Abstract
Automatic processing of magnetic resonance images is a vital part of neuroscience research. Yet even the best and most widely used medical image processing methods will not produce consistent results when their input images are acquired with different pulse sequences. Although intensity standardization and image synthesis methods have been introduced to address this problem, their performance remains dependent on knowledge and consistency of the pulse sequences used to acquire the images. In this paper, an image synthesis approach that first estimates the pulse sequence parameters of the subject image is presented. The estimated parameters are then used with a collection of atlas or training images to generate a new atlas image having the same contrast as the subject image. This additional image provides an ideal source from which to synthesize any other target pulse sequence image contained in the atlas. In particular, a nonlinear regression intensity mapping is trained from the new atlas image to the target atlas image and then applied to the subject image to yield the particular target pulse sequence within the atlas. Both intensity standardization and synthesis of missing tissue contrasts can be achieved using this framework. The approach was evaluated on both simulated and real data, and shown to be superior in both intensity standardization and synthesis to other established methods.
Keywords: magnetic resonance imaging, brain, synthesis, pulse sequence
1. Introduction
Magnetic resonance (MR) imaging (MRI) is the preferred diagnostic and research imaging modality for soft tissue contrast. It is a very versatile modality in part because MR pulse sequences allow for diverse manipulations of the nuclear magnetic spins, thus providing a vast amount of complementary information in neuroimaging. For example, T1-weighted (T1-w) images are typically preferred for tissue segmentation (Styner et al., 2000; Prastawa et al., 2004; Roy et al., 2012) and cortical reconstruction (Dale and Fischl, 1999; Han et al., 2004; Shiee et al., 2014), whereas Double Spin Echo (DSE) and FLAIR (Fluid Attenuated Inversion Recovery) sequences are useful to delineate tissue abnormalities like white matter lesions (Shiee et al., 2010).
The versatility of MRI also poses a problem. Patient scanner time is finite and thus decisions are made about which subset of the many available pulse sequences to acquire. Another factor in this decision is the expense of scanning and the long scan time associated with various sequences, T2-weighted (T2-w) images for example. A confound in the acquired data is the inhomogeneous quality of the data due to the different imaging requirements of each sequence. For example, the DSE sequence is generally acquired at a much lower resolution than a T1-w magnetization prepared rapid gradient echo (MPRAGE) sequence because of the longer repetition time (TR). Sequences like FLAIR suffer from artifacts that are not present in other sequences (Stuckey et al., 2007), contributing to problems with their use in multimodal analysis. More fundamentally, the imaging data can be corrupted due to patient motion or inappropriate parameter settings. Our work is focused on improving the utility of multimodal data through image synthesis, either by restoring corrupt data or by standardizing the intensity of existing data.
By image synthesis we mean learning and applying an intensity transformation to MR images in order to produce images that perform better in various image processing tasks. The resultant synthetic images could belong to any pulse sequence we choose to synthesize. Image synthesis, in the broadest sense, is already used in nearly every image processing pipeline—for example in intensity inhomogeneity correction or intensity scaling. Intensity standardization transforms the intensities of a given subject image to a reference image, typically of the same (or similar) pulse sequence. It is a special case of image synthesis, in which the synthesized image is of the same (or similar) pulse sequence. Synthesized images are not meant to be used for diagnostic purposes or to replace scanning subjects. Rather, they are intended to facilitate image analysis for the extraction of clinical or scientific information.
Intensity standardization has long been an important problem for MR image processing, and many solutions have been proposed. Unlike x-ray computed tomography (CT), MRI does not have a consistent image intensity scale for different tissues. Though this does not pose a problem for diagnostic purposes, automatic image processing algorithms such as segmentation algorithms are known to be inconsistent as a result (Nyúl and Udupa, 1999). Since there is no consistent anatomical meaning to the numerical value of an intensity, it is difficult to directly compare MR data acquired at different sites and on different scanners. Even data acquired in the same machine for the same subject can differ in intensity characteristics. Intensity standardization using ideas of image synthesis can assist in consistent processing of such data (Roy et al., 2013a).
Image synthesis of an alternative modality has been shown to be useful in many image analysis tasks. Iglesias et al. (2013) showed that it is better to register a T2-w image to a synthetic T2-w image created from a T1-w image, than to directly register a T2-w image to the T1-w image. A similar application to correct geometric distortion in diffusion tensor images using synthetic T2-w images was demonstrated by Roy et al. (2013a). In Jog et al. (2014a) it was shown that datasets with a missing FLAIR image could be augmented by creating a synthetic FLAIR image. Lesion segmentation using synthetic FLAIRs was shown to be equivalent to and more consistent than the lesion segmentation using real FLAIRs. The consistency was attributed to the fact that the synthetic FLAIRs were produced using a common set of atlas or training images. The lesion segmentation markedly improved when real FLAIR images with significant imaging artifacts were replaced with synthetic FLAIRs. In Jog et al. (2014b), a method to create super-resolution images using image synthesis ideas was presented. This significantly improved the image quality and resolution of inherent low resolution data acquired using the DSE or FLAIR pulse sequences.
Image synthesis in general has gained significant attention in the medical imaging community in the last seven years (Rousseau, 2008; Roy et al., 2010a,b; Rousseau, 2010; Roy et al., 2011; Roohi et al., 2012; Jog et al., 2013a; Rousseau and Studholme, 2013; Konukoglu et al., 2013; Burgos et al., 2013; Ye et al., 2013; Iglesias et al., 2013; Roy et al., 2013a,b, 2014). A classical registration-based solution to this problem was presented in (Miller et al., 1993): Given a subject image b1 with contrast
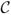 1 and a pair of co-registered atlas images a1 and a2 of contrasts
1 and a pair of co-registered atlas images a1 and a2 of contrasts
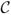 1 and
1 and
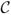 2, respectively, a1 is registered to b1 using a deformable registration algorithm, and the transformation is then applied to a2 to produce the synthetic image b̂2 with contrast
2, respectively, a1 is registered to b1 using a deformable registration algorithm, and the transformation is then applied to a2 to produce the synthetic image b̂2 with contrast
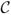 2. The method demonstrated synthesis of positron emission tomography images from MR images. An extension of this approach, described by Burgos et al. (2013), uses multiple, aligned pairs of MR and CT images as atlases and performs multi-atlas deformable registration to the subject with intensity fusion to synthesize a subject CT image from a subject MR. This approach produces synthetic CT images from which the attenuation coefficients are learned and used for PET reconstruction. A more general, intensity transformation-based approach to image synthesis is described by Hertzmann et al. (2001), wherein an image transformation, which can be defined as an application of an image filter, was learned. Given training data the filtered image was synthesized from the unfiltered one by applying the learned transformation. This approach, known as image analogies, has seen use in MRI synthesis (Iglesias et al., 2013). More recently, the work by Roy et al. (2011, 2013a) handles image synthesis using a sparse reconstruction of a dictionary of atlas image patches.
2. The method demonstrated synthesis of positron emission tomography images from MR images. An extension of this approach, described by Burgos et al. (2013), uses multiple, aligned pairs of MR and CT images as atlases and performs multi-atlas deformable registration to the subject with intensity fusion to synthesize a subject CT image from a subject MR. This approach produces synthetic CT images from which the attenuation coefficients are learned and used for PET reconstruction. A more general, intensity transformation-based approach to image synthesis is described by Hertzmann et al. (2001), wherein an image transformation, which can be defined as an application of an image filter, was learned. Given training data the filtered image was synthesized from the unfiltered one by applying the learned transformation. This approach, known as image analogies, has seen use in MRI synthesis (Iglesias et al., 2013). More recently, the work by Roy et al. (2011, 2013a) handles image synthesis using a sparse reconstruction of a dictionary of atlas image patches.
While each of these methods produces a useful, synthetic image under different initial assumptions, they each have certain shortcomings. For instance, deformable registration may fail to yield an accurate registration throughout the whole brain. If the subject images show pathology in normal tissue regions and the atlas images do not, then registration is unable to recreate these differences. Methods based on intensity transformation like image analogies and the sparse reconstruction approach are dependent on the b1 image being correctly intensity standardized with the training data (atlas image a1). These methods also do not take into account the MR image formation process while synthesizing the new image. We would like to improve upon these approaches by using the atlas-based intensity transformation ideas while incorporating the MR image formation process.
Jog et al. (2013a) presented a patch-based regression approach using random forest regression for synthesizing MR images. The synthesis is modeled as a nonlinear regression in terms of a patch in a1 transforming to the corresponding central voxel in a2. This approach, though extremely fast, also requires the subject image to be intensity standardized to the atlas image prior to applying the learned regression and does not account for the MR physics of acquisition. In this paper, we have built upon this random forest regression by synthesizing a new subject image from the available subject image(s) and a set of atlas images. We create a preprocessing framework in which we synthesize images using knowledge of both the imaging physics and information in the atlas. Voxel intensity in MRI is primarily dependent on intrinsic nuclear magnetic resonance (NMR) parameters such as proton density (PD), transverse relaxation time (T2), longitudinal relaxation time (T1) and on pulse sequence parameters such as scanner gain (G0), repetition time (TR), echo times (TE), etc. Given an unseen subject image (and the pulse sequence) we estimate the pulse sequence parameters using information derived solely from the image intensities. We apply this pulse sequence with the estimated parameters to quantitative PD, T1, and T2 images in our atlas. Thus, a new atlas image with the same imaging characteristics as the subject image is synthesized. We then learn a nonlinear regression (Jog et al., 2013a) between this standardized atlas image and the desired, target atlas contrast. This learned regression can then be applied to the subject image, thereby synthesizing a subject image with the desired contrast.
The core idea of estimating tissue parameters was previously used in Fischl et al. (2004). However, that approach required the acquisition of very specific pulse sequences—a requirement that our approach does not share. We refer to our method as Pulse Sequence Information-based Contrast Learning On Neighborhood Ensembles (PSI-CLONE)—which we stylize as Ψ-CLONE. We describe our four-step algorithm to perform image synthesis in Section 2. Results on phantom data are in Section 3 and on real data in Section 4; these experiments include scanner intensity standardization and synthesis of T2-w images. In Section 5, we present additional synthesis applications: super-resolution and FLAIR synthesis. In Section 6, there is a discussion about the potential impact of this work and concluding remarks.
2. Method
Let
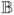 = {b1, b2, … , bm} be the given subject image set, imaged with pulse sequences Γ1, … , Γm. This set can contain images from different pulse sequences such as MPRAGE, spoiled gradient recalled (SPGR), and DSE. Let
= {b1, b2, … , bm} be the given subject image set, imaged with pulse sequences Γ1, … , Γm. This set can contain images from different pulse sequences such as MPRAGE, spoiled gradient recalled (SPGR), and DSE. Let
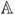 = {a1, a2, … , an} be the atlas collection, with images of contrasts
= {a1, a2, … , an} be the atlas collection, with images of contrasts
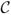 1,
1,
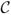 2, … ,
2, … ,
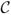 n, generated by pulse sequences Ψ1, … , Ψn, respectively. By pulse sequence we mean particular, named procedures like MPRAGE. By contrast we mean the tissue contrast produced by a pulse sequence, say a T1-weighted contrast or a T2-weighted contrast. For instance, the MPRAGE pulse sequence produces a T1-w contrast image. Since in most cases our data consists of specific pulse sequences producing specific contrasts, these terms can be used interchangeably.
n, generated by pulse sequences Ψ1, … , Ψn, respectively. By pulse sequence we mean particular, named procedures like MPRAGE. By contrast we mean the tissue contrast produced by a pulse sequence, say a T1-weighted contrast or a T2-weighted contrast. For instance, the MPRAGE pulse sequence produces a T1-w contrast image. Since in most cases our data consists of specific pulse sequences producing specific contrasts, these terms can be used interchangeably.
The pulse sequence sets Γ1, … , Γm and Ψ1, … , Ψn need not intersect, which represents an important distinction between Ψ-CLONE and all other atlas-based image synthesis methods. The atlas also contains quantitative PD, T1, and T2 maps, denoted aPD, aT1, and, aT2. Our goal is to synthesize the subject image b̂r, r ∈ {1 … , n}, which is how the subject brain would look had it been imaged with pulse sequence Ψr used to acquire the atlas image ar. The steps of our algorithm are as follows: 1) Estimate the pulse sequence parameters used to acquire bi, i ∈ {1, … , m}; 2) With this estimate, generate abi the atlas image with the same contrast,
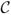 i, as bi; 3) From the expanded atlas collection
i, as bi; 3) From the expanded atlas collection
 ∪ {abi} we learn the nonlinear intensity transformation between contrast
∪ {abi} we learn the nonlinear intensity transformation between contrast
 i and the target contrast image ar (of contrast
i and the target contrast image ar (of contrast
 r), using patch-based random forest regression; 4) The intensity transformation is then applied to bi, generating the subject image b̂r, of the desired contrast. These steps are outlined graphically in Fig. 1 and detailed in the following sections.
r), using patch-based random forest regression; 4) The intensity transformation is then applied to bi, generating the subject image b̂r, of the desired contrast. These steps are outlined graphically in Fig. 1 and detailed in the following sections.
Figure 1.
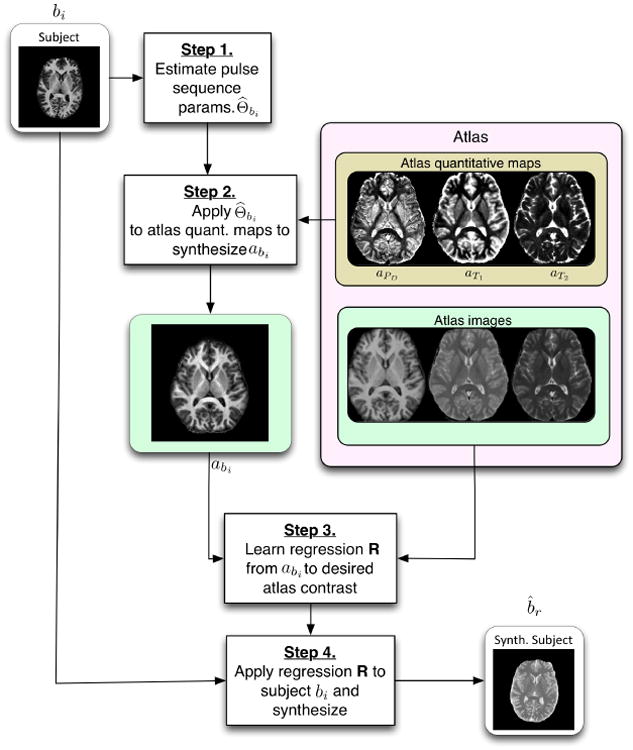
A flow chart of the Ψ-CLONE algorithm.
2.1. Estimation of Subject Pulse Sequence Parameters
The intensity observed at voxel location x in bi is assumed to be a result of the underlying tissue parameters—proton density PD, longitudinal relaxation time T1, and transverse relaxation time T2—denoted by β(x) = [PD(x), T1(x), T2(x)]. The intensity is also a result of the pulse sequence used, Γi, and its imaging parameters denoted Θbi. Thus we denote the imaging equation as,
| (1) |
For the DSE pulse sequence, the equation is
| (2) |
where ΘDSE = {TR, TE1, TE2, GDSE} consists of repetition time TR, two echo times TE1 and TE2 and scanner gain GDSE (Glover, 2004). For the T1-w SPGR sequence the imaging equation is
| (3) |
where ΘS = {TR, TE, θ, GS} consists of repetition time TR, echo time TE, and flip angle θ (Glover, 2004). The imaging equation for the MPRAGE sequence can be approximated from the mathematical formulation calculated by Deichmann et al. (2000) as
| (4) |
where ΘM = {TI, TD, τ, GM} consists of inversion time TI, delay time TD, and the slice imaging time τ (Deichmann et al., 2000). We assume that we know one of these parameters from the image header and estimate the rest by fitting the imaging equation to average tissue intensities.
Given an input subject image, bi, we want to estimate a subset (such as scanner gain, flip angle, echo times) of pulse sequence parameters Θbi of Γbi. We make certain assumptions about the tissues being imaged thereby simplifying the system of equations we need to solve. As the human brain is dominated by three primary tissues, cerebrospinal fluid (CSF), gray matter (GM), and white matter (WM), we can use the known average values of β to solve for the imaging parameters. The mean values of β for CSF, GM, and WM denoted by β̄C, β̄G, and β̄W, respectively have been reported previously for 1.5 T (Kwan et al., 1999) and 3 T MRI (Wansapura et al., 1999). To identify the three tissue classes, we run a simple three-class fuzzy c-means (Bezdek, 1980) algorithm on the T1-w image (bS or bM), and choose voxels with high class memberships (≥0.8) to compute the mean intensities of CSF, GM, and WM in bi as b̄iC, b̄iG, and b̄iW, respectively for i = 1, … , m.
We make the assumption that these mean intensities are a result of the mean tissue parameter values. This relationship is written as,
| (5) |
The only unknown is Θbi, which for DSE type pulse sequences is parametrized by four terms. Similarly, MPRAGE and SPGR pulse sequences (Deichmann et al., 2000; Glover, 2004) have four parameters. Thus, we have three equations (Eqn. 5) and four unknowns, and we can solve this system of equations using Newton's method after assuming knowledge of one of the unknowns. For example, in the SPGR pulse sequence, we assume that the repetition time, TR is known from the image header and the unknowns that are often not well-calibrated in an MR scanner—e.g, flip angle and scanner gain—are estimated. For the given subject image set
 = {b1, b2, … , bm}, we can thus estimate {Θ̂b1, … , Θ̂bm} for each of the subject images at the end of Step 1.
= {b1, b2, … , bm}, we can thus estimate {Θ̂b1, … , Θ̂bm} for each of the subject images at the end of Step 1.
There are many factors that affect the accuracy of a pulse sequence equation, and more unknowns are likely to be actually involved than we may know about. To address this, we have simplified the problem by using theoretical equations or approximations of theoretical equations that describe the basic, functional relationship between the NMR parameters (PD, T1, T2) and the signal intensity. For instance, the signal intensity equation for the MPRAGE pulse sequence, given in Eqn. 4, is an approximation derived from a complex theoretical derivation of a simple MPRAGE sequence by Deichmann et al. (2000). In practice, the MPRAGE sequence implemented on the scanner has many additional parameters that are not accounted for by this derivation. Therefore, in most cases, our estimates of the pulse sequence parameters are not particularly close to the parameters recorded in the image headers. It turns out that this is not important for the problem at hand, since we do not need to know the exact imaging parameters of any given pulse sequence. Instead, we need to be able to generate a realistic synthetic image from our atlas that has the same intensity characteristics as the subject image using the estimated parameters and approximate pulse sequence. Approximate imaging equations and their estimated parameters are sufficient for this purpose.
2.2. Synthesizing a New Atlas Image with Subject Pulse Sequence Estimates
Now we describe Step 2 of the process, as illustrated in Fig. 1. It is unlikely that our atlas would contain an image with the exact pulse sequence parameters Θbi estimated from bi, which is why we synthesize an atlas image with the same parameters. Using the estimated imaging parameters, Θ̂bi, we apply the pulse sequence to the atlas β. The atlas
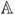 = {a1, a2, … , an} consists of a set of co-registered brain MR images of a single brain with different pulse sequences. It also consists of aPD, aT1, and aT2, the quantitative PD, T1, and T2 maps for the atlas. Thus, we can directly apply the subject pulse sequence, Γi, and its estimated Θ̂bi to synthesize a new atlas image abi. We thereby create an atlas image that looks as if the atlas brain was imaged with the pulse sequence Γi with parameters Θ̂bi. We need this intermediate step so that we can learn the intensity transformation between the subject pulse sequence Γi and the reference pulse sequence Ψr in a common image space, which is the atlas image space.
= {a1, a2, … , an} consists of a set of co-registered brain MR images of a single brain with different pulse sequences. It also consists of aPD, aT1, and aT2, the quantitative PD, T1, and T2 maps for the atlas. Thus, we can directly apply the subject pulse sequence, Γi, and its estimated Θ̂bi to synthesize a new atlas image abi. We thereby create an atlas image that looks as if the atlas brain was imaged with the pulse sequence Γi with parameters Θ̂bi. We need this intermediate step so that we can learn the intensity transformation between the subject pulse sequence Γi and the reference pulse sequence Ψr in a common image space, which is the atlas image space.
In practice, the atlas collection
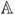 may lack the quantitative PD, T1, and T2 maps—the relaxometry sequence data is generally not available for most clinical data. We can approximately estimate these maps from the images present in the atlas collection by solving for PD, T1, and T2 at each voxel. Since we are estimating three quantities in β(x) = [PD(x), T1(x), T2(x)], we require at least three atlas images, au, av, and aw. From the method described in Section 2.1, we can estimate Θ̂au, Θ̂av, and Θ̂aw. For each voxel x, we have three intensity values from three images, thus leading to three equations,
may lack the quantitative PD, T1, and T2 maps—the relaxometry sequence data is generally not available for most clinical data. We can approximately estimate these maps from the images present in the atlas collection by solving for PD, T1, and T2 at each voxel. Since we are estimating three quantities in β(x) = [PD(x), T1(x), T2(x)], we require at least three atlas images, au, av, and aw. From the method described in Section 2.1, we can estimate Θ̂au, Θ̂av, and Θ̂aw. For each voxel x, we have three intensity values from three images, thus leading to three equations,
| (6) |
This system of simultaneous nonlinear equations can be solved by Newton's method for each voxel to provide us with an estimate β̂(x). The component parts of β̂(x) are [β̂1(x), β̂2(x), β̂3(x)] that are our estimates of [PD(x), T1(x), T2(x)]. Thus, we add the images aβ̂1, aβ̂2, and aβ̂3 to our atlas to represent the PD, T1, and T2 quantitative maps, respectively. This calculation needs to be done only once, during the construction of a suitable atlas. A variant of this approach was used in Jog et al. (2013b) as a step in intensity standardization.
2.3. Learning and Applying Nonlinear Regression on Image Patches
Having synthesized the atlas image abi that has the pulse sequence characteristics of the subject image bi, we next learn the intensity transformation that will convert the intensities in abi to the corresponding intensities in the target atlas image ar. We depict this as Step 3 in Fig. 1. This is achieved through a nonlinear regression by considering the image patches of abi together with the corresponding central voxel intensities in ar. We extract p × q × r sized patches from abi, centered at the vth voxel—in our experiments p = q = r = 3. We stack the 3D patch into a d × 1 = 27 × 1 vector and denote it by fv ∈ ℝd, which we refer to as a feature vector of the vth voxel. The corresponding intensity at the vth voxel of ar is denoted by yv and acts as the dependent variable in our regression; we denote these training data pairs as 〈fv, yv〉. We use patches as intensity features to learn this transformation. A small patch captures the local context at a voxel and ensures spatial smoothness. We could use other synthesis methods like MIMECS (Roy et al., 2013a) to learn this transformation, however we chose to use the random forest regression as it was shown to produce better quality synthetic images at an order of magnitude lower computation time (Jog et al., 2013a).
We use a bagged ensemble of regression trees to learn this nonlinear regression (Breiman, 1996). This standalone regression ensemble for synthesis was previously explored in Jog et al. (2013a). A single regression tree learns a nonlinear regression by partitioning the d-dimensional space. This is done by performing binary comparison splits at each node of the tree, based on a particular attribute value which is compared to a threshold. The tree is built by minimizing the least squares criterion during training. The growth of the tree is limited by fixing the maximum number of vectors allowed at each leaf, in our experiments this was limited to five data vectors. This stops a tree from becoming too deep and hence over-fitting the training data. A single regression tree is considered a weak learner and in general has higher error (Breiman, 1996), therefore we use a bagged ensemble of regression trees (30 in our experiments), which reduces errors by bootstrap aggregation (Breiman, 1996). To create a bootstrapped data set, a training sample is picked at random with replacement N times, where N is the size of the training data, ∼106 in our experiments.
Once the training is complete, the trained regression ensemble transforms intensities of abi to those of ar. This ensemble is used to synthesize the subject image b̂r by extracting image patches from bi and applying the trained regression ensemble to each patch to synthesize the corresponding b̂r voxel intensities, which is the last step, Step 4 in Fig. 1.
Thus, starting with a subject image and a set of atlas images, we estimate the pulse sequence parameters of the subject image, create an additional atlas image by applying those parameters to the atlas quantitative images, learn an intensity transformation from the additional atlas image to the target atlas contrast image using random forest regression, and lastly apply the regression to the given subject image to create a synthetic image of the required contrast. In the following sections, we will describe validation experiments and applications of Ψ-CLONE in different image analysis contexts.
3. Computational Phantom Experiments
The goal of our method is to produce synthetic images that are useful substitutes for real images for image processing tasks. Thus, one aspect of algorithm performance evaluation consists of using image quality metrics to compare synthetic images with known ground truth images. The ground truth images are simulated with known pulse sequence parameters on brain voxels with known NMR parameters. We compare our synthetic images with these known simulated images to validate our method in a controlled experimental setting. In this section, we used the Brainweb image phantom for intensity standardization and synthesis of T2-w images from T1-w images.
3.1. Brainweb SPGR: Estimating abi
In this validation experiment the atlas set
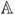 consisted of images from the Brainweb (Cocosco et al., 1997) phantom, consisting of:
consisted of images from the Brainweb (Cocosco et al., 1997) phantom, consisting of:
a1: SPGR image (1.5 T, TR = 18 ms, α = 30°, TE = 10 ms) with 0% noise,
aT1: Quantitative T1 map derived from two different SPGR images, with two different flip angles (TR = 100 ms, TE = 15 ms, α1 = 15°, and α2 = 30°) using the dual flip angle method (Bottomley and Ouwerkerk, 1994),
aT2: Quantitative T2 map derived from a DSE sequence (TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms) by the two point method (Landman et al., 2011),
aPD: Quantitative PD map derived from the reference SPGR and the DSE images using the method described in Section 2.2.
The aim of this experiment is to validate Step 1 and Step 2 of Ψ-CLONE. We do this by taking several images as a potential subject image and carry out just the first two steps of Ψ-CLONE which results in the image abi. Specifically the subject images were:
b1: SPGR image (1.5 T, TR = 18 ms, α = {15°, 30°, 45°, 60°, 75°, 90°}, TE = 10 ms) with 0% noise. (Parameters, excepting TR, not provided to the algorithm)
We note that the subject imaging parameters are shown for the sake of the reader and are not provided to the algorithm (except TR).
The first step of Ψ-CLONE estimates the imaging parameters from the subject image. As both the atlas and subject images come from the same phantom—they have the same phantom anatomy and NMR parameters—this allows us to validate Step 2, in which we create a synthetic atlas image using the pulse sequence parameters of the subject image. Since the anatomy of subject and atlas is the same in this special case, we can directly compare the synthetic atlas image with the subject image to validate if the pulse sequence parameters are producing an identical image. We would prefer to compare the estimated pulse sequence parameters with the known true parameters. Unfortunately, this comparison is not suitable for all parameters because we use theoretical equations or their approximations to estimate the pulse sequence parameters and the actual simulation or scanner implementation can be more complex with a larger number of parameters. As a small example, we carried out an experiment on the Brainweb phantom data to measure the error between the true parameters and the estimated parameters. We simulated different SPGR images by keeping the repetition time TR = 18 ms fixed, and varying the flip angle. We next estimated the imaging parameters of these images by the method described above. The estimated flip angles for these images and the true flip angles used to simulate these images are recorded in Table 1. As can be seen, these estimates are close to but not equal to the truth. This error increases in realistic settings.
Table 1.
Flip angles used in Brainweb SPGR simulation vs estimated Flip angles after fitting.
| True flip angle (°) | Estimated flip angle (°) |
|---|---|
| 30 | 32.08 |
| 45 | 48.70 |
| 60 | 65.08 |
| 75 | 84.79 |
| 90 | 106.57 |
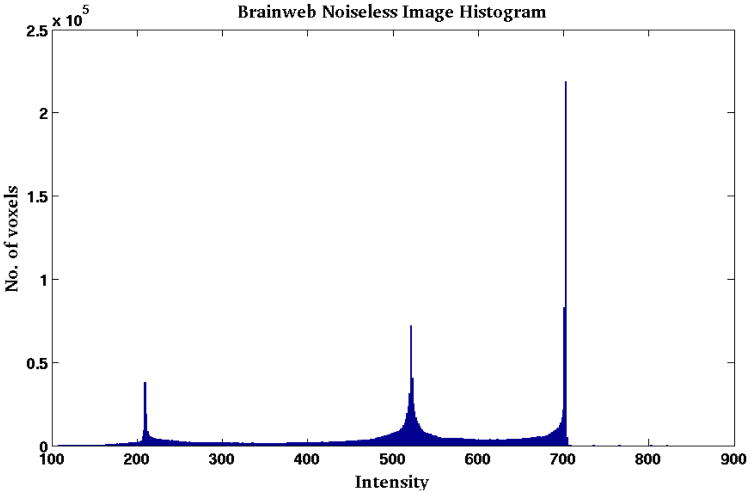
Hence we focus on the images that these parameter estimates create when applied to the estimated PD, T1, and T2 values. We compute the root mean squared error (RMSE) and peak signal to noise ratio (PSNR) between the new atlas image (abi) and the subject image (bi), as shown in Table 2. Figure 2 shows the input b1 image and the estimated ab1, along with the difference image for the case of α= 45° (third column of Table 2). The images are very similar to each other with small differences at tissue boundaries. The differences are only visible at the boundaries because, as the histogram in Fig. 4 shows, Brainweb phantom images with no noise typically have very homogenous intensity distribution in each of the tissue classes (WM, GM, CSF). Hence the error inside the tissues is very close to zero, but is slightly higher at the intermediate intensity voxels that are closer to the boundaries. We note that RMSE and PSNR are computed over the non-zero voxels in the image. RMSE is reported in terms of percentage error with respect to the maximum intensity in the image. The high PSNR values in Table 2 along with the visual result in Fig. 2 confirm that the theoretical pulse sequence equations and the underlying quantitative PD, T1, and T2 maps produce images that are close to the ground truth images, thus validating the first two steps of our algorithm.
Table 2.
RMSE between ab1 and b1 (as a % w.r.t max intensity) and PSNR (dB) values for Brainweb T1-w SPGR atlas synthesis for subject pulse sequences with varying flip angles.
| Flip angle | 15° | 30° | 45° | 60° | 75° | 90° |
|---|---|---|---|---|---|---|
|
|
|
|||||
| RMSE % | 5.09 | 2.49 | 1.63 | 1.18 | 0.88 | 0.68 |
| PSNR | 29.82 | 34.24 | 35.42 | 35.76 | 35.88 | 35.94 |
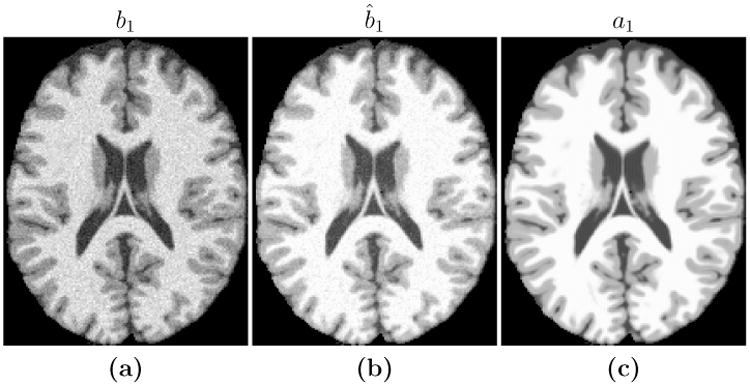
Figure 2.
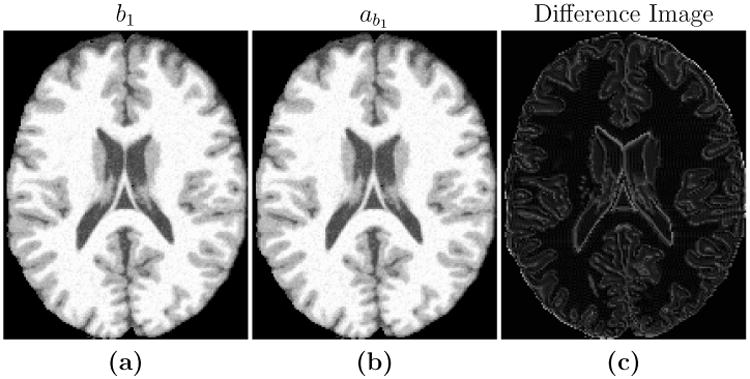
Shown are (a) an SPGR image (TR = 18 ms, TE = 10 ms, α= 45° with 0% additive noise) which we use as our subject image b1, the maximum intensity value is 1080, the (b) new atlas image, ab1, with pulse sequence parameters estimated from b1, and (c) the difference image |ab1 − b1|, the maximum value is 5.
Figure 4. Histogram of a typical noiseless Brainweb phantom.
3.2. Brainweb SPGR Intensity Standardization
The goal of this experiment is to standardize a Brainweb SPGR subject image to an atlas target SPGR image. Different subject SPGR images were simulated using different input pulse sequence parameters. Using the same Brainweb atlas collection described in Section 3.1, we evaluated the results of Steps 3 and 4 of our method, the regression based image synthesis. Our subject images are:
b1: SPGR image (1.5 T, TR = 18 ms, α= {15°, 30°, 45°, 60°, 75°}, TE = 10 ms) with {0%, 3%} noise levels. (Parameters, excepting TR, not provided to the algorithm)
As the target atlas and subject images are both from the SPGR pulse sequence, this is a special case of synthesis, normally referred to as intensity standardization or normalization. We can compare the standardized subject image to the target atlas image directly as they have the same anatomy. We also compared the performance of our standardization with a landmark-based piecewise linear scaling method (UPL) (Nyúl et al., 2000), reporting PSNR for the input subject image in Table 3. UPL estimates the landmarks in the images for each of the three tissue classes (CSF, GM, & WM) and then uses a piecewise linear scaling between the target and subject histogram to normalize the images.
Table 3.
PSNR (dB) values between a1 and b̂1, for standardization of Brainweb phantoms with varying flip angles (°) and noise levels are shown for UPL and Ψ-CLONE. In the noise free case, UPL is better, however the introduction of noise causes UPL results to deteriorate.
| 0% Noise | 3% Noise | |||
|---|---|---|---|---|
| Flip Angle | UPL | Ψ-CLONE | UPL | Ψ-CLONE |
|
|
|
|
||
| 15° | 37.95 | 27.95 | 27.27 | 26.00 |
| 30° | 62.66 | 35.90 | 28.93 | 30.96 |
| 45° | 49.26 | 37.32 | 28.88 | 31.48 |
| 60° | 46.94 | 37.67 | 28.81 | 31.39 |
| 75° | 46.08 | 37.79 | 28.78 | 31.52 |
The histogram of a noise free Brainweb SPGR phantom is shown in Fig. 4. The histogram landmarks consist of very sharp peaks, indicating that a large number of voxels have very similar intensities. A piecewise linear transform can map exactly between two (subject and atlas) such histograms of noiseless Brainweb phantoms. This explains why the UPL method performs better than our method in the 0% noise case. However, with the introduction of noise our method outperforms UPL in four out of the five cases, as shown in Table 3.
3.3. Brainweb T2-w Synthesis
Ψ-CLONE was next used to synthesize a T2-w image from a subject SPGR image, using the Brainweb atlas collection described in Section 3.1 with the addition of:
a2: T2-w image from the second echo of a DSE (1.5 T, TR = 3000 ms, TE1 = 17 ms, TE2 = 80 ms) with 0% noise.
We use the following input subject Brainweb SPGR images:
b1: SPGR image (1.5 T, TR = 18 ms, α= {15°, 30°, 45°, 60°}, TE = 10 ms) with {0%, 1%, 3%, 5%} noise levels. (Parameters, excepting TR, not provided to the algorithm)
The UPL method is unable to synthesize a T2-w image from an SPGR, as it is primarily an intensity standardization approach; thus, for this experiment we compare Ψ-CLONE to MIMECS (Roy et al., 2011, 2013a). MIMECS is a state-of-the-art MR contrast synthesis approach that uses an example-based sparse reconstruction from image patches to perform intensity standardization and missing tissue contrast recovery. MIMECS, unlike Ψ-CLONE, is blind to the MR physics and solves the synthesis problem based on patch similarity between the subject and the atlases. As the atlas and the subject have the same phantom anatomy, an ideal synthesis would result in an image that is exactly equal to the atlas T2-w image. Thus, we evaluate the quality of synthesis by calculating the PSNR between the atlas image and the synthesized subject image. We also evaluate the quality of synthesis using the universal quality index (UQI) (Wang and Bovik, 2002), which is an image quality metric that models how the human visual system would perceive the similarity of two images. If the images are perceptually identical, their UQI is 1; otherwise it lies between 0 and 1.
The results for this experiment are shown in Table 4. The top half of the table shows results for changing noise levels with a fixed flip angle (30°) for the input SPGR, while the bottom half shows the results for changing flip angles with 0% noise in the input SPGR. For both metrics, Ψ-CLONE outperforms MIMECS. An example of an input SPGR used in this experiment, the true T2-w, image and the outputs of both MIMECS and Ψ-CLONE are shown in Fig. 5.
Table 4.
PSNR (dB) and UQI values for Brainweb T2-w synthesis with varying noise levels (%) and flip angles (°) are shown for MIMECS and our algorithm (Ψ-CLONE).
| Noise (%) | 0 | 1 | 3 | 5 | |
|---|---|---|---|---|---|
|
|
|
||||
| PSNR | MIMECS | 26.25 | 19.96 | 19.09 | 19.21 |
| Ψ-CLONE | 30.33 | 30.71 | 29.09 | 26.63 | |
|
|
|
||||
| UQI | MIMECS | 0.93 | 0.88 | 0.83 | 0.82 |
| Ψ-CLONE | 0.95 | 0.94 | 0.91 | 0.88 | |
|
| |||||
| Flip Angle (°) | 15 | 30 | 45 | 60 | |
|
|
|
||||
| PSNR | MIMECS | 24.51 | 26.25 | 25.27 | 24.82 |
| Ψ-CLONE | 25.99 | 30.33 | 30.94 | 31.06 | |
|
|
|
||||
| UQI | MIMECS | 0.90 | 0.93 | 0.92 | 0.92 |
| Ψ-CLONE | 0.90 | 0.95 | 0.96 | 0.96 | |
Figure 5.
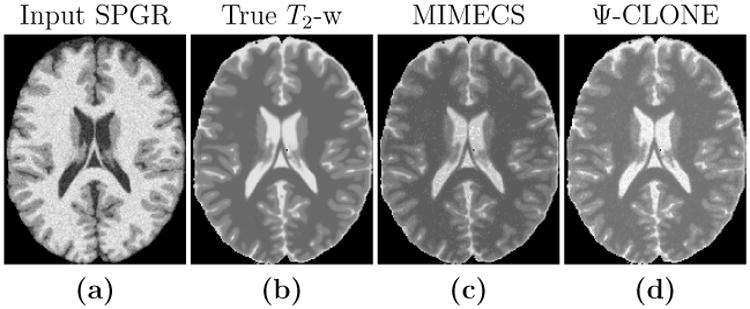
(a) An example input subject SPGR from which we synthesize a T2-w image. (b) The true T2-w image and the outputs of synthesis produced by (c) MIMECS and (d) Ψ-CLONE.
4. Real Data Experiments
In this section, we present intensity standardization and image synthesis experiments performed on real datasets.
4.1. Human Stability Data
A normal, healthy human subject was imaged at weekly intervals using the same scanner and pulse sequence for nine weeks. We demonstrate that image segmentation is more consistent on data which is intensity standardized using Ψ-CLONE. To do this we standardize each time point to an atlas consisting of:
a1: MPRAGE image (3 T, TR = 10.3 ms, TE = 6 ms, 0.82 × 0.82 × 1.17 mm3 voxel size),
aT1: Quantitative T1 map computed as described in Section 2.2,
aT2: Quantitative T2 map derived as described in Section 2.2,
aPD: Quantitative PD map derived as described in Section 2.2,
and each of the nine subject images is:
b1: MPRAGE image (3 T, TR = 10.3 ms, TE = 6 ms, 0.82 × 0.82 × 1.17 mm3 voxel size. (Parameters, excepting TR, not provided to the algorithm)
We compare the segmentations of the images pre- and post-standardization using Ψ-CLONE, based on segmentations generated by TOADS (Bazin and Pham, 2007). We specifically compare the relative tissue volumes (relative to the intra-cranial volume (ICV)), over the nine weeks. Ideally, a normal healthy subject should not present any changes in tissue volumes over such a short period of time.
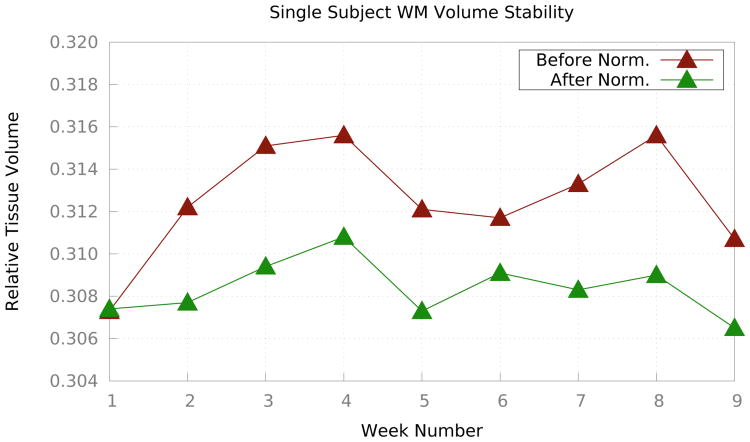
Fig. 6 shows the relative WM volumes before and after Ψ-CLONE was applied. Visually, it is apparent that the WM volumes change less when standardized images are segmented. Table 5 shows the coefficient of variation of relative tissue volumes for CSF, cortical GM, subcortical GM, ventricles, and cortical WM. The coefficients of variation from segmentation after standardization by Ψ-CLONE are smaller than those without standardization for cortical WM, cortical and subcortical GM, and ventricles, indicating that the segmentation is more stable. As there are only nine time-points there is insufficient data to determine significance.
Figure 6.
The brown plot illustrates relative WM volumes (with respect to the ICV) over nine weeks before any standardization while the green plot illustrates the same values after intensity standardization using Ψ-CLONE.
Table 5.
Coefficient of Variation (CV) of the relative tissue volumes (× 10−3) over the nine weeks on the original data and after intensity standardization with Ψ-CLONE for each of white matter (WM), cortical gray matter (Cort. GM), subcortical gray matter (Sub. GM), cerebrospinal fluid (CSF), and the ventricles (Vent.). Table 6 shows further breakdown of Sub. GM structures.
| Classes | |||||
|---|---|---|---|---|---|
|
|
|||||
| Vol. CV | WM | Cort. GM | Sub. GM | CSF | Vent. |
| Original | 8.6 | 6.4 | 20.4 | 11.9 | 30.7 |
| Ψ-CLONE | 4.3 | 5.7 | 19.4 | 12.0 | 23.1 |
The subcortical gray matter class consists of the thalamus, caudate and putamen structures. We compared the volumes of these structures before and after standardization and calculated the coefficient of variation in both cases. Results are shown in Table 6. The thalamus volumes for Ψ-CLONE standardized images are most stable as evinced by the reduced coefficient of variation. The coefficient of variation decreases slightly for putamen and increases slightly for caudate. However the accuracy of LesionTOADS segmentations for subcortical structures is not as high as it is for the cortical GM or WM, hence these numbers may not be reliable indicators of segmentation consistency. Additionally, these are only nine time-points of a single subject, so we cannot claim statistical significance in these measurements at this point.
Table 6.
Coefficient of Variation (CV) of the relative tissue volumes (× 10−3) over the nine weeks on the original data and after intensity standardization with Ψ-CLONE for each of caudate, putamen, and thalamus.
| Structures | |||
|---|---|---|---|
|
|
|||
| Vol. CV | Caudate | Putamen | Thalamus |
| Original | 9.9 | 9.9 | 51.9 |
| Ψ-CLONE | 10.3 | 9.7 | 45.3 |
4.2. MR Intensity Scale Standardization for MS Patients
To allow us to have statistical power in our exploration of MR intensity scale standardization, we employ a cohort of 15 Multiple Sclerosis (MS) patients with 57 scans. Each patient has at least three scans (mean # of scans per subject is 3.8) acquired approximately a year apart. Preprocessing of the images included skull-stripping (Carass et al., 2011) and bias field inhomogeneity correction (Sled et al., 1998). For this experiment our atlas consisted of:
a1: MPRAGE image (3 T, TR = 10.3 ms, TE = 6 ms, 0.82 × 0.82 × 1.17 mm3 voxel size),
a2: T2-w image from the second echo of a DSE (3 T, TR = 4177 ms, TE1 = 12.31 ms, TE2 = 80 ms, 0.82 × 0.82 × 2.2 mm3 voxel size),
a3: PD-w from the first echo of a DSE (3 T, TR = 4177 ms, TE1 = 12.31 ms, TE2 = 80 ms, 0.82 × 0.82 × 2.2 mm3 voxel size),
a4: FLAIR (3 T, TI = 11000 ms, TE = 68 ms, 0.82 × 0.82 × 2.2 mm3 voxel size)
aT1: Quantitative T1 map computed as described in Section 2.2,
aT2: Quantitative T2 map derived as described in Section 2.2,
aPD: Quantitative PD map derived as described in Section 2.2,
and our subject image was:
b1: MPRAGE image (3 T, TR = 10.3 ms, TE = 6 ms, 0.82 × 0.82 × 1.17 mm3 voxel size). (Parameters, excepting TR, not provided to the algorithm)
We use an atlas MPRAGE as the target pulse sequence to which we standardize the 57 data sets. We treat each data set independently, handling the intensity standardization as a cross-sectional task. To validate the intensity standardization, we segmented (Shiee et al., 2010) the original MPRAGE datasets giving us ten labeled structures: ventricles, sulcal CSF, cerebellar GM (Cereb. GM), cortical GM (Cort. GM), thalamus, putamen, cerebellar WM (Cereb. WM), cortical WM (Cort. WM), and lesions. Using these structures as reference, we compared the mean intensity within these structures prior to standardization and after standardization with UPL—using the target MPRAGE in our atlas as a standardization target—and our method (Ψ-CLONE). We note that the atlas images did not have lesions. For applications like T1-w intensity standardization, we observed that the presence or absence of lesion samples in the training data did not affect the synthesis result. The primary reason is that white matter lesion intensities are similar to GM (and rarely CSF) in T1-w contrasts. Thus, the database has a large number of normal appearing GM and CSF samples available to reconstruct the lesion intensities.
Results are shown in Table 7. The mean intensity values of the original images and the Ψ-CLONE standardized images recorded in Table 7, demonstrate that our method is moving the MS data intensities closer to the atlas intensities, as desired by standardization. A one tailed F-test on the mean structure intensities after standardization shows that the standard deviation of the mean structure intensities across the 57 datasets for Ψ-CLONE is significantly smaller in comparison to UPL for seven of the ten structures. We also note that the statistical significance does not change if the segmentation is done on the original data or on the standardized versions.
Table 7.
The mean intensity value for the atlas used in the MS standardization experiment are shown for ten structures. We also show the mean (× 104) and std (× 104) (over 57 images) of the average intensity value for each structure, based on the original unnormalized data (Original) and after standardization with both UPL (Nyúl et al., 2000) and our method (Ψ-CLONE).
| Atlas | Original | UPL | Ψ-CLONE | ||||
|---|---|---|---|---|---|---|---|
| Mean | Mean | Std | Mean | Std | Mean | Std | |
|
|
|||||||
| Ventricles | 3.92 | 5.41 | 1.815 | 4.25 | 0.664 | 4.11† | 0.271* |
| Sulcal CSF | 2.50 | 3.95 | 1.132 | 3.11 | 0.543 | 2.97 | 0.478 |
| Lesions | — | 18.54 | 5.880 | 14.62 | 1.293 | 14.51 | 1.516 |
| Cereb. GM | 12.27 | 15.86 | 5.295 | 12.68 | 1.013 | 12.27† | 0.433* |
| Cort. GM | 10.47 | 13.40 | 4.722 | 10.62 | 1.199 | 10.27† | 0.260* |
| Caudate | 12.93 | 16.98 | 5.866 | 13.53 | 1.130 | 13.10† | 0.526* |
| Thalamus | 14.99 | 21.20 | 7.036 | 16.96 | 1.010 | 16.63† | 0.548* |
| Putamen | 15.53 | 20.26 | 7.286 | 16.23 | 1.196 | 15.77† | 0.485* |
| Cereb. WM | 20.79 | 27.66 | 9.333 | 21.05 | 0.209 | 22.02 | 0.266 |
| Cort. WM | 20.53 | 25.77 | 8.778 | 20.22 | 0.266 | 20.41† | 0.186* |
Difference between atlas mean and normalized mean is significantly smaller than UPL (α level of 0.05) using a two-sample one-tailed T-test.
Standard deviation is significantly smaller than UPL (α level of 0.05) based on a two-sample F-test.
Next, we ran Lesion TOADS segmentation on images before and after standardization. These segmentations were used to calculate image contrast between neighboring structures to indicate the effect of synthesis-based standardization on subsequent segmentation. We looked at the following structure boundaries, Cortical CSF-Cortical GM, Cortical GM-WM, WM-lesions, WM-Ventricles, WM-Caudate, WM-Putamen, WM-Thalamus, Caudate-Ventricles, and Thalamus-Ventricles and have tabulated the results in Table 8. We can show that on average these contrast values for synthetic images are higher than the real images, significantly in most cases. The contrast values for synthetic images are also closer to the reference contrast values for the same structures, than the original images. We have defined contrast between two neighboring structures f and g as , where μ(f) is the mean intensity of the brighter structure f and μ(g) is the mean intensity of the structure g. The higher the contrast, the easier it is to differentiate structures.
Table 8.
Contrast values between neighboring structures for original, synthetic, and atlas images.
| Structure Boundary Contrasts | |||
|---|---|---|---|
|
|
|||
| Struct. Boundaries | Original | Ψ-CLONE | Atlas |
| Cort CSF–Cort GM | 0.594 | 0.604* | 0.625 |
| Cort GM–WM | 0.480 | 0.482 | 0.489 |
| WM–Lesions | 0.279 | 0.300 | — |
| WM–Caudate | 0.341 | 0.367* | 0.370 |
| WM–Putamen | 0.214 | 0.219 | 0.240 |
| WM–Thalamus | 0.176 | 0.208* | 0.269 |
| WM–Ventricles | 0.846 | 0.863* | 0.878 |
| Ventricles–Caudate | 0.765 | 0.784* | 0.804 |
| Ventricles–Thalamus | 0.814 | 0.827* | 0.834 |
indicates that the contrast in synthetic images higher than the original images (statistically significant using Student's one-tailed T test with p < 0.05).
4.3. T2-w Synthesis from Real MPRAGE Data
To demonstrate T2-w synthesis from MPRAGE on real data, we used the 21 subjects from the publicly available multimodal reproducibility data (Landman et al., 2011). We held out a single subject as the atlas:
a1: MPRAGE image (3 T, TR = 6.7 ms, TE = 3.1 ms, TI = 842 ms, 1.0 × 1.0 × 1.2 mm3 voxel size),
a2: T2-w image from the second echo of a DSE (3 T, TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms, 1.5 × 1.5 × 1.5 mm3 voxel size),
a3: PD-w image from the first echo of a DSE (3 T, TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms, 1.5 × 1.5 × 1.5 mm3 voxel size),
aT1: Quantitative T1 map computed from two flip angles (3 T, TR = 100 ms, TE = 15 ms, α1 = 15°, α2 = 60°, 1.5 × 1.5 × 1.5 mm3 voxel size),
aT2: Quantitative T2 map derived from a two-point method (3 T, TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms, 1.5 × 1.5 × 1.5 mm3 voxel size),
aPD: Quantitative PD map derived from the MPRAGE and the DSE images using the method described in Section 2.2.
Our subject image is:
b1: MPRAGE image (3 T, TR = 6.7 ms, TE = 3.1 ms, TI = 842 ms, 1.0 × 1.0 × 1.2 mm3 voxel size). (Parameters, excepting TR, not provided to the algorithm)
We note that the atlas image did not have lesions. For an application like T2-w synthesis, we again observed that the presence or absence of lesion samples in the training data did not affect the synthesis result. The primary reason is that white matter lesion intensities are similar to GM (and rarely CSF) in T1-w and T2-w contrasts. Thus, the database has a large number of normal appearing GM and CSF samples available to reconstruct the lesion intensities.
The remaining 20 available subjects each have two MPRAGE acquisitions and two corresponding DSE images which are co-registered to the MPRAGE. These images were acquired on the same scanner within a short duration of each other. For each of these 40 images (20 subjects × 2 MPRAGE scans) we synthesized a T2-w image. As the atlas was imaged on the same scanner we directly compare the synthesized T2-w image with the true T2-w image from the same scanning session, using PSNR and UQI. We compared to MIMECS (Roy et al., 2011) and to a deformable registration-based synthesis.
To carry out synthesis using deformable registration, the atlas image is registered deformably to the subject image of the same contrast. The same deformation is then applied to the atlas image of the desired contrast to produce the synthetic subject image. We use the state-of-the-art registration method SyN (Avants et al., 2008) for this synthesis and refer to this method as D-SyN. Table 9 shows the PSNR and UQI for these three methods. We observe that our method (Ψ-CLONE) provides a significantly (α < 0.01, using the right-tailed two sample t-test) better quality synthesis in comparison to both MIMECS and D-SyN.
Table 9.
Mean and standard deviation (Std. Dev.) of the PSNR and UQI values for synthesis of T2-w images from 40 MPRAGE scans.
| PSNR | UQI | |
|---|---|---|
| Mean (Std. Dev.) | Mean (Std. Dev.) | |
|
|
|
|
| D-SyN | 16.59 (1.35) | 0.64 (0.06) |
| MIMECS | 15.01 (0.84) | 0.78 (0.03) |
| Ψ-CLONE | 18.59 (1.09)* | 0.79 (0.02)* |
Statistically significantly better than either of the other two methods (α level of 0.01) using a right-tailed test.
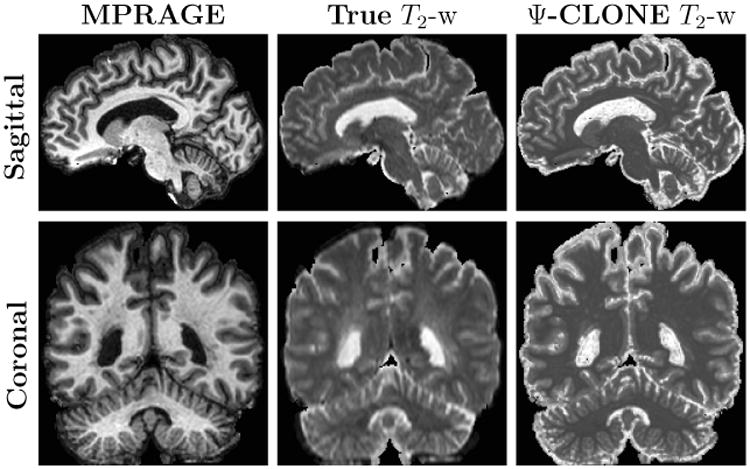
Figure 7 shows the results for each synthesis approach in comparison to the ground truth image. Though the PSNR values for D-SyN are better than MIMECS (see Table 9), the D-SyN synthesis result is anatomically incorrect—the ventricle boundary is incorrect and lesions posterior to the ventricles are not synthesized. As the output of D-SyN is based on deformably registering the atlas to the subject, if the atlas does not contain certain tissue features—lesions, for example—then the synthesized subject will not contain them as well. The lesion in Fig. 7 is a white matter lesion. We know that lesion boundaries appear slightly different in the MPRAGE than in the real T2-w image, and hence it cannot be perfectly reproduced in the synthetic T2-w image. However we aim to synthesize it as correctly as possible and do a better job than currently available synthesis algorithms. The result of MIMECS is quite noisy and Ψ-CLONE yields an image that is most visually similar to the true image.
Figure 7.
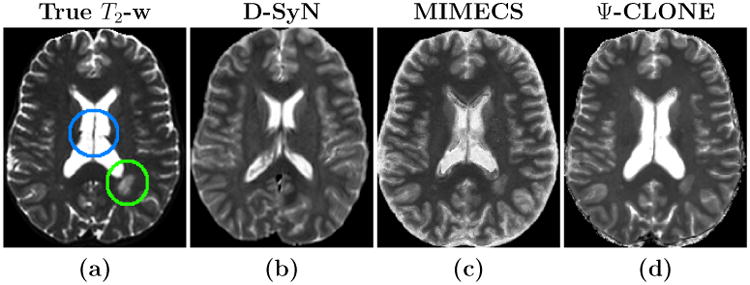
Shown are (a) the true T2-w image, and the synthesis results from the MPRAGE for each of (b) D-SyN, (c) MIMECS, and (d) Ψ-CLONE (our method). The lesion (in the green circle) and the ventricles (in the blue circle) in the true image are synthesized by MIMECS and Ψ-CLONE, but not by D-SyN.
We also used the atlas images of this dataset to evaluate our estimation procedure for the intrinsic parameters T2 and T1. The median T2 values calculated by the two point method (Landman et al., 2011) using the DSE images were 76 ms for WM, 85 ms for GM, and 175 ms for CSF. We used our estimation procedure (as described in Section 2.2) and the estimated median T2 values obtained were 76 ms for WM, 91 ms for GM, and 762 ms for CSF. Both CSF distributions have a very large standard deviation (∼104), due to numerical errors. The intensities determined by the imaging equations also plateau off after a certain T2 value due to their inverse exponential nature (see Eq. 2). Thus the intensities produced for high enough T2 values are very close to each other. The T1 map was estimated via two flip angle spoiled gradient images, as mentioned in the atlas description. This is not an ideal approach as the images acquired were noisy and the flip angle calibration is not considered accurate enough. The median T1 values thus calculated using the dual flip angle image were 775 ms for WM, 1074 ms for GM, and 1616 ms for CSF. Our estimation procedure returned the following median T1 values of 779 ms for WM, 1151 ms for GM, and 2916 ms for CSF. As with T2 values, the intensities produced by high T1 values plateau off after a certain point due to the inverse exponential dependence on T1 (see Eqs. 2, 3, 4). Thus, despite being slightly different from the expected values, our estimated T2 and T1 values are good enough to provide a realistic synthesis.
5. Further Synthesis Applications
In this final experimental section, we present additional results that demonstrate potential uses of Ψ-CLONE.
5.1. Synthesizing Higher Resolution T2-w Data
Example-based synthesis of high resolution brain MR images has been explored in many recent works (Rousseau, 2008; Manjón et al., 2010a,b; Konukoglu et al., 2013). We applied Ψ-CLONE to synthesize higher resolution T2-w images than those acquired on the scanner. Our atlas collection
 is:
is:
a1: MPRAGE image (3 T, TR = 6.7 ms, TE = 3.1 ms, TI = 842 ms, 1.1 × 1.1 × 1.1 mm3 voxel size),
a2: T2-w image from the second echo of a DSE (3 T, TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms, 1.1 × 1.1 × 1.1 mm3 voxel size),
and our subject images come from our MS cohort:
b1: MPRAGE image (3 T, TR = 10.3 ms, TE = 6 ms, 0.82 × 0.82 × 1.17 mm3 voxel size). (Parameters, excepting TR, not provided to the algorithm)
Pulse sequences like the DSE or FLAIR tend to have large TR or TI values to achieve the right contrast. To reduce the scan time while imaging patients, these pulse sequences are usually acquired at a lower resolution than a T1-w sequence such as MPRAGE. Multimodal analysis of such datasets requires all images to exist in the same coordinate system at the same digital resolution. This is usually achieved by upsampling the low resolution scans to the high resolution ones using interpolation, which results in blurring of the image data. Using Ψ-CLONE, we can synthesize a T2-w image from the high resolution MPRAGE. This synthetic image will have the same resolution as that of the MPRAGE and hence can replace the acquired low resolution image. As we have no ground truth for the higher resolution T2-w image we visually compare it with the acquired T2-w image. The acquired T2-w image has a through-plane resolution of 2.2 mm whereas the subject MPRAGE, and consequently the synthetic T2-w image have a through-plane resolution of 1.1 mm. Both are shown in Fig. 8. The quality and resolution of the newly synthesized image is visually superior to the original acquisition.
Figure 8.
The MPRAGE has a through-plane resolution of 1.1 mm, while the original T2-w has through-plane resolution of 2.2 mm. This is evident as the true interpolated T2-w image shows blurring while the Ψ-CLONE synthesized image is crisp.
5.2. FLAIR Synthesis
FLAIR is the pulse sequence of choice when identifying white matter lesions present in MS patients. The lesions appear hyperintense with respect to the rest of the tissue which makes delineating them easier. Most leading lesion segmentation algorithms rely on the FLAIR image to provide intensity information for accurate classification (Lecoeur et al., 2009; Shiee et al., 2010; Forbes et al., 2010). FLAIR images are prone to certain artifacts for a variety of reasons (Stuckey et al., 2007). The long inversion times make it difficult to acquire high resolution scans in a short time. We demonstrate that if we acquire T1-w, PD-w, and T2-w images of a subject, we have enough information to generate a synthetic FLAIR using Ψ-CLONE. The lesion intensity signature in FLAIR images is very distinct from the rest of the tissues. Hence the presence of lesion samples in the atlas set is essential in order to learn to reproduce it correctly. For this experiment, the atlas brain we use has a moderate lesion load. The atlas contained the following images:
a1: MPRAGE image (3 T, TR = 6.7 ms, TE = 3.1 ms, TI = 842 ms, 1.0 × 1.0 × 1.2 mm3 voxel size),
a2: T2-w image from the second echo of a DSE (3 T, TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms, 1.5 × 1.5 × 1.5 mm3 voxel size),
a3: PD-w image from the first echo of a DSE (3 T, TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms, 1.5 × 1.5 × 1.5 mm3 voxel size),
aT1: Quantitative T1 map computed as described in Section 2.2,
aT2: Quantitative T2 map derived as described in Section 2.2,
aPD: Quantitative PD map derived as described in Section 2.2.
Our subject images are:
b1: MPRAGE image (3 T, TR = 6.7 ms, TE = 3.1 ms, TI = 842 ms, 1.0 × 1.0 × 1.2 mm3 voxel size), (Parameters, excepting TR, not provided to the algorithm)
b2: T2-w image from the second echo of a DSE (3 T, TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms, 1.5 × 1.5 × 1.5 mm3 voxel size), (Parameters, excepting TR, not provided to the algorithm)
b3: PD-w image from the first echo of a DSE (3 T, TR = 6653 ms, TE1 = 30 ms, TE2 = 80 ms, 1.5 × 1.5 × 1.5 mm3 voxel size). (Parameters, excepting TR, not provided to the algorithm)
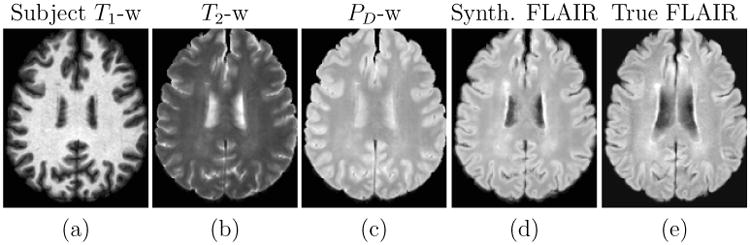
We would like to stress that the set of subject pulse sequence parameters (except TR) are unknown and these are extracted using the first step of Ψ-CLONE. Next, the new atlas T1-w, PD-w, and T2-w atlas images are generated by applying the respective pulse sequence equations to the atlas PD, T1, and T2 values. The following step of learning a patch-based regression is slightly different from the previous experiments. The feature vector bi for a voxel i, is created by concatenating the corresponding 3 × 3 × 3-sized patches centered on voxel i, from all three images. The dependent variable ri is the corresponding target atlas FLAIR intensity at voxel i. Thus, the training data consists of pairs of 〈bi, ri〉 from the extracted synthetic atlas images and the atlas FLAIR image. A nonlinear regression is learned using random forests and the trained regression is then applied to the extracted patches from the subject images to synthesize the subject FLAIR. Figure 9 displays the subject input images and synthetic FLAIR with the true FLAIR for visual comparison.
Figure 9.
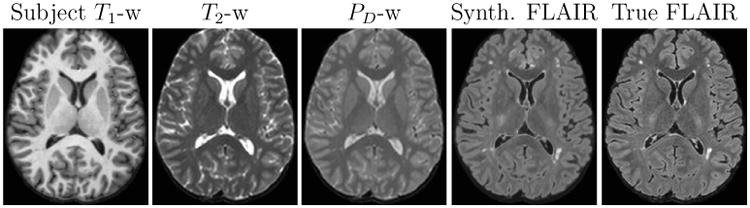
Subject input images along with the synthetic and true FLAIR images.
We ran LesionTOADS on the real and synthetic FLAIR images shown in Fig. 9. The resulting segmentations are shown in Fig. 10. The lesion volume obtained from the real FLAIR using LesionTOADS was 3107.5 mm3 whereas that obtained from a synthetic FLAIR was 5900.3 mm3. The excess seems to come from slightly enlarged regions with lesion-like intensities in the synthetic FLAIR. Our result, though visual in nature, is still a large improvement on the FLAIR synthesis result demonstrated in Roy et al. (2013a). Synthesizing FLAIR images is especially useful when the original FLAIR has artifacts, which can lead to erroneous tissue segmentation. In the next experiment, we looked at data where the acquired FLAIR was of bad quality due to motion artifacts and created a synthetic FLAIR image for visual comparison. The atlas brain for this experiment also has lesion voxels, which are essential for training. The atlas set was:
a1: MPRAGE image (3 T, TR = 10.3 ms, TE = 6 ms, 0.82 × 0.82 × 1.17 mm3 voxel size),
a2: T2-w image from the second echo of a DSE (3 T, TR = 4177 ms, TE1 = 3.41 ms, TE2 = 80 ms, 0.82 × 0.82 × 2.2 mm3 voxel size),
a3: PD-w from the first echo of a DSE (3 T, TR = 4177 ms, TE1 = 3.41 ms, TE2 = 80 ms, 0.82 × 0.82 × 2.2 mm3 voxel size),
a4: FLAIR (3 T, TI = 11000 ms, TE = 68 ms, 0.82 × 0.82 × 2.2 mm3 voxel size)
aT1: Quantitative T1 map computed as described in Section 2.2,
aT2: Quantitative T2 map derived as described in Section 2.2,
aPD: Quantitative PD map derived as described in Section 2.2.
Figure 10.
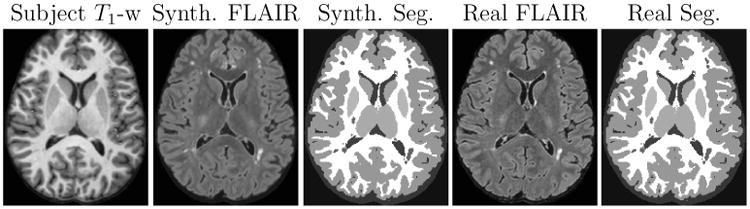
Real T1-w image along with the corresponding real and synthetic FLAIR images and their resulting segmentations.
The subject set consisted of:
b1: MPRAGE image (3 T, TR = 10.3 ms, TE = 6 ms, 0.82 × 0.82 × 1.17 mm3 voxel size), (Parameters, excepting TR, not provided to the algorithm)
b2: PD-w image from the first echo of a DSE (3 T, TR = 4177 ms, TE1 = 3.41 ms, TE2 = 80 ms, 0.82 × 0.82 × 2.2 mm3 voxel size), (Parameters, excepting TR, not provided to the algorithm)
b3: T2-w image from the second echo of a DSE (3 T, TR = 4177 ms, TE1 = 3.41 ms, TE2 = 80 ms, 0.82 × 0.82 × 2.2 mm3 voxel size). (Parameters, excepting TR, not provided to the algorithm)
The results of this experiment are shown in Fig. 11. The synthetic FLAIR shown in Fig. 11(d) does not possess the motion artifacts present in the true FLAIR in Fig. 11(e), since these are not present in the input T1-w, T2-w, and PD-w images.
Figure 11.
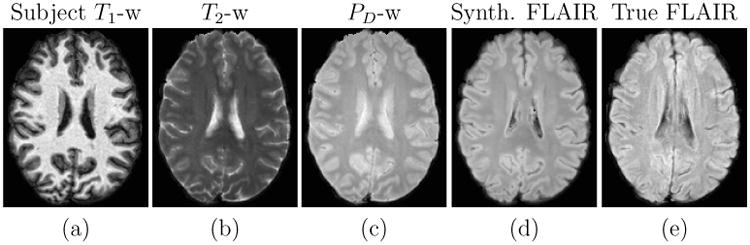
Input T1-w, T2-w, and PD-w images followed by the synthetic FLAIR and the true FLAIR. The true FLAIR shows motion artifacts, which are not present in the synthetic FLAIR.
Segmentation errors can also stem from misalignment of multimodal images. In the next experiment we will demonstrate the potential benefit of using Ψ-CLONE generated synthetic FLAIR images in multimodal analysis. We use the same set of atlas and subject image sets as above, but for a different individual subject in this experiment. The original FLAIR image (Fig. 12(e)) for this experiment has a voxel size of 0.82 × 0.82 × 4.4 mm3, which is much larger than the MPRAGE voxel size of 0.82 × 0.82 × 1.17 mm3. Registering the original FLAIR to the MPRAGE requires an upsampling by a factor of four in the through-plane direction. Upsampling via interpolation (trilinear, in this case) results in blurring in the through-plane direction. This blurring is clearly visible in the original FLAIR image in Fig. 12(e), especially in the ventricles. It is also visible to some extent in right posterior ventricle region of Fig 11(e). The blurring is also a cause of gross misalignment between the high resolution images and the low resolution FLAIR images. Such misalignment has the potential to adversely affect segmentation results. The synthetic FLAIR image is created by applying Ψ-CLONE on a high resolution T1-w image (1.17 mm slice thickness) and intermediate resolution PD-w and T2-w images (2.2 mm slice thickness). Thus, it is better aligned to the high resolution images and is at a higher resolution than the acquired FLAIR image. Fig. 12(d) shows the synthetic FLAIR along with the true FLAIR in Fig. 12(e). Visually it is apparent that it is better aligned to the rest of the images than the true FLAIR.
Figure 12.
Input T1-w, T2-w, PD-w images with the synthetic FLAIR and the true FLAIR. The true FLAIR shows blurring because of interpolation in order to match the high resolution T1-w
6. Discussion and Conclusion
We have proposed an MR image pre-processing framework, Ψ-CLONE, which allows us to perform MR image synthesis and scanner standardization. Ψ-CLONE represents a new direction in image synthesis. It works by estimating relevant information about the imaging parameters of a given image and incorporating these into a synthesis that respects MR image formation resulting in more correct synthetic images then previous synthesis methods. Prior work in MR image synthesis has ignored the image acquisition process when solving the synthesis problem. Our use of imaging equation approximations and the underlying NMR tissue parameters to construct atlases that are adaptive to the subject image is a unique feature. In Sections 3 and 4, we demonstrated the significantly higher quality of image standardization and synthesis than the existing state-of-the-art methods. In addition to this in Section 5, we showcased advanced capabilities of our synthesis approach—specifically FLAIR synthesis—which cannot be accomplished by other methods (Roy et al., 2013a).
Application of Ψ-CLONE for image synthesis can be used to enhance and expand multimodal datasets for better image processing. Improved resolution for modalities like FLAIR and DSE pulse sequences which are often acquired at a low resolution can prove useful for tasks such as segmentation and registration. In addition, the ability to replace an artifact-ridden image with a synthetic one for better and more consistent processing of the entire dataset will help in providing more usable subject data, which should in turn help improve the statistical power of any derived scientific results.
In addition to improving the quality and capabilities of MR image synthesis, Ψ-CLONE is also quite fast, taking less than five minutes to synthesize a new image. In comparison, state-of-the-art methods like MIMECS take around 2–3 hours on the same computational resources. This makes Ψ-CLONE well-suited as a quick pre-processing tool or as an MR intensity standardization that can be done on the scanner prior to any other processing. The nonlinear simultaneous equation solver can sometimes lead to local minima in the absence of a good initialization. The robustness of estimation is something we want to work on in the immediate future, by incorporating more accurate models of the pulse sequence equations. This will help improve the pulse sequence parameter estimation. The atlases used in the experiments are obviously of critical importance in the quality of the synthesis that can be performed. As such we are working to acquire high resolution data from a small cohort of subjects on multiple scanners to have a complete picture of the NMR properties at various field strengths as is technically feasible.
Ψ-CLONE has certain limitations which we would like to address in the future. First, it requires a segmentation of the input image(s) to estimate the imaging equation parameters. Specifically, the pulse sequence parameter estimation depends on tissue class means provided by a fuzzy k-means algorithm on T1-w images. For typical T1-w sequences like MPRAGE, the fuzzy k-means algorithm is fairly robust in providing the class mean intensities. In the rare case that the algorithm fails, the estimated imaging parameters tend to have very large errors, which results in the formation of inferior synthetic atlas images that can be easily spotted as inaccurate and rectified at the end of Step 2. Some of the earlier image synthesis methods (Rousseau, 2008; Roy et al., 2010b, 2011) had similar drawbacks that have since been overcome by dictionary selection techniques and use of a higher-dimensional space to normalize image patches (Roy et al., 2013a). Incorporation of these ideas is feasible, but we have yet to explore them. However, unlike Roy et al. (2013a) we do not require a WM peak normalization step.
Second, our pulse sequence parameter estimation method allows us to generate images of high quality and similar characteristics to the ground truth images (see Section 3 for experiments). However, the estimated parameters are not identical to the truth for various reasons including the use of theoretical and approximate pulse sequence equations. This is not ideal, as even though the metrics we use to measure the similarity of the images are commonplace; they may have a subtle deficiency that could only be revealed by a far larger and more rigorous study. Our estimation of PD, T1, and T2 maps also requires (a) 3 different types of pulse sequence images, preferably T1-w, PD-w, and T2-w images, of an acceptable resolution (the worst we have worked with is 1 × 1 × 5 mm3 voxel-size in experiments described in Jog et al. (2013b) for a different application), (b) known pulse sequence name (for example SPGR or DSE), and (c) known imaging equation or approximation for each of the three. These can be potentially restrictive in some clinical scenarios and we are working towards relaxing these requirements.
A third deficiency is our chosen regression model (random forests), it is being used because of it expediency and its ability to handle nonlinear intensity transformations. When predicting, random forest regression takes the mean of all the training data that accumulates in a leaf node during model training. In addition, when using the random forest the output value from each tree is averaged to give the final prediction from the regression. Clearly all these averages diminish the quality of the results, which can be seen in Fig. 11 where the synthetic result appears smoother than the truth. This could be addressed by modifying the random forest to do a linear fit of the data in the leaf nodes or through the use of a different regression approach. Finally, the influence of the patch size on the synthesis results has been explored—not reported here—though a complete understanding and rationale of the optimality of small patch size, i.e. 3 × 3 × 3, is not completely understood at present.
In summary, we have described a new MR synthesis approach which incorporates principles based on the pulse sequence equations. The framework is validated on synthetic and real data demonstrating its superior synthesis to state-of-the-art approaches. In addition, we have demonstrated the capability to synthesize the FLAIR pulse sequence, which is a noted deficiency of the MIMECS algorithm Roy et al. (2013a). Our estimation of pulse sequence parameters to generate a better atlas image could be used by any synthesis approach (Rousseau, 2008; Rousseau and Studholme, 2013; Roy et al., 2013a,b; Jog et al., 2014a,b) to help improve results immediately.
Figure 3.
Shown are (a) an SPGR image (TR = 18ms, TE = 10ms, α= 45° with 3% additive noise) which we use as our subject image b1, the (b) reconstruction, b̂1, of the subject image with the same pulse sequence as used to image (c) the atlas target image a1 (TR = 18ms, α= 30°, TE = 10ms with 0% noise).
Highlights.
We describe an MR image synthesis algorithm that works by estimating pulse sequence parameters of given image
We create subject specific atlas (training) images to learn an intensity mapping using patch-based random forest regression
We demonstrate superior image synthesis and intensity standardization and compare to the state-of-the-art
We also demonstrate advanced capabilities of image synthesis via super-resolution and FLAIR synthesis experiments
Our algorithm is computationally fast and is a powerful image preprocessing tool
Acknowledgments
This work was supported by the NIH/NIBIB under grant R21 EB012765 and R01 EB017743, and by the NIH/NINDS through grant R01 NS070906.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avants B, Epstein C, Grossman M, Gee J. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin PL, Pham DL. Topology-preserving tissue classification of magnetic resonance brain images. IEEE Trans Med Imag. 2007;26:487–498. doi: 10.1109/TMI.2007.893283. [DOI] [PubMed] [Google Scholar]
- Bezdek JC. A convergence theorem for the fuzzy ISO-DATA clustering algorithms. IEEE Trans on Pattern Anal Machine Intell. 1980;20:1–8. doi: 10.1109/tpami.1980.4766964. [DOI] [PubMed] [Google Scholar]
- Bottomley P, Ouwerkerk R. The dual-angle method for fast, sensitive T1 measurement in vivo with low-angle adiabatic pulses. Journal of Magnetic Resonance, Series B. 1994;104:159–167. [Google Scholar]
- Breiman L. Bagging predictors. Machine Learning. 1996;24:123–140. [Google Scholar]
- Burgos N, Cardoso M, Modat M, Pedemonte S, Dickson J, Barnes A, Duncan J, Atkinson D, Arridge S, Hutton B, Ourselin S. Attenuation Correction Synthesis for Hybrid PET-MR Scanners. In: Mori K, Sakuma I, Sato Y, Barillot C, Navab N, editors. Medical Image Computing and Computer-Assisted Intervention MICCAI 2013 volume 8149 of Lecture Notes in Computer Science. 2013. pp. 147–154. [DOI] [PubMed] [Google Scholar]
- Carass A, Cuzzocreo J, Wheeler MB, Bazin PL, Resnick SM, Prince JL. Simple paradigm for extra-cerebral tissue removal: Algorithm and analysis. NeuroImage. 2011;56:1982–1992. doi: 10.1016/j.neuroimage.2011.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocosco CA, Kollokian V, Kwan RKS, Evans AC. BrainWeb: Online interface to a 3D MRI simulated brain database. NeuroImage. 1997;5:S425. [Google Scholar]
- Dale AM, Fischl B. Cortical surface-based analysis: Segmentation and surface reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Deichmann R, Good CD, Josephs O, Ashburner J, Turner R. Optimization of 3-D MP-RAGE sequences for structural brain imaging. NeuroImage. 2000;12:112–127. doi: 10.1006/nimg.2000.0601. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Segonne F, Quinn BT, Dale AM. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23:S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Forbes F, Doyle S, García-Lorenzo D, Barillot C, Dojat M. Adaptive weighted fusion of multiple MR sequences for brain lesion segmentation. 7th International Symposium on Biomedical Imaging (ISBI 2010) 2010:69–72. [Google Scholar]
- Glover GH. Handbook of MRI pulse sequences. Vol. 18. Elsevier Academic Press; 2004. [Google Scholar]
- Han X, Pham DL, Tosun D, Rettman RE, Xu C, Prince JL. CRUISE: Cortical reconstruction using implicit surface evolution. NeuroImage. 2004;23:997–1012. doi: 10.1016/j.neuroimage.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Hertzmann A, Jacobs CE, Oliver N, Curless B, Salesin DH. Image analogies. Proceedings of the 28th annual conference on Computer graphics and interactive techniques. 2001:327–340. [Google Scholar]
- Iglesias JE, Konukoglu E, Zikic D, Glocker B, Van Leemput K, Fischl B. Is synthesizing MRI contrast useful for inter-modality analysis? 16th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2013) 2013:631–638. doi: 10.1007/978-3-642-40811-3_79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog A, Carass A, Pham DL, Prince JL. Random forest FLAIR reconstruction from T1, T2, and PD-weighted MRI. Biomedical Imaging (ISBI), 2014 IEEE 11th International Symposium on. 2014a:1079–1082. doi: 10.1109/ISBI.2014.6868061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog A, Carass A, Prince JL. Improving magnetic resonance resolution with supervised learning. Biomedical Imaging (ISBI), 2014 IEEE 11th International Symposium on. 2014b:987–990. doi: 10.1109/ISBI.2014.6868038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog A, Roy S, Carass A, Prince JL. Magnetic resonance image synthesis through patch regression. 10th International Symposium on Biomedical Imaging (ISBI 2013) 2013a:350–353. doi: 10.1109/ISBI.2013.6556484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog A, Roy S, Carass A, Prince JL. Pulse sequence based multi-acquisition MR intensity normalization; Proceedings of SPIE Medical Imaging (SPIE-MI 2013); Orlando, FL. February 9-14, 2013; 2013b. pp. 86692H–86692H–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konukoglu E, van der Kouwe A, Sabuncu MR, Fischl B. Example-based restoration of high-resolution magnetic resonance image acquisitions. 16th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2013) 2013:131–138. doi: 10.1007/978-3-642-40811-3_17. [DOI] [PubMed] [Google Scholar]
- Kwan RKS, Evans AC, Pike GB. MRI simulation-based evaluation of image-processing and classification methods. IEEE Trans Med Imag. 1999;18:1085–1097. doi: 10.1109/42.816072. [DOI] [PubMed] [Google Scholar]
- Landman BA, Huang AJ, Gifford A, Vikram DS, Lim IAL, Farrell JAD, Bogovic JA, Hua J, Chen M, Jarso S, Smith SA, Joel S, Mori S, Pekar JJ, Barker PB, Prince JL, van Zijl P. Multi-parametric neuroimaging reproducibility: A 3-T resource study. NeuroImage. 2011;54:2854–2866. doi: 10.1016/j.neuroimage.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoeur J, Ferré J, Barillot C. Optimized supervised segmentation of MS sesions from multispectral MRIs. MICCAI workshop on Med Image Anal on Multiple Sclerosis. 2009:1–11. [Google Scholar]
- Manjón JV, Coupé P, Buades A, Collins DL, Robles M. MRI superresolution using self-similarity and image priors. International Journal of Biomedical Imaging. 2010a;425891:11. doi: 10.1155/2010/425891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjón JV, Coupé P, Buades A, Fonov V, Collins DL, Robles M. Non-local MRI upsampling. Medical Image Analysis. 2010b;14:784–792. doi: 10.1016/j.media.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Miller MI, Christensen GE, Amit Y, Grenander U. Mathematical textbook of deformable neuroanatomies. Proc Natl Acad Sci. 1993;90:11944–11948. doi: 10.1073/pnas.90.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyúl LG, Udupa JK. On standardizing the MR image intensity scale. Mag Reson Med. 1999;42:1072–1081. doi: 10.1002/(sici)1522-2594(199912)42:6<1072::aid-mrm11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imag. 2000;19:143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- Prastawa M, Gilmore J, Lin W, Gerig G. Automatic segmentation of neonatal brain MRI. 7th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2004) 2004:10–17. [Google Scholar]
- Roohi S, Zamani J, Noorhosseini M, Rahmati M. Super-resolution MRI images using Compressive Sensing. Electrical Engineering (ICEE), 2012 20th Iranian Conference on. 2012:1618–1622. [Google Scholar]
- Rousseau F. Brain hallucination. 2008 European Conference on Computer Vision (ECCV 2008) 2008:497–508. [Google Scholar]
- Rousseau F. A non-local approach for image super-resolution using intermodality priors. Medical Image Analysis. 2010;14:594–605. doi: 10.1016/j.media.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F, Studholme C. A supervised patch-based image reconstruction technique: Application to brain MRI super-resolution. 10th International Symposium on Biomedical Imaging (ISBI 2013) 2013:346–349. [Google Scholar]
- Roy S, Carass A, Bazin PL, Resnick SM, Prince JL. Consistent segmentation using a Rician classifier. Medical Image Analysis. 2012;16:524–535. doi: 10.1016/j.media.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Carass A, Prince JL. Synthesizing MR contrast and resolution through a patch matching technique; Proceedings of SPIE Medical Imaging (SPIE-MI 2010); San Diego, CA. February 14-17, 2010; 2010a. p. 7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Carass A, Prince JL. 22nd Inf Proc in Med Imaging (IPMI 2011) 2011. A compressed sensing approach for MR tissue contrast synthesis; pp. 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Carass A, Prince JL. Magnetic Resonance Image Example Based Contrast Synthesis. IEEE Trans Med Imag. 2013a doi: 10.1109/TMI.2013.2282126. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Carass A, Shiee N, Pham DL, Prince JL. MR contrast synthesis for lesion segmentation. 7th International Symposium on Biomedical Imaging (ISBI 2010) 2010b:932–935. doi: 10.1109/ISBI.2010.5490140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Jog A, Carass A, Prince JL. Atlas based intensity transformation of brain MR images. 3rd International Workshop on Multimodal Brain Image Analysis at the 16th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2013) 2013b:51–62. [Google Scholar]
- Roy S, Wang WT, Carass A, Prince JL, Butman JA, Pham DL. PET attenuation correction using synthetic CT from Ultrashort Echo-time MR. Journal of Nuclear Medicine. 2014;55:2071–2077. doi: 10.2967/jnumed.114.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiee N, Bazin PL, Cuzzocreo JL, Ye C, Kishore B, Carass A, Calabresi PA, Reich DS, Prince JL, Pham DL. Robust reconstruction of the human brain cortex in the presence of the WM lesions: Method and validation. Human Brain Mapping. 2014;35:3385–3401. doi: 10.1002/hbm.22409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiee N, Bazin PL, Ozturk A, Reich DS, Calabresi PA, Pham DL. A Topology-Preserving Approach to the Segmentation of Brain Images with Multiple Sclerosis Lesions. NeuroImage. 2010;49:1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imag. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Stuckey S, Goh TD, Heffernan T, Rowan D. Hyperintensity in the subarachnoid space on FLAIR MRI. American Journal of Roentgenology. 2007;189:913–921. doi: 10.2214/AJR.07.2424. [DOI] [PubMed] [Google Scholar]
- Styner M, Brechbuhler C, Szekely G, Gerig G. Parametric estimate of intensity inhomogeneities applied to MRI. IEEE Trans Med Imag. 2000;19:153–165. doi: 10.1109/42.845174. [DOI] [PubMed] [Google Scholar]
- Wang Z, Bovik AC. A universal image quality index. IEEE Signal ProcLetters. 2002;9:81–84. [Google Scholar]
- Wansapura JP, Holland SK, Dunn RS, Ball WS. NMR relaxation times in the human brain at 3.0 Tesla. Jrnl of Mag Reson Imaging. 1999;9:531–538. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Ye DH, Zikic D, Glocker B, Criminisi A, Konukoglu E. Modality propagation: Coherent synthesis of subject-specific scans with data-driven regularization. 16th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2013) 2013:606–613. doi: 10.1007/978-3-642-40811-3_76. [DOI] [PubMed] [Google Scholar]