWAS approaches but it adds relevant information (Debnath et al., 2013; Drago et al., 2014; Schizophrenia Working Group of the Psychiatric Genomics, 2014). Although further tests are needed to completely validate this approach, the consistency we found between the data obtained with this method compared with more classic analyses, suggests this strategy as a possible useful complement in the study of the genetics of psychiatric disorders in independent samples.
Supplementary Material
Highlights.
A biomolecular network related to schizophrenia was identified from literature
The network was clustered in sub-pathways. Each pathway was independently analyzed
“Inflammation and neuronal pruning” pathway was associated with psychotic phenotype
Acknowledgments
Thanks to the NIMH Center for Collaborative Genetic Studies on Mental Disorders and the Psychiatric GWAS Consortium (PGC) for providing their full sample GWAS results, individual quality controlled genotypes, and individual level dosage data for distribution to qualified investigators. We are deeply grateful to the NIMH, NARSAD, and the Netherlands Genetic Computing Cluster for their sponsorship of the PGC.
Footnotes
Conflict of Interest
The Authors declare that they have no conflicts of interest.
Contributors
All authors contributed to and have approved the final manuscript. In particular: Authors Antonio Drago, Concetta Crisafulli and Marco Calabrò designed the study. Author Marco Calabrò managed the literature searches and the analyses and wrote the first draft of the manuscript. Authors Antonio Drago and Concetta Crisafulli managed the Statistical analyses and wrote the protocol. Author Alessandro Serretti supervised the study.
Additional information about methods used in this paper may be found on supplementary material.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res. 1999;117 (2):145–152. doi: 10.1016/s0165-3806(99)00113-3. [DOI] [PubMed] [Google Scholar]
- Amital H, Shoenfeld Y. Autoimmunity and schizophrenia: an epiphenomenon or an etiology? Isr J Med Sci. 1993;29 (9):593–597. [PubMed] [Google Scholar]
- Axelsson R, Martensson E, Alling C. Impairment of the blood-brain barrier as an aetiological factor in paranoid psychosis. Br J Psychiatry. 1982;141:273–281. doi: 10.1192/bjp.141.3.273. [DOI] [PubMed] [Google Scholar]
- Beers MH, Berkow R. The Merck Manual of Diagnosis and Therapy. Whitehouse Station, NJ: Merck Research Laboratories; 2004. Psychiatric Emergencies. [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86 (1):6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics, C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381 (9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath M, Cannon DM, Venkatasubramanian G. Variation in the major histocompatibility complex [MHC] gene family in schizophrenia: associations and functional implications. Prog Neuropsychopharmacol Biol Psychiatry. 2013;42:49–62. doi: 10.1016/j.pnpbp.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Drago A, Giegling I, Schafer M, Hartmann AM, Konte B, Friedl M, Serretti A, Rujescu D. Genome-wide association study supports the role of the immunological system and of the neurodevelopmental processes in response to haloperidol treatment. Pharmacogenet Genomics. 2014;24 (6):314–319. doi: 10.1097/FPC.0000000000000052. [DOI] [PubMed] [Google Scholar]
- Faissner A, Pyka M, Geissler M, Sobik T, Frischknecht R, Gundelfinger ED, Seidenbecher C. Contributions of astrocytes to synapse formation and maturation - Potential functions of the perisynaptic extracellular matrix. Brain Res Rev. 2010;63 (1–2):26–38. doi: 10.1016/j.brainresrev.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Fourgeaud L, Boulanger LM. Role of immune molecules in the establishment and plasticity of glutamatergic synapses. Eur J Neurosci. 2010;32 (2):207–217. doi: 10.1111/j.1460-9568.2010.07342.x. [DOI] [PubMed] [Google Scholar]
- Gilvarry CM, Sham PC, Jones PB, Cannon M, Wright P, Lewis SW, Bebbington P, Toone BK, Murray RM. Family history of autoimmune diseases in psychosis. Schizophr Res. 1996;19 (1):33–40. doi: 10.1016/0920-9964(95)00045-3. [DOI] [PubMed] [Google Scholar]
- Hosak L, Silhan P, Hosakova J. Genome-wide association studies in schizophrenia, and potential etiological and functional implications of their results. Acta Medica (Hradec Kralove) 2012;55 (1):3–11. doi: 10.14712/18059694.2015.67. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5 (6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia, C. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460 (7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva EI, Morris DW, Moates AF, Suppes T, Thaker GK, Tamminga CA. Genetics and intermediate phenotypes of the schizophrenia--bipolar disorder boundary. Neurosci Biobehav Rev. 2010;34 (6):897–921. doi: 10.1016/j.neubiorev.2009.11.022. [DOI] [PubMed] [Google Scholar]
- Karanikas EP. Psycho-immunological mechanisms in schizophrenia. Psychiatriki. 2011;22 (1):43–52. [PubMed] [Google Scholar]
- Krause D, Wagner J, Matz J, Weidinger E, Obermeier M, Riedel M, Gruber R, Schwarz M, Mueller N. Monocytic HLA DR antigens in schizophrenic patients. Neurosci Res. 2012;72 (1):87–93. doi: 10.1016/j.neures.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Kreutzberg GW. Microglia: a sensor for pathological events in the CNS. Trends Neurosci. 1996;19 (8):312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- Li T, Li Z, Chen P, Zhao Q, Wang T, Huang K, Li J, Li Y, Liu J, Zeng Z, Feng G, He L, Shi Y. Common variants in major histocompatibility complex region and TCF4 gene are significantly associated with schizophrenia in Han Chinese. Biol Psychiatry. 2010;68 (7):671–673. doi: 10.1016/j.biopsych.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39 (7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Modrego PJ, Ferrandez J. Familial multiple sclerosis with repetitive relapses of manic psychosis in two patients (mother and daughter) Behav Neurol. 2000;12 (4):175–179. doi: 10.1155/2000/685948. [DOI] [PubMed] [Google Scholar]
- Muller N, Riedel M, Ackenheil M, Schwarz MJ. The role of immune function in schizophrenia: an overview. Eur Arch Psychiatry Clin Neurosci. 1999a;249(Suppl 4):62–68. doi: 10.1007/pl00014187. [DOI] [PubMed] [Google Scholar]
- Muller N, Riedel M, Hadjamu M, Schwarz MJ, Ackenheil M, Gruber R. Increase in expression of adhesion molecule receptors on T helper cells during antipsychotic treatment and relationship to blood-brain barrier permeability in schizophrenia. Am J Psychiatry. 1999b;156 (4):634–636. [PubMed] [Google Scholar]
- O’Connell G, Lawrie SM, McIntosh AM, Hall J. Schizophrenia risk genes: Implications for future drug development and discovery. Biochem Pharmacol. 2011;81 (12):1367–1373. doi: 10.1016/j.bcp.2010.11.009. [DOI] [PubMed] [Google Scholar]
- O’Keefe GM, Nguyen VT, Benveniste EN. Regulation and function of class II major histocompatibility complex, CD40, and B7 expression in macrophages and microglia: Implications in neurological diseases. J Neurovirol. 2002;8 (6):496–512. doi: 10.1080/13550280290100941. [DOI] [PubMed] [Google Scholar]
- Paus T, Keshavan M, Giedd JN. Why do many psychiatric disorders emerge during adolescence? Nat Rev Neurosci. 2008;9 (12):947–957. doi: 10.1038/nrn2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Smyth GK. Permutation P-values should never be zero: calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol. 2010;9:Article39. doi: 10.2202/1544-6115.1585. [DOI] [PubMed] [Google Scholar]
- Pont-Lezica L, Bechade C, Belarif-Cantaut Y, Pascual O, Bessis A. Physiological roles of microglia during development. J Neurochem. 2011;119 (5):901–908. doi: 10.1111/j.1471-4159.2011.07504.x. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81 (3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing 2014 [Google Scholar]
- Rothermundt M, Arolt V, Bayer TA. Review of immunological and immunopathological findings in schizophrenia. Brain Behav Immun. 2001;15 (4):319–339. doi: 10.1006/brbi.2001.0648. [DOI] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, Picchioni M, Vassos E, Ettinger U, Bramon E, Murray R, Ruggeri M, Tosato S, Bonetto C, Steinberg S, Sigurdsson E, Sigmundsson T, Petursson H, Gylfason A, Olason PI, Hardarsson G, Jonsdottir GA, Gustafsson O, Fossdal R, Giegling I, Moller HJ, Hartmann AM, Hoffmann P, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Djurovic S, Melle I, Andreassen OA, Hansen T, Werge T, Kiemeney LA, Franke B, Veltman J, Buizer-Voskamp JE, Investigators G, Sabatti C, Ophoff RA, Rietschel M, Nothen MM, Stefansson K, Peltonen L, St Clair D, Stefansson H, Collier DA. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18 (5):988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruutiainen J, Arnadottir T, Molnar G, Salmi A, Frey H. Myelin basic protein antibodies in the serum and CSF of multiple sclerosis and subacute sclerosing panencephalitis patients. Acta Neurol Scand. 1981;64 (3):196–206. doi: 10.1111/j.1600-0404.1981.tb04399.x. [DOI] [PubMed] [Google Scholar]
- Saetre P, Emilsson L, Axelsson E, Kreuger J, Lindholm E, Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9 (11):1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study, C. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43 (10):969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511 (7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Hasan A, Gruber O, Falkai P. Schizophrenia as a disorder of disconnectivity. Eur Arch Psychiatry Clin Neurosci. 2011;261(Suppl 2):S150–154. doi: 10.1007/s00406-011-0242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz MJ, Riedel M, Ackenheil M, Muller N. Decreased levels of soluble intercellular adhesion molecule-1 (sICAM-1) in unmedicated and medicated schizophrenic patients. Biol Psychiatry. 2000;47 (1):29–33. doi: 10.1016/s0006-3223(99)00206-1. [DOI] [PubMed] [Google Scholar]
- Sequeira PA, Martin MV, Vawter MP. The first decade and beyond of transcriptional profiling in schizophrenia. Neurobiol Dis. 2012;45 (1):23–36. doi: 10.1016/j.nbd.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Vawter MP. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol Psychiatry. 2008;64 (2):89–97. doi: 10.1016/j.biopsych.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460 (7256):753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA Genetic, R., Outcome in, P. Common variants conferring risk of schizophrenia. Nature. 2009;460 (7256):744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AH, Barres BA, Stevens B. The complement system: an unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- Strakowski SM, Adler CM, Almeida J, Altshuler LL, Blumberg HP, Chang KD, DelBello MP, Frangou S, McIntosh A, Phillips ML, Sussman JE, Townsend JD. The functional neuroanatomy of bipolar disorder: a consensus model. Bipolar Disord. 2012;14 (4):313–325. doi: 10.1111/j.1399-5618.2012.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous RD, Shoenfeld Y. Schizophrenia, autoimmunity and immune system dysregulation: a comprehensive model updated and revisited. J Autoimmun. 2006;27 (2):71–80. doi: 10.1016/j.jaut.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60 (12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Tian L, Ma L, Kaarela T, Li Z. Neuroimmune crosstalk in the central nervous system and its significance for neurological diseases. J Neuroinflammation. 2012;9:155. doi: 10.1186/1742-2094-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RM, Christoforou A, Thomson PA, McGhee KA, Maclean A, Muhleisen TW, Strohmaier J, Nieratschker V, Nothen MM, Rietschel M, Cichon S, Morris SW, Jilani O, Stclair D, Blackwood DH, Muir WJ, Porteous DJ, Evans KL. Association analysis of Neuregulin 1 candidate regions in schizophrenia and bipolar disorder. Neurosci Lett. 2010;478 (1):9–13. doi: 10.1016/j.neulet.2010.04.056. [DOI] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38 (Web Server issue):W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzba-Bobrowicz T, Lewandowska E, Lechowicz W, Stepien T, Pasennik E. Quantitative analysis of activated microglia, ramified and damage of processes in the frontal and temporal lobes of chronic schizophrenics. Folia Neuropathol. 2005;43 (2):81–89. [PubMed] [Google Scholar]
- Williams HJ, Norton N, Dwyer S, Moskvina V, Nikolov I, Carroll L, Georgieva L, Williams NM, Morris DW, Quinn EM, Giegling I, Ikeda M, Wood J, Lencz T, Hultman C, Lichtenstein P, Thiselton D, Maher BS, Malhotra AK, Riley B, Kendler KS, Gill M, Sullivan P, Sklar P, Purcell S, Nimgaonkar VL, Kirov G, Holmans P, Corvin A, Rujescu D, Craddock N, Owen MJ, O’Donovan MC Molecular Genetics of Schizophrenia Collaboration International Schizophrenia Consortium, S.-p.G. Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol Psychiatry. 2011;16 (4):429–441. doi: 10.1038/mp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, Morris Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41 (Web Server issue):W115–122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.