Abstract
Biomedical ontologies are a critical component in biomedical research and practice. As an ontology evolves, its structure and content change in response to additions, deletions and updates. When editing a biomedical ontology, small local updates may affect large portions of the ontology, leading to unintended and potentially erroneous changes. Such unwanted side effects often go unnoticed since biomedical ontologies are large and complex knowledge structures. Abstraction networks, which provide compact summaries of an ontology’s content and structure, have been used to uncover structural irregularities, inconsistencies and errors in ontologies. In this paper, we introduce Diff Abstraction Networks (“Diff AbNs”), compact networks that summarize and visualize global structural changes due to ontology editing operations that result in a new ontology release. A Diff AbN can be used to support curators in identifying unintended and unwanted ontology changes. The derivation of two Diff AbNs, the Diff Area Taxonomy and the Diff Partial-area Taxonomy, is explained and Diff Partial-area Taxonomies are derived and analyzed for the Ontology of Clinical Research, Sleep Domain Ontology, and Eagle-I Research Resource Ontology. Diff Taxonomy usage for identifying unintended erroneous consequences of quality assurance and ontology merging are demonstrated.
Keywords: ontology version change, summarizing ontology change, ontology quality assurance, summarizing ontology evolution, visualizing ontology evolution, abstraction networks, ontology diff
Graphical abstract
1 Introduction
Ontologies are becoming increasingly important in the domain of biomedicine. Biomedical ontologies support interdisciplinary research [1, 2], information retrieval [1, 2], knowledge management [3–6], natural language processing (NLP) [7–9], and annotation [10], among other applications [1, 11]. A biomedical ontology needs to cover the knowledge, expressed as classes, relationships, attributes, and axioms, of its domain. The structure of a biomedical ontology continually evolves as its content goes through cycles of editing, e.g., adding new domain-specific knowledge or importing additional knowledge from other ontologies. Classes, relationships, etc., are added, deleted, or updated. Each of these modifications affects the knowledge represented in the ontology.
A typical ontology will go through several stages of evolution. The early stage involves the initial design of the ontology, which may include importing one or more upper level ontologies, e.g., the Basic Formal Ontology (BFO) [12]. The later stages involve its maintenance, including periodic updates, which incorporate newly available knowledge into the ontology. Two or more ontologies may also be merged together, creating a new ontology that includes the knowledge from all of the source ontologies. Ontology merging is a complicated process which can lead to many different kinds of problems, including entity redundancy, entity name conflicts, class hierarchy redundancy, and dangling references [13], in addition to severe errors, e.g., wrong or missing parents, incorrect domains or ranges in object properties. To ensure the correctness of the merging process an ontology curator needs a high level view of all of the changes that occur. During its evolution, an ontology may also go through stages of quality assurance (QA), where errors and inconsistencies are identified and corrected. During each of the various stages, the ontology goes through numerous release cycles, where changes are made from one release to the next.
The problem is that while such changes are intended to extend the ontology’s knowledge or to correct previously discovered problems, they may have unintended, and potentially erroneous, consequences. In particular, a QA phase may introduce new errors, while old errors are fixed. Such errors are typically not detected, due to the perception that the change is fulfilling its desired purpose of correcting errors. Sometimes, undesired changes may have broad effects, yet they still might go undetected because the curator “cannot see the forest for the trees.”
Not all editing operations affect an ontology in the same way. While adding a new leaf class will have no global impact, changing the domain of an object property may affect the definition of hundreds of classes. Similarly, modifying superclass axioms may lead to unintended object property inheritance. Having a global view of all of the changes that result from a series of editing operations is important for ontology maintenance. Ontology editing tools, such as Protégé [14], typically show an ontology as an indented hierarchy of classes. A curator can see only a few classes, or one class with its properties, at a time. It is difficult for a curator to identify the overall impact of an editing phase. To find all of the changes, a curator would have to check every potentially affected class, which is impractical for large ontologies.
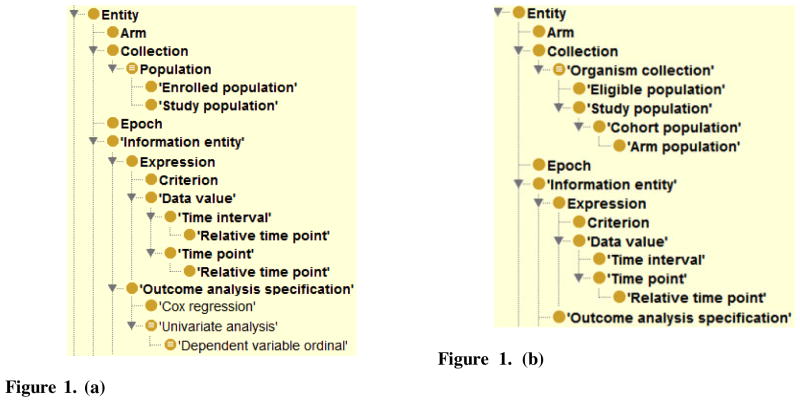
Figure 1(a) illustrates an indented hierarchy for an excerpt of 18 classes from the Entity hierarchy of the Ontology of Clinical Research (OCRe), Release 244 [15]. Figure 1(b) shows the same excerpt, from a later release. Clearly, a series of editing operations were applied between these two releases. While the hierarchical changes are easy to identify in this small example, it is not possible to see other changes, e.g., changes in object property inheritance. To identify unwanted changes, a curator would have to directly compare each version’s class definitions, which is a time-consuming process. If there are dozens or hundreds of classes in the ontology then this manual comparison process is not practical.
Figure 1.
Figure 1. (a) A subhierarchy of 18 classes taken from OCRe Version 244, as shown in Protégé.
Figure 1. (b) The subhierarchy from OCRe Version 258, after several editing operations were applied to the classes of (a).
Whenever working with different versions of a document, whether it contains a diagram, plain text or an ontology, it is important to be able to identify changes between them. UNIX-based operating systems have the “diff” tool for this purpose [16]. For ontologies, the problem of identifying individual changes between two ontology versions has been extensively studied. PromptDiff [17], OWLDiff [18], and ContentCVS [19], among others, identify individual ontology changes in support of collaborative development and version control [20]. However, these tools show individual differences as a list or in an indented hierarchy. If there are hundreds of changes (both explicit and implicit) between two ontology versions, then the amount of difference information becomes overwhelming and unintended changes will likely remain undiscovered. In the Background section we illustrate an excerpt of an ontology diff, using Protégé’s [14] “Compare Ontologies” tool, for the two releases of OCRe shown in Figure 1.
By summarizing, in a compact way, the changes that occur between any two releases, either consecutive or not, of an ontology we may be able to detect unintended consequences of changes, due to the compact representation of the summary diff, and take steps to correct erroneous or undesired side effects of those changes.
In this paper, we introduce a new innovative structural diff technique called a Diff Abstraction Network (“Diff AbN”), for summarizing and visualizing differences between two versions of an ontology. A Diff AbN summarizes the difference in structure and content between two ontology releases. Unlike traditional ontology diff methods, which typically identify axiom changes for individual classes and properties, a Diff AbN shows the overall impact on the whole ontology, summarizing many explicit and implicit structural changes in a compact visualization. Thus, using a compact Diff AbN, an ontology curator can identify the global changes that result from her editing operations. By identifying unintended consequences of changes during the ontology development process, fewer errors will be introduced into the released ontology. Two types of Diff AbNs, the Diff Area Taxonomy and the more refined Diff Partial-area taxonomy, are described and applied to three OWL-format ontologies: the Ontology of Clinical Research (OCRe) [15], the Sleep Domain Ontology (SDO) [21], and the Eagle-I Research Resource Ontology (ERO) [22]. These ontologies are used to illustrate how diff taxonomies can be applied in different stages of ontology evolution. OCRe and SDO both previously [23, 24] underwent a QA phase which identified several errors. ERO was recently merged [25] with the VIVO ontology [26]. Observations by the ontology curators about how the diff taxonomies reflect occurrences and unintended consequences in the QA and merging phases are reported and the DPATs are compared to the output of a traditional ontology diff.
2 Background
2.1 Ontology Diff Approaches
A “diff” is a comparison method that identifies the differences between two versions of a file. Difference detection is important for tracking content evolution and version control. Hunt and McIlroy [16] developed the diff utility for detecting differences between text files. However, the textual diff approach generally does not work well for identifying structural changes between ontology versions. While the OWL [27] and OBO [28] formats define a structure for individual ontology elements (e.g., classes, properties) they do not specify an order in which instances of each element will appear. For example, an OWL file that defines a class A and then another class B represents the same ontology as an OWL file that defines class B and then class A. Thus, the same ontology can be defined using two or more different textual representations. Noy et al. [20] discuss the importance of detecting changes during ontology evolution.
To overcome this problem, various structural diff approaches have been developed. Instead of identifying the textual changes in OWL files, a structural diff identifies individual axiom changes between two ontology versions. Noy and Musen [17] developed PromptDiff, a fixed point algorithm that uses heuristic matchers to compare the axioms of two ontologies. Kremen et al. [18] developed OWLDiff, an open source application for comparing OWL ontologies. Jiménez-Ruiz et al. [19] describe a structural diff approach in support of collaborative ontology development. Goncalves et al. [29] discuss Ecco, a diff tool that uses structural and semantic techniques. Redmond and Noy [30] discuss the OWL Difference Engine, an open source tool for comparing OWL ontologies.
Figure 2 provides an example of a structural diff created using Protégé’s “Compare Ontologies” tool, which is based on the OWL Difference Engine. Entities (e.g., classes or object properties) that have been added, removed or modified are shown on the left. Clicking on an entity shows which axioms were changed. In the example of Figure 2, on the right, the domain of the object property duration in the Ontology of Clinical Research (OCRe) changed from Time interval to Relative time point or Time interval. Additionally, an annotation associated with the object property was also changed.
Figure 2.
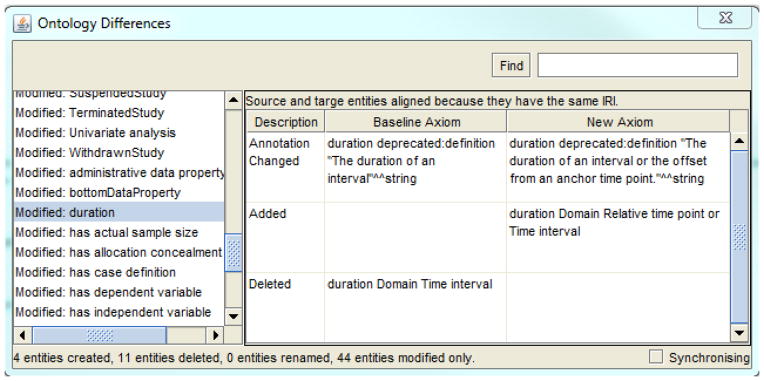
An example of an ontology diff taken from Protégé’s “Compare Ontologies” tool, with the modified object property duration selected.
The view provided by Protégé’s compare ontologies tool, and similar ontology diff tools, enables the review of individual local changes. However, this view does not allow a curator to see the global impact of each change, which is important for discovering unintended and undesired modifications.
2.2 Abstraction Networks
An abstraction network (“AbN”) is a compact network that summarizes the knowledge in an ontology. An AbN is a hierarchical network (allowing multiple parents), which consists of nodes and links. Nodes summarize groups of “similar” classes, where the definition of similar depends on the ontology and the type of AbN being derived. Every class in an ontology is summarized by at least one node. Links between nodes summarize the hierarchical relationships between the groups of similar classes. For a review on AbNs, see [31].
To better support ontology QA for the biomedical field, we introduced a family-based QA approach [32] for the ontologies in the NCBO BioPortal [33]. BioPortal is currently the largest repository of biomedical ontologies. Most ontologies in BioPortal are released in OWL [27] or OBO [28] format, which provide a standard framework for ontology development. He et al. [32] categorized 186 BioPortal ontologies into families according to the types of structural features used to define their classes. The structural features used included object properties, data properties, and subclass configurations, all of which are utilized in the definitions of some ontologies.
We have developed different kinds of AbNs by focusing on common structural features. Each AbN is applicable to many ontologies that have similar structures. Using object properties, we developed the area taxonomy AbN and the more detailed partial-area taxonomy AbN for OWL and OBO format ontologies. We successfully used these taxonomies [23, 24, 32, 34] to:
summarize the Ontology of Clinical Research (OCRe) [15], the Sleep Domain Ontology (SDO) [21], the Drug Discovery Investigations Ontology (DDI) [35], and the Cancer Chemoprevention Ontology (CanCo) [36]
support QA for these ontologies
An object property defines a directed binary relationship between two sets of classes, enabling (but not requiring) their respective instances to be related. Using an example from OCRe, the object property hasMember has a domain Organization and a range Person. This means that when an instance A is related to another instance B via a hasMember object property, then A is an instance of Organization, and B is an instance of Person.
One way of determining an object property’s domain is analyzing its rdfs:domain axiom(s). In OWL, rdfs:domain links an object property to an OWL class. The OWL class(es) within the rdfs:domain are the domain of the object property. For OCRe [15] and CanCo [36] we used this approach to derive domain-defined partial-area taxonomies. While rdfs:domain axioms may contain any kind of class expression (e.g., unions of classes), in our current study we consider each class used in the domain individually.
Another way of determining the domain of an object property is its use in property restrictions. In OWL, an owl:Restriction axiom consists of a property, a constraint, e.g., value constraints like someValuesFrom or allValuesFrom, and range class(es). In OWL, restrictions are treated as anonymous classes. When an OWL class c is a subclass of a restriction r, then c can be treated as belonging to the domain of r’s object property. For SDO [21] and DDI [34] we used restrictions to derive restriction-defined partial-area taxonomies. In the current study we consider all restrictions, regardless of their constraint type.
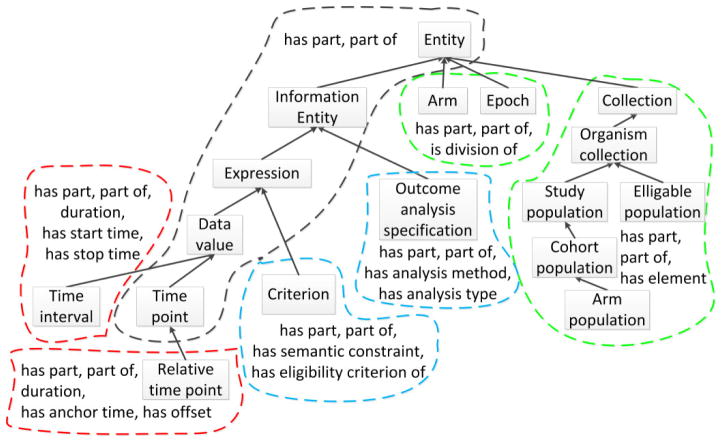
Figure 3(a) illustrates the hierarchy of classes from Figure 1(a), along with the object properties that are used to define these classes. The complete Entity hierarchy consists of 120 classes and uses 75 different object property types. Classes are drawn as labeled boxes and superclass relationships are shown as upward directed arrows. Classes that are within the domain of a particular set of object properties (explicitly or implicitly) are shown in a colored, dashed bubble. For example, the classes Entity, Information Entity, Expression Data value, and Time point are in the domain of the object properties has part and part of, and are enclosed by a dark gray bubble. The classes Arm and Epoch are within the domain of has part and part of by inheritance from Entity and they also introduce another object property, is division of. Thus, they are grouped separately (by the green bubble) from the classes that are in the domain of only has part and part of.
Figure 3.
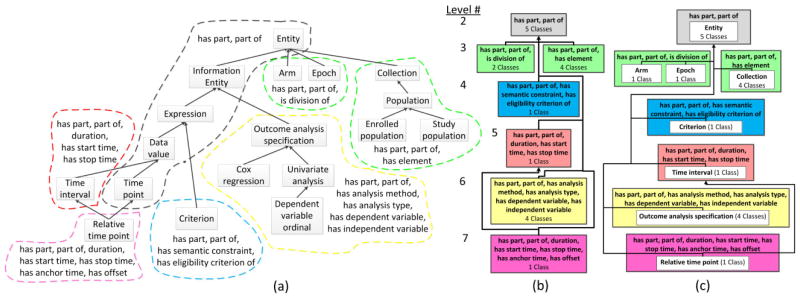
(a) The excerpt of 18 classes from Figure 1(a) shown as a diagram, using bubbles to identify sets of classes with the same object properties. (b) The area taxonomy for the hierarchy of classes in (a). (c) The partial-area taxonomy for the hierarchy of classes in (a).
We define an area as the set of all classes that are in the domain of a given set of object properties. Classes in an area may explicitly be in the domain of an object property (e.g., Organization is explicitly in the domain of hasMember) or may be a descendant of a class which is explicitly in the domain, meaning the class is implicitly in the property’s domain due to inheritance. The set of object property labels is used to name the area. Areas serve as the nodes in an area taxonomy, which provides an object-property-focused summary of an ontology. A root of an area is a class that has no superclasses in the same area; none of a root’s parents are in the same set of object property domains as the root itself. Areas may be multi-rooted. Within an area taxonomy, areas are hierarchically connected by child-of links that are derived from the ontology’s subclass hierarchy. An area A is a child-of another area B if a root class in A has a superclass in B. Areas are disjoint; every class in the ontology is summarized by exactly one area.
Figure 3(b) shows the area taxonomy for the classes in Figure 3(a). The five classes within the domain of has part and part of are now summarized by an area named after that object property set. Furthermore, each area is labeled with the number of classes it summarizes. Child-of links are shown as arrows between areas. Areas are organized into color-coded levels based on their numbers of object properties. Areas with more object properties are at lower levels in the diagram. Levels are numbered according to the number of object properties for the areas in the level, thus lower levels in the diagram have higher level numbers.
Each root of an area defines a partial-area, which is the set of classes consisting of this root and all of its descendants in that area. Partial-areas serve as the nodes for the partial-area taxonomy, which summarizes classes that are both structurally and semantically similar. Partial-areas are connected by child-of links based on the underlying superclass relationships. A partial-area A is a child-of another partial-area B if a parent of A’s root is summarized by the partial-area B. Partial-areas are not necessarily disjoint. If a class has multiple parents it may be summarized by several partial-areas.
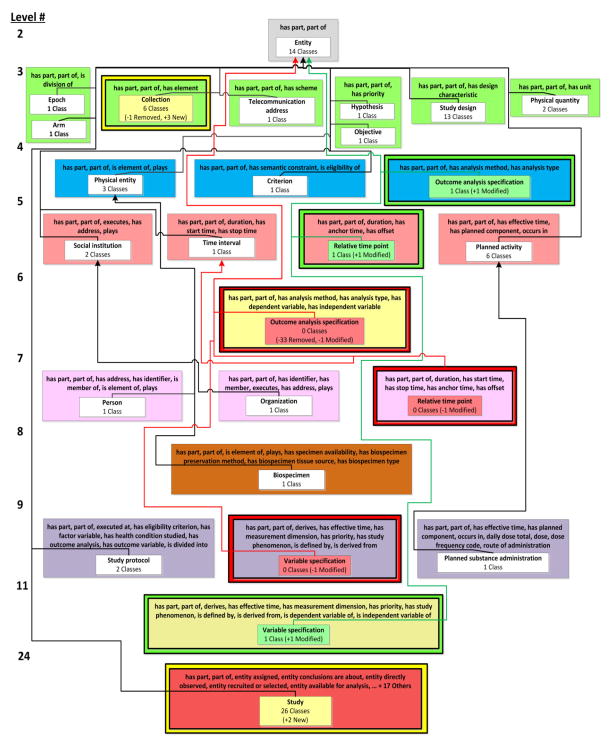
Figure 3(c) illustrates the partial-area taxonomy for the excerpt of classes in Figure 3(a). Partial-areas are represented as white boxes within the colored area boxes and each partial area is labeled using the defining root’s name. Child-of links are shown as thin arrows. The number of classes summarized by a partial-area is shown below the name of the partial area. For example, the multi-rooted area {has part, part of, is division of} has two partial-areas, Arm and Epoch, each summarizing one class. OCRe’s structure is characterized by singly-rooted areas. The complete partial-area taxonomy for OCRe, with 23 partial-areas in 21 areas was derived [23].
2.3 Abstraction Networks in Support of Ontology Quality Assurance
In previous studies [23, 24, 32, 34, 37, 38] we demonstrated how partial-area taxonomies can support the quality assurance of ontologies. As described in Ochs et al. [23], taxonomy-based quality assurance involves reviewing a partial-area taxonomy to see if it conforms to the original conception that the designer of the ontology had. For example, one can see if the various partial-areas indeed have the correct sets of object properties. Such a review can be done by an individual who is familiar with the content and structure of the ontology. This methodology was successfully used to uncover and correct errors in, e.g., OCRe [15], SDO [21], GO [39], NCIt [40], CanCo [36], and DDI [35].
Due to their importance for this paper, we now briefly describe some results from the taxonomy-based QA reviews of OCRe and SDO. For OCRe, we reviewed the Entity hierarchy, which consisted of 120 classes. The taxonomy-based review of this hierarchy identified several significant modeling errors [23]. Two examples include the erroneous inclusion of 33 statistical classes due to incorrect domains for the object properties has dependent variable and has independent variable and an erroneous subclass relationship between Relative time point and Time interval. These errors were corrected by OCRe’s curator (and a co-author) (ST) and a new version of OCRe was released.
In [24] we performed a preliminary QA review of the SDO’s Entity hierarchy, together with the curator of the SDO (SA), also a co-author of the current paper. The partial-area taxonomy for the hierarchy was reviewed and several modeling errors were identified, e.g., duplicate classes and incorrectly assigned object property domains. For example, we identified two pairs of duplicate classes, two clinical finding classes (both imported, one from OGMS [41] and the other from BioTop [42]) and the classes clinical diagnosis and diagnosis. To remove the duplicate classes, equivalence was established between the classes of each pair.
Correcting the various errors led to significant structural changes in the SDO. While a relatively small number of axioms were edited to fix these errors, hundreds of classes were implicitly affected due to these changes. When we manually compared the taxonomy for SDO before the errors were corrected and the taxonomy after the corrections were in place, (SA) was surprised at the extent of modifications to the partial-area taxonomy and could not obtain an adequate display, focusing on those changes, by using the diff view provided in Protégé [14].
3 Methods
Given two releases of an ontology, Ofrom and Oto, we define a new kind of abstraction network, called a Diff Abstraction Network (“Diff AbN”), to summarize and visualize, in a compact way, the global structural changes that occurred when moving from Ofrom to Oto due to editing operations. While previously developed AbNs focused on summarizing the content and structure of a single ontology release, the Diff AbN approach supports the reflection of which structural changes occurred between two ontology releases, and which classes in the ontology were affected by each change, by summarizing the changes that affect groups of structurally similar classes.
We will now describe, in detail, the derivation of two Diff AbNs: the Diff Area Taxonomy (DAT) and the Diff Partial-area Taxonomy (DPAT). A diff area taxonomy summarizes and visualizes the structural changes between Ofrom and Oto. A diff partial-area taxonomy refines the diff area taxonomy by summarizing and visualizing both structural and semantic changes to the subhierarchies of classes in each area. Object properties are an important structural feature used in the definition of many ontologies’ classes [32], thus, it is important to identify the changes that occurred to the sets of object properties used to define the ontology’s classes.
Various types of editing operations can alter the structure of an ontology, and thus, alter the area taxonomy and partial-area taxonomy derived from it. Any editing operation that affects object property introduction or inheritance for a set of classes will affect the taxonomies derived for the ontology. Some examples (labeled E1-E4) include: (E1) Adding or removing a class from an object property’s domain; (E2) Adding or removing an object property from the ontology; (E3) Adding or removing a class from an ontology; (E4) Adding or removing a superclass axiom from a class. Multiple editing operations may be applied to a given class.
As discussed in Background, we previously [23] performed a QA review of OCRe’s Entity hierarchy using a partial-area taxonomy. The QA review identified errors in OCRe’s modeling. To fix the identified errors, OCRe’s curators made significant changes and a new version of OCRe was released. To illustrate the derivation of the diff taxonomies, we will use an excerpt of classes from the version of OCRe we reviewed for errors (Version 244, Figure 3(a)) and the corresponding excerpt for the version released after all of the uncovered errors were corrected (Version 258, Figure 4).
Figure 4.
The excerpt of classes, after corrections (from Release 258) corresponding to the excerpt of Figure 1(b), shown as a diagram.
Figure 4 illustrates the corresponding class hierarchy of Figure 1(b), obtained from Figure 3(a) after several editing operations. Four classes have been removed from the hierarchy: Population, Cox regression, Univariate analysis, and Dependent variable ordinal. Three classes have been added: Organism collection, Cohort population, and Arm population. Outcome analysis specification was removed from the domain of two object properties and Relative time point is no longer a subclass of Time interval, thus it is no longer in the domain of has start time and has stop time. Note that these object property changes, responsible for the removal of the four classes, are not reflected in Figure 1(b).
3.1 Diff Area Taxonomy (DAT)
A Diff Area Taxonomy (DAT) is a Diff AbN that summarizes the structural changes between two different versions of an ontology (i.e., additions, deletions, and modifications to sets of classes with the same set of object properties). The input of a DAT consists of two ontologies Ofrom and Oto and the output consists of a compact, visual summary of the structural changes that occurred between Ofrom to Oto. To detect the changes to all inferable axioms in the ontology a reasoner, e.g., HermiT [43], should be applied to both Ofrom and Oto.
DAT derivation starts with identifying the set of object properties (both introduced and inherited) used to define each class in Ofrom and Oto. Classes and object properties that are added or removed between Ofrom and Oto are also identified. The sets of object properties used to define each class in Ofrom and Oto are then compared. Four kinds of Diff Areas are created based on the identified differences, as follows. These diff areas are used to summarize the structural changes that occurred between Ofrom and Oto.
An Introduced Area indicates a set of object properties for which there exists a set of one or more classes in their domains Oto but no such class exists in Ofrom. The classes summarized by an introduced area display a new object property structure in the ontology. An introduced area may summarize a set of classes that previously were in the domains of a different set of object properties in Ofrom, or they are newly added classes, or both.
A Removed Area indicates a particular set of object properties for which a non-empty set of classes exists in their domains in Ofrom but no such class exists in Oto. The classes that were previously summarized by a removed area are now in the domains of a different set of object properties in Oto or were removed from the ontology.
The third kind is a Modified Area. A modified area exists for both versions of the ontology, i.e., there is a set of object properties A in both versions of the ontology whose domains contain some set of classes (though the set is not the same and one set is not necessarily a subset of the other). If the set of classes in the domain of A in Ofrom is different the set of classes in the domain of A in Oto, then the area named after A is said to be a modified area. Classes that were originally summarized by a modified area in Ofrom may be summarized by different areas in Oto if they were added or removed from A, or the classes may have been removed from the ontology.
If the set of classes in the domains of A is the same in Ofrom and Oto then it is an Unmodified Area.
Given two releases of an ontology, Ofrom and Oto, we define ATfrom as the area taxonomy derived for Ofrom and PATfrom as the partial-area taxonomy derived for Ofrom. ATto and PATto are similarly defined for Oto. These definitions are used to simplify the definitions of DAT (and later, DPAT) elements. For example, an introduced area can alternatively be defined as an area A which exists in ATto but not in ATfrom and a modified area can be defined as an area A which exists in both ATfrom and ATto but summarizes a different set of classes in each version. We will use these definitions to shorten and simplify the descriptions throughout the remainder of this paper.
In regards to the child-of links between areas that summarize the class hierarchy, a child-of is called an introduced child-of if it exists between two areas in ATto but not in ATfrom. Similarly a child-of is called a removed child-of if it exists between two areas in ATfrom but not in ATto. A child-of is an unmodified child-of if it exists between the same two areas in ATfrom and in ATto. Additionally, we define the following rules: (1) All of the child-of links sourced at an introduced area are introduced child-ofs; (2) All of the child-of links sourced from a removed area are removed child-ofs. We note that modified areas and unmodified areas may have introduced, removed, or unmodified child-ofs. Note that child-of links cannot be modified because a child-of link either existed or did not exist in ATfrom.
A DAT is represented as a compact hierarchical network of diff area nodes connected by child-of links based on the subclass hierarchies in Ofrom and Oto. In a DAT, all areas are shown, including removed areas which summarize no classes in ATto.
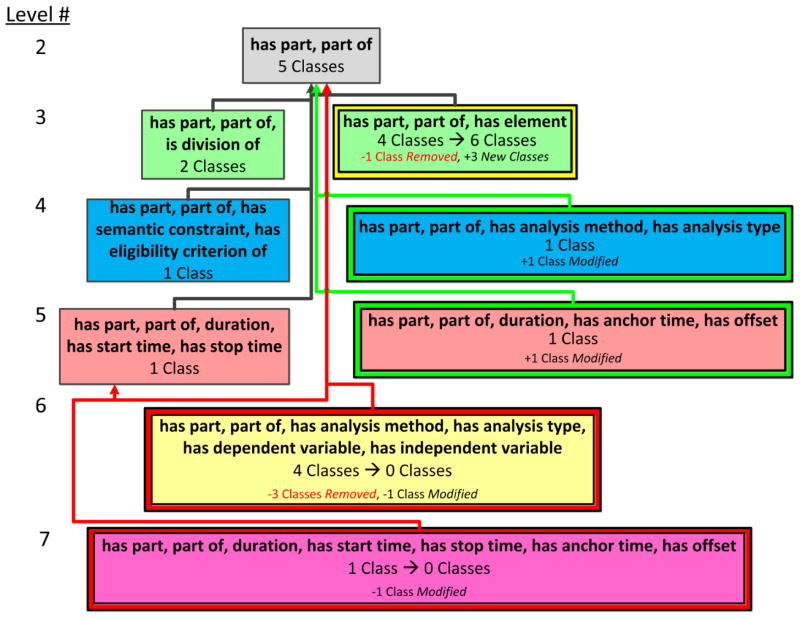
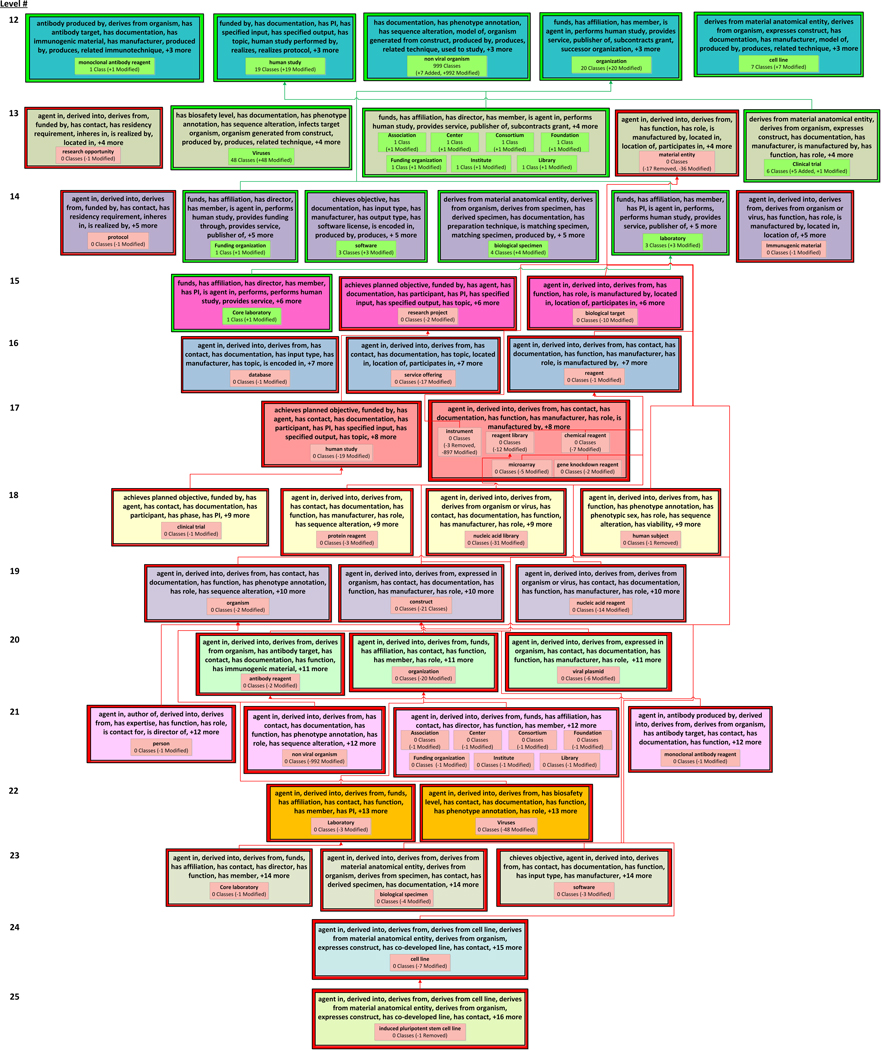
The OCRe DAT, shown in Figure 5, captures the structural changes between the ontology excerpt of Figure 3(a) and Figure 4.
Figure 5.
The visualization of the diff area taxonomy between the ontology excerpts in Figure 3(a) and Figure 4. The diff areas are organized into color coded levels according to the number of their object properties. The level numbers appear at the left edge of the figure. Diff areas are shown with differed colored borders to indicate the type of diff area. Modified areas, introduced areas, and removed areas are drawn with a yellow border, green border, and red border, respectively. Unmodified areas are shown with no border. Child-of links are colored red, green, or black if they were removed, introduced, or unmodified, respectively. (Child-of links cannot be modified.)
If the number of classes summarized by an area changes between Ofrom and Oto, e.g. the area {has part, part of, has element} summarizes four classes in ATfrom but six in ATto, then the change is noted using an arrow from the old number to the new number (i.e., 4 Classes → 6 Classes). A brief textual summary of the modifications in the area is shown under the number of classes summarized by the area. For example, the area {has part, part of, has element} indicates that one class was removed from the ontology (“−1 Class Removed”) and three classes were added to the ontology (“+3 New Classes”) (see right green box in Figure 5). In addition to “new” and “removed,” a third label, “modified”, indicates if one or more classes that existed in both Ofrom and Oto were modified and moved from one area to another, e.g., Relative time point went from the removed area {has part, part of, duration, has start time, has stop time, has anchor time, has offset} (“−1 Class Modified”) to the introduced area {has part, part of, duration, has anchor time, has offset} (“+ 1 Class Modified”).
Ontology editing operations have various effects. For example, removing the superclass axiom (E4) between Relative time point and Time interval resulted in Relative time point being summarized by a different area, {has part, part of, duration, has anchor time, has offset} (Level 5).
The diff areas {has part, part of, has analysis method, has analysis type} (Level 4) and {has part, part of, duration, has anchor time, has offset} (Level 5) are introduced areas marked with a green border; they exist in ATto but did not exist in ATfrom. In this example, the introduced areas in Figure 5 summarize classes that were summarized by different areas in ATfrom (Figure 3(b)). This indicates a change in the object property structure of these classes (due to E1and E4, respectively) and they are now defined differently.
In Figure 5, the single diff area on Level 6 and the single diff area on Level 7 are removed areas, as indicated by their red borders; these areas existed in ATfrom but no longer exist in ATto. It is important to display the removed areas in the DAT figure, even though these areas no longer exist in ATto, to capture the important change(s) that resulted in their removal. For example, several editing operations led to the yellow Level 6 area being removed: three classes (e.g., Cox regression) were removed from the Entity hierarchy (E3) and the class Outcome analysis specification is summarized by a different introduced area, {has part, part of, has analysis method, has analysis type} (Level 4) due to (E1).
In Figure 5 {has part, part of, has element} is a modified area (with a yellow border), because the class Population was removed from the ontology (E3) and three new classes, Organism collection, Cohort population, and Arm population, with the modified area’s object property set, were added to the ontology (also E3). The new classes inherited their object property set, because they are descendants of Collection and they introduce no new object properties to the subhierarchy. The unmodified areas are {has part, part of}, {has part, part of, is division of}, {has part, part of, has semantic constraint, has eligibility criterion}, and {has part, part of, duration, has start time, has stop time}.
3.2 Diff Partial-area Taxonomy (DPAT)
In the past, the partial-area taxonomy has been used to support QA of ontologies [23, 24, 32, 34] for various ontologies. A Diff Partial-area Taxonomy (DPAT) summarizes the changes to the subhierarchies of classes in each DAT area. Just as a partial-area taxonomy is a refinement of an area taxonomy into partial-areas (i.e., semantically similar subgroups within the structurally similar area groups), a DPAT refines a DAT by summarizing subhierarchy changes, represented as changes to the partial-areas in each area.
The derivation of the DPAT starts from the already derived DAT. For each diff area A in the DAT, the changes to the subhierarchies of classes in A, as named after the roots, are summarized. The set of root classes of A in ATfrom is compared to the set of root classes of A in ATto, in cases where A exists in both. If the two root sets are not equal this indicates that partial-areas have been introduced or removed (or both) from the area. Based on the identified changes, four kinds of Diff Partial-areas are created.
We define an Introduced Partial-area as a partial-area that exists in area A in PATto but did not exist in A in PATfrom. A partial-area is introduced to an area A whenever a root class is added to A or stopped being a root of A. Partial-areas can be introduced to any diff area that is not a removed area. All partial-areas in an introduced area are by definition introduced partial-areas.
We define a Removed Partial-area as a partial-area that exists in area A in PATfrom but not in A in PATto. A partial-area is removed from an area whenever a root class is removed from A. Partial-areas can be removed from any diff area that is not an introduced area. All partial-areas in a removed area are by definition removed partial-areas.
If area A has one or more of the same root classes in both PATfrom and PATto then the subhierarchies of classes from both versions are compared. A Modified Partial-area is a partial-area that exists in A in both PATfrom and PATto and summarizes a different set of classes in PATto than in PATfrom.
An Unmodified Partial-area is a partial-area that summarizes the same set of classes in area A in PATfrom and in area A in PATto.
We note that an unmodified area can contain modified, introduced, and removed partial-areas. This occurs when the set of classes summarized by the unmodified area remains the same between Ofrom and Oto but the subhierarchies of classes change within the diff area. For example, if a descendant of a root class in A is made a sibling of the root class then a partial-area is introduced within the unmodified area. Similarly, if a class is summarized by two partial-areas in PATfrom (which are, thus, not disjoint) but only one partial-area in PATto, the diff area can still be unmodified. We also note that the definition of child-ofs between diff partial-areas follows that of the child-ofs between diff areas.
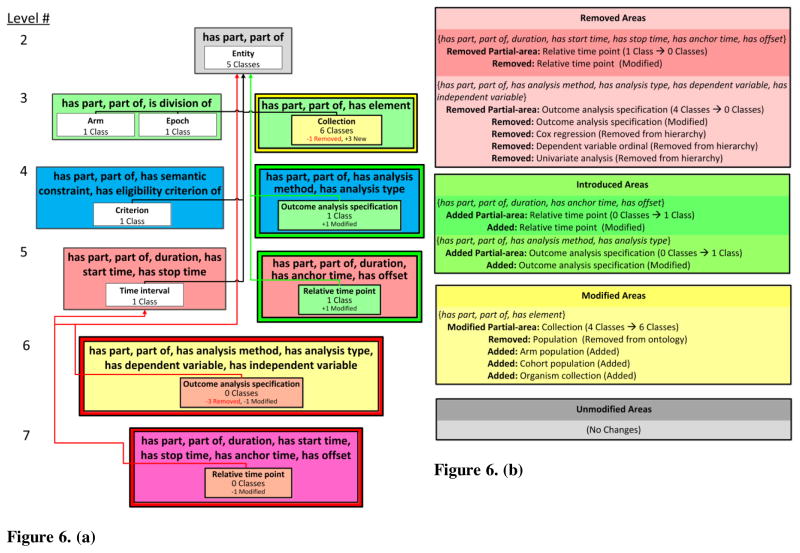
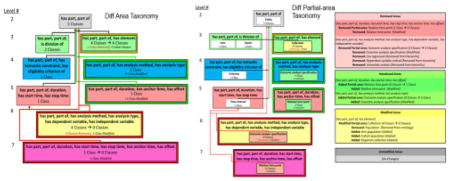
The DPAT consists of a visualization and a textual list of differences. The visualization of a DPAT is composed of a refined DAT visualization where the DPAT partial-areas are shown within their respective DAT areas. Figure 6(a) shows the visualization of the DPAT capturing the changes from the ontology version shown in Figure 3(a) to the new version in Figure 4.
Figure 6.
Figure 6. (a) The visualization of the DPAT between Figure 3(a) and Figure 4. Modified, introduced, and removed partial-areas are shown with a light yellow, light green, and light red background, respectively. A summary of changes is shown below the number of classes summarized by each partial-area. Unmodified partial-areas are shown with white background. Child-of links between partial-areas are colored red, green, and black if they are removed, introduced, or unmodified, respectively.
Figure 6. (b) Color-coded text output for the DPAT between Figure 3(a) and Figure 4. The text output is composed of changes grouped by area change type (e.g., removed or modified area). Within each type, the list of affected areas is shown. Indented under each area is a list of modifications to the partial-areas within the area. The modifications to the set of classes summarized by each partial-area are listed indented under the partial-area root (which is its name).
Figure 6(b) shows a colored example of text-based output for the DPAT between the ontologies in Figure 3(a) and Figure 4. The background color alternates between brighter and darker shades, in order to visually separate different areas. The text-based output is combined with the visualization to provide more details about the modification of each affected taxonomic element.
In Figure 6(a), the introduced partial-area Outcome analysis specification appears in the area {has part, part of, has analysis method, has analysis type} (Level 4) and the introduced partial-area Relative time point in the area {has part, part of, duration, has anchor time, has offset} (Level 5). Both of these diff areas are introduced areas, as indicated by their green borders. Note that the green, red, and yellow colors of the areas in levels 3, 5, and 6, respectively, do not communicate changes to the areas, but are the colors of the different levels. At the same time, Outcome analysis specification and Relative time point are removed partial-areas in the removed areas of Levels 6 and 7. Occurrences of identically named introduced and removed partial-areas reflect the changes in object properties of their root class in the DPAT. In both of these cases the classes were removed from the domains of the object properties as a result of the errors discovered in Ochs et al. [23].
Collection in {has part, part of, has element} is a modified partial-area because one class was removed from the ontology and three new classes were added to the ontology as descendants of Collection. Entity, Arm, Epoch, Criterion, and Time interval are unmodified partial-areas.
3.3 DPATs Ontology Evolution Phases
DPATs highlight groups of similar classes that underwent some kind of structural change between two ontology releases. By deriving a DPAT for two ontology releases, a curator has access to a simplified view that allows her to focus on the important changes that occurred. During the various stages of ontology evolution, these changes may occur for different reasons. During the initial development stages of an ontology, and during expansion stages, significant amounts of new knowledge, e.g., classes and properties, will be added. In a DPAT, this type of evolution will be reflected by many introduced and modified partial-areas. If an ontology undergoes a phase of quality assurance, then existing content will be modified. In such a case, there may be relatively more removed areas and removed partial-areas than in an expansion phase. When two or more ontologies are merged together, many different kinds of editing operations are applied, including the addition and removal of entities from the source ontologies. For example, redundant classes and properties may be removed.
During each phase of ontology evolution, an ontology curator should review a DPAT after one or more editing operations are applied. In particular, the curator should review classes that are summarized by added, removed, and modified partial-areas. For example, if the domain of an object property is changed, then the curator should review the classes in the added and removed partial-areas to ensure they have the correct sets of object properties. Similarly, if a subclass relationship is established or removed between two classes, then the curator should review all of the diff partial-areas that contain the descendants of the modified class to ensure that the inheritance of object properties is still correct. During the merging process, the integrated content should be reviewed to ensure the classes are within the domain of the correct sets of object properties. This can be accomplished by reviewing the added and removed partial-areas. To ensure that there is no redundancy in a subhierarchy of classes, modified partial-areas can be reviewed.
4 Results
To illustrate the utility of diff taxonomies we derive DPATs for ontologies that have gone through different kinds of evolutionary phases. For the quality assurance phase we illustrate DPATs for the Ontology of Clinical Research (OCRe) and the Sleep Domain Ontology (SDO). OCRe also underwent additional expansion unrelated to the QA phase. For the case of two ontologies being merged, we illustrate a DPAT for the eagle-I Research Resource Ontology (ERO), which was recently merged [25] with the VIVO Ontology for Researcher Discovery (VIVO) [44]. OCRe, SDO, and ERO are publicly available on the NCBO BioPortal [33] ontology repository.
For each ontology we investigate the DPAT’s ability to visualize and summarize the changes between releases, compare the DPAT to the output of a traditional ontology diff tool, and review portions of the DPAT to determine if the editing operations led to any inconsistent or incorrect modeling in the ontology.
As described in Background, OCRe and SDO both underwent a phase of QA. We identified, together with their curators, several errors and inconsistencies that were confirmed and corrected. In both cases we derived taxonomies before and after our QA review [23, 24] and manually compared pairs of taxonomies. In this study we derived DPATs for these ontologies to examine the reflection of the exact changes that were implemented due to our QA reports (i.e., traced evolution [45]). This allows us to illustrate how different changes are captured by the DPAT. Additionally, following the methodology of Section 3.3, we review the diff partial-areas in the in the OCRe and SDO DPATs to ensure that the corrections did not result in any unintended or erroneous changes. We use the ontology release before the QA review and the ontology release immediately after the errors uncovered during the QA review were corrected
ERO represents a case of ontology evolution due to merging. In comparison to OCRe and SDO, ERO underwent a significantly more complex series of editing operations. For this study, this represents a case of untraced evolution [45]; we were unaware of the specific changes which occurred when deriving the DPAT. To obtain insight into how the ERO DPAT captures various design decisions made during the merging process we collaborated with (MH) and (MB), ERO’s curators and co-authors on this paper.
To illustrate how DPATs summarize changes between two ontology releases, and how they may capture more changes than a regular structural diff, we compare the information provided by the DPAT (including the textural output) to a diff created using the “Compare Ontologies” feature in Protégé [14], which is based on the OWL Difference Engine [30]. For short, we’ll refer to this tool as “Protégé diff.”
To derive the OCRe, SDO, and ERO diff taxonomies, we have built a prototype software tool (see Future Work) that produces DATs and DPATs. The tool is implemented in Java using the OWL API [46]. The output of the tool is used to create DAT and DPAT visualizations (e.g., Figure 5 and 6) and to explore which changes affected the classes summarized by each diff taxonomy element. Additionally, the tool provides a list of editing operations which affected each DAT/DPAT element.
4.1 Ontology of Clinical Research DPAT
The OCRe Entity hierarchy DPAT (Figure 7) captures the structural changes that occurred due to the corrections implemented by OCRe’s curator and a co-author (ST). OCRe also underwent additional editing unrelated to our QA review. The complete DPAT has two modified partial-areas, three deleted partial-areas, and three added partial-areas, summarizing the changes to 32 classes (see diff areas with yellow, red, and green borders, respectively). Eighteen partial-areas are unmodified. The details of the editing operations that affected each DPAT element are provided in the textual output of the DPAT tool.
Figure 7.
The complete diff partial-area taxonomy for OCRe.
Consider the two errors mentioned in background: the erroneous inclusion of 33 statistical classes and the incorrect subclass relationship between Relative time point and Time interval. To correct these errors, OCRe’s curator applied several editing operations, the results of which are summarized by the elements of the DPAT. The removal of the 33 statistical classes is highlighted by the removed partial-area Outcome analysis specification on Level 6. From the removed partial-area one can see that the 33 classes were removed from the hierarchy (“−33 Removed”) and one class (Outcome analysis specification) was modified, as it now is summarized by the introduced partial-area on Level 4. From this view, one can see that Outcome analysis specification now is in the domain of fewer object properties; specifically it is no longer in the domain of has dependent variable and has independent variable. The changes to Relative time point are similarly reflected.
In parallel to our QA review, OCRe’s curators made small modifications to other parts of the Entity hierarchy. These changes are reflected by the modified partial-areas Collection and Study, which both had new classes added under their root. From the DPAT, one can see which object property domains these newly added classes are in.
In comparison, the Protégé diff identified 27 modified entities (classes, properties, etc.). However, since OCRe underwent additional development outside of our QA review [23], eleven of these entities did not have any structural changes (only changes to annotations, e.g., class labels). Four modified classes had restrictions removed, which is not captured by the DPAT shown in Figure 7, but is captured by a DPAT derived using property restrictions (like for SDO’s DPAT). The remaining 12 modified entities relate to the addition and removal of classes and changes to object property domains, e.g., duration. Without a DPAT, identifying the 12 structural changes requires a user to manually review each change in the standard diff. Furthermore, the standard diff did not provide a view that shows the definition of the classes impacted by the change, e.g., Relative time point, which already had the has offset object property.
In comparison with the Protégé diff, the DPAT provided a more accurate and concise view of the implicit structural changes that occurred. The Protégé diff did not explicitly identify the removal of the 33 statistical classes from the hierarchy, which was a major change. The only differences identified, that were related to this change, were the modifications to the domains of object properties has dependent variable and has independent variable. The removal of the classes from the hierarchy is only apparent after applying a reasoner (e.g., Hermit [43]) to the ontology and performing a manual comparison of the output and the input.
Following the recommendations in Section 3.3, we reviewed the added and removed areas and partial-areas in the OCRe DPAT to determine if their classes are in the domain of the correct set of object properties. In Figure 7, we find the introduced partial-area Relative time point in the introduced area {has part, part of, duration, has anchor time, has offset}. This diff area and this diff partial-area were introduced due to the removal of an incorrect subclass relationship to Time interval [23], which corrected the erroneous inheritance of two object properties (has start time, has stop time) by Relative time point.
After reviewing this introduced area in the DPAT, we identified that Relative time point has another incorrect object property: duration, since a time point has no duration. Indeed, this object property was determined to be redundant with has offset. When correcting the Relative time point class, the domain of duration was changed from only Time interval to Time interval or Relative time point, due to the removal of the subclass relationship between Relative time point and Time interval. Hence, the duration object property was no longer inherited by the class Relative time point. Upon investigation, it was found that Duration was previously used to express offsets for relative time points but this should have changed when the object property has offset was introduced to the ontology. (ST) confirmed the error and Relative time point was consequently removed from the domain of the duration object property.
4.2 Sleep Domain Ontology DPAT
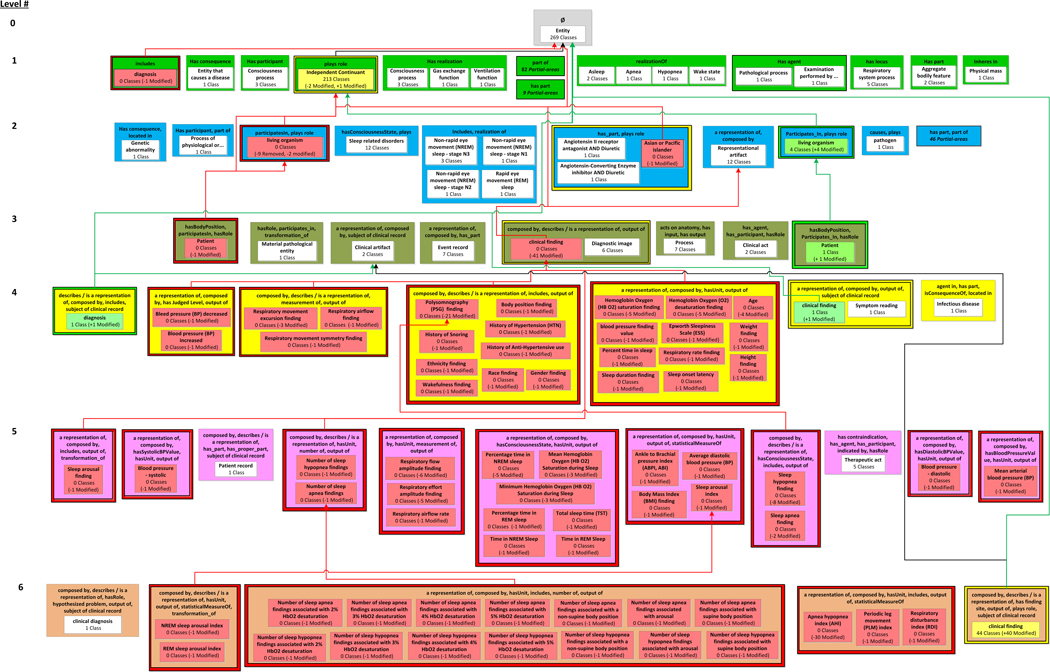
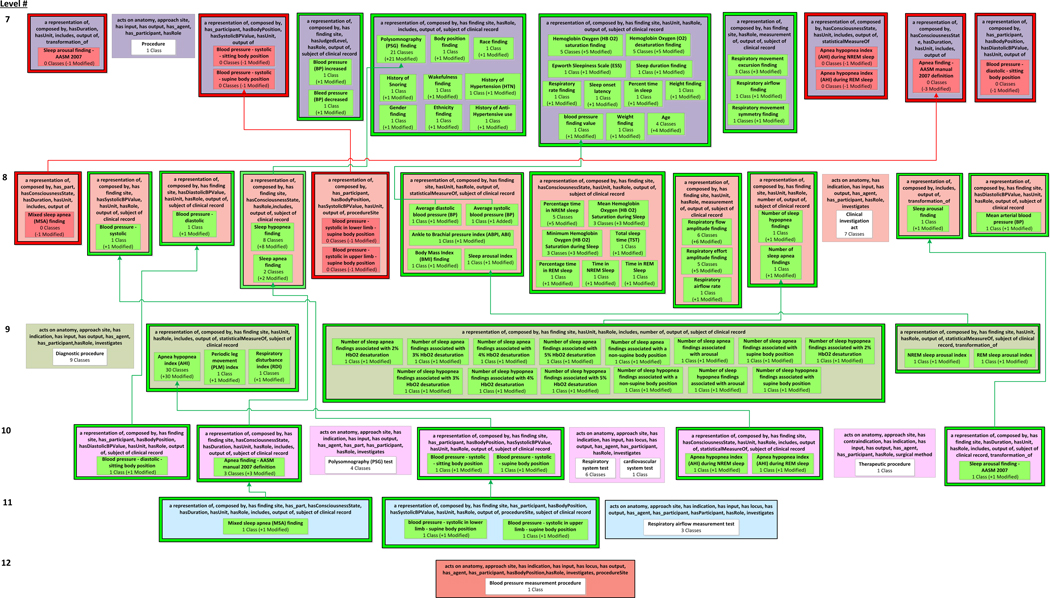
The corrections that were implemented during the QA review of SDO resulted in many classes’ object property sets changing, as captured by the 25 removed areas, 25 introduced areas, and four modified areas (along with all of their diff partial-areas) in the SDO DPAT in Figure 8 and Figure 9 (derived using the inferred versions of SDO). When there is this much structural change between two releases of an ontology, in terms of the sets of object properties and their domains, there is a greater chance of a class being in the domain of incorrect set of object properties.
Figure 8.
The top half of the Sleep Domain Ontology’s diff partial-area taxonomy. An excerpt of the child-of links between added/removed diff partial-areas is shown.
Figure 9.
The bottom half of the Sleep Domain Ontology’s diff partial-area taxonomy.
By reviewing the introduced partial-areas in the SDO’s DPAT, we identified several problems with the object properties for the equivalent classes. Even though the clinical finding partial-area on Level 3 was (correctly) removed and 42 of its classes are now summarized by the clinical finding modified partial-area in the Level 6 modified area {a representation of, composed by, has finding site, hasRole, output of, subject of clinical record}, we found an introduced partial-area clinical finding (with one class) on Level 4 in the modified area {a representation of, composed by, output of, subject of clinical record}. Similarly, we still found diagnosis introduced at Level 4 (and removed from Level 1) in {composed by, describes/is a representation of, includes, subject of clinical record} (the object properties in bold are extra).
But the equivalent class clinical diagnosis is in {composed by, describes/is a representation of, hasRole, hypothesized problem, output of, subject of clinical record}. The object properties for equivalent classes should be equivalent. However, as shown in bold, they are not. For diagnosis, one is not even a subset of the other. By reviewing the added and removed partial-areas that contain the classes that were edited, several inconsistencies were identified. Both equivalent classes should have the union of the two sets of object properties, as confirmed by SDO’s curator.
The Protégé diff for the SDO identified one added class, ten removed classes, and seven other structural changes (e.g., the equivalences described above). Unlike OCRe, which underwent development unrelated to our QA review, the SDO only changed due to the error corrections described by us [24]. However, the Protégé diff did not provide a complete picture of the changes that occurred, particularly in regards to inheritance of object properties.
For example, while the Protégé diff identified the added equivalence axioms between the two clinical finding classes, it did not capture how this change affected their many descendent classes. Furthermore, the Protégé diff did not provide a way of directly comparing the properties for the classes that were declared equivalent. Additionally, it did not uncover that the object property sets for these equivalent classes were not equivalent, as found in the DPAT. Uncovering these changes without a DPAT requires a manual comparison of the SDO before and after the QA review.
4.3 eagle-I Research Resource Ontology DPAT
The eagle-i Research Resource Ontology (ERO) [47] was developed as part of the eagle-i project [48], which enables biomedical researchers to discover scientific resources via a searchable network of resource repositories. These repositories are curated by over 20 different research institutions [48]. Like the SDO, ERO imports the content of several external ontologies, including BFO and OCRe. However, ERO differs from OCRe and SDO in that it is used to drive applications for data entry and search. ERO is composed of several modules. Notably, the representation of research resource data is in a separate module from the representation of application specific data used to control the appearance and behavior of the user interface. Many of ERO’s classes and properties in the application module were designed to drive eagle-i’s user interface and the various data collection tools used in the eagle-i project.
Unlike OCRe and SDO, which had a relatively small number of local editing operations applied to correct modeling errors uncovered during our quality assurance reviews, ERO underwent a significantly more complex sequence of editing operations. ERO was merged with the VIVO ontology, which covers the orthogonal but overlapping domain of researcher interests, activities, and accomplishments. We derived a DPAT for the version of ERO before the merge (August 2013 release on BioPortal) and the version after the merge (available at [49]), with the goal of summarizing the major structural changes that occurred due to the merge.
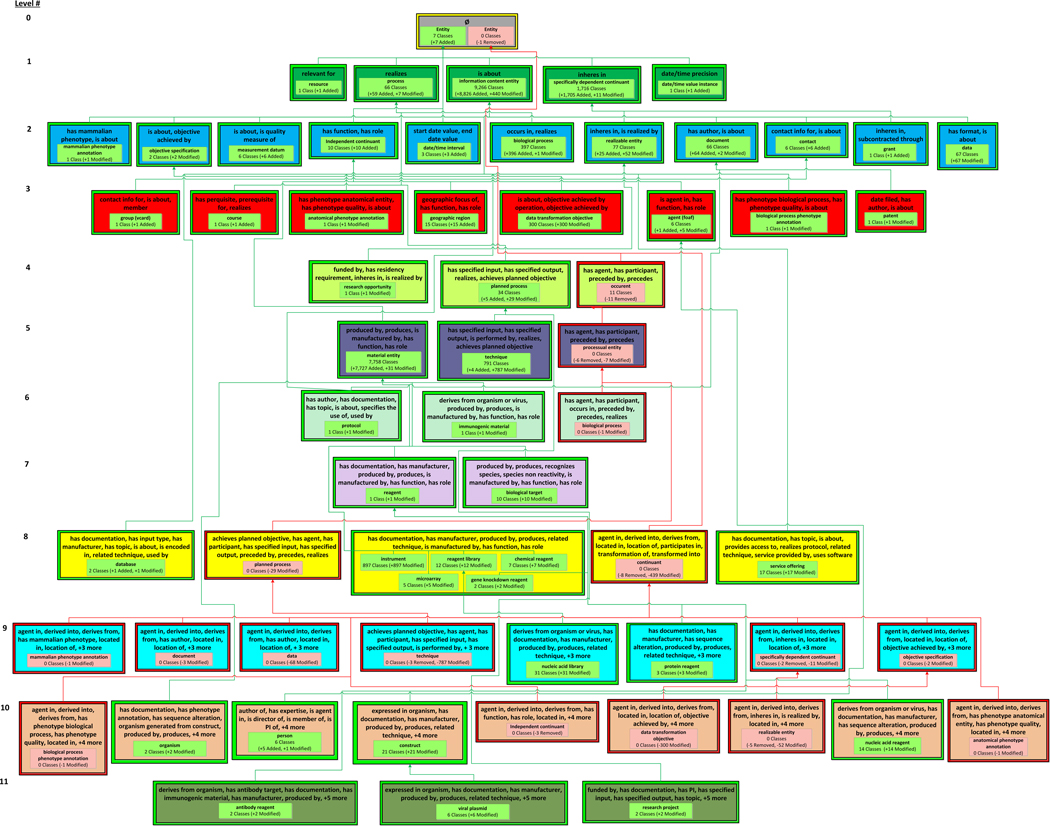
The ERO DPAT, which has 26 levels, is shown in Figure 10 and Figure 11. Child-of links from diff partial-areas in Figure 11 that have a parent diff partial-area in Figure 10 are not shown. The structural changes resulting from the merge are summarized by the 57 introduced areas, 48 removed areas, and one modified area (the root area) of ERO’s DPAT. Like OCRe, most of ERO’s areas are singly-rooted, meaning there is only one partial-area in most areas.
Figure 10.
The top portion of the ERO diff partial-area taxonomy, summarizing all classes with 0–11 object properties.
Figure 11.
The bottom portion of the ERO diff partial-area taxonomy. Most of the diff areas at lower levels are removed areas due to the reduction in ERO’s complexity.
The most significant structural change highlighted by ERO’s DPAT is the highly desirable overall reduction in complexity, in terms of number of object properties used to define ERO’s classes. This change is reflected in the large number of removed areas at the bottom of the DPAT (Figure 11) (since the levels are listed in increasing order according to the number of object properties of the areas) and the large number of introduced areas at the top (Figure 10). For example, before the merge, the class cell line (Level 24 in Figure 11) and its six descendants were in the domain of 24 object properties (16 by inheritance from reagent (1), eight introduced explicitly at cell line). After the merge, cell line (the rightmost introduced partial-area in the top level (Level 12) in Figure 11) and its descendants are in the domain of only 12 object properties (7 inherited, 5 introduced).
A combination of changes led to this reduction in complexity. First, the ontology’s set of object properties was significantly changed. A total of 36 object properties were removed and 91 object properties were added. This affected many classes. For example, cell line was implicitly in the domain of the object property agent in, whose domain was defined as continuant. The object property agent in was removed from the ontology. Some of these removals happened because a newer version of the Relations Ontology (RO) [50] was imported.
Prior to the merge, eight object properties imported from RO had continuant assigned as a domain. All of these object properties are no longer in the ontology after the merge. Thus, the many descendants of continuant are no longer implicitly in their domains. Several of these object properties were replaced with object properties from a newer version of RO that had no domains (e.g., has participant). However, their sub properties (e.g., has specified input, a sub property of has participant), retained the same domains after the merge.
In regards to the 91 newly added object properties, from the DPAT one can see that these newly introduced properties have domains that are mostly disjoint, since there are few classes that are in the domain of many object properties. There are a total of 13 introduced areas in the top four levels of Figure 11 but 26 removed areas in levels 16–25. After the merge, the most complex class is core laboratory, with 15 object properties, as compared to the most complex class before the merge, induced pluripotent stem cell line, with 25 object properties.
The DPAT view shows that by adding more object properties than were removed, ERO became richer in terms of types of properties used to define classes, but also simpler in its model, as the number of object properties per class was reduced.
The second kind of change that led to a reduction in complexity was the modification of various object property domains. For example, before the merge, the domain of the object property has sequence alteration was (cell line or protein reagent or nucleic acid reagent or human subject or organism). After the merge, the introduced partial-area cell line is no longer in the object property’s domain.
By comparing the object properties of the removed partial-area cell line and the introduced partial-area cell line, we found that no new object properties were added to cell line, five removed object properties were removed from RO (agent in, derived into, derives from, located in, and location of), one RO object property’s domain was modified (participates in), three ERO object properties were removed (derives from cell line, has co-developed line, and has contact), and one ERO object property was modified (has sequence alteration). (MB), an ERO curator and a coauthor, reviewed a sample of added/removed diff area pairs that contained classes defined by ERO (e.g., Document, Organization, Person, and Technique) and confirmed that the classes are in the correct set of object property domains.
Another major structural change for ERO is evident from the very large introduced partial-areas in the DPAT, e.g., the information content entity introduced partial-area in the introduced area {is about} summarizes 9266 classes. Over 8,800 of these classes are new to ERO. Most were imported from other ontologies, e.g., the Mammalian Phenotype Ontology (MP) [51] and the Software Ontology (SWO) [52]. Similarly, most of the 7,727 added classes in the introduced partial-area material entity (7758) are imported from UBERON [53]. From the DPAT, one can see the property structure of the classes imported from these ontologies.
The Protégé diff of ERO was several orders of magnitude larger than the Protégé diffs for OCRe and SDO. A total of 19,256 entities (mostly classes) were identified as created, 159 were deleted, 27 were renamed, and 609 were modified. Reviewing each of these changes (20,051 in total) is impractical. In comparison with the DPAT, the Protégé diff is overwhelming to the point of being practically unusable.
5 Discussion
In this paper we introduced the innovative idea of Diff Abstraction Networks. The need for a Diff AbN methodology emerged out of feedback obtained from the ontology curators we worked with in our previous studies [23, 24]. Diff AbNs summarize and visualize the structural changes that occurred between two ontology releases, an idea not found in any other publication during our literature review. A curator can inspect the change summary provided by the DAT and DPAT to review global changes, as well as determine if the changes have any unintended side-effects (e.g., incorrectly assigned or inferred object property domains). In particular, due to the summary, the curator could quickly determine if the classes in the various areas and partial-areas have the intended object properties.
Such a detection of unintended consequences is less likely if the curator needs to review an OWL-based structural diff between ontologies [17–19] since the amount of information would be overwhelming, as detailed for the SDO audit [24]. Furthermore, unintended and erroneous changes may be identified by reviewing a Diff AbN for nonconsecutive releases, since some unintended changes may not be detected for consecutive pairs of releases, but may be detected between releases that are farther apart, due to the cumulative impact of the changes made between consecutive releases.
In comparison with standard ontology diff approaches, which generally only identify individual changes per-entity (e.g., class or property), the Diff AbN approach shows the global impact of an editing operation. With the Diff AbN a user does not have to manually scan through potentially hundreds or thousands of entries to identify important structural changes. Furthermore, the Diff AbN approach shows the implicit changes that occur due to inheritance of properties within an ontology, e.g., for the many descendants of Clinical finding in the SDO.
Notably, even when comparing diagrams of the complete before taxonomy and after taxonomy of two releases it is difficult for a curator to notice the differences between them. She would have to manually compare the classes and object properties of these two taxonomies and detect the changes. This task is overwhelming for the curator. Thus, we introduced the DPAT, which summarizes the changes. As a matter of fact, the researchers of the current paper failed to detect the above mentioned unintended changes, even though they reviewed the before and after taxonomies of OCRe and SDO and published the results [23, 24].
Another potential use of Diff AbNs is to compare the stated and inferred versions of an ontology to determine if the inferred axioms are correct or have unintended consequences. An error may not be easily detectable in the stated view but may become apparent after a reasoner has been applied. The DAT and the DPAT would show the structural differences between these two views. By creating a DPAT between the stated version of OCRe and the inferred version of OCRe (before our QA review), it would be easier to identify the incorrect object property domains and the erroneous inclusion of 33 statistical classes into OCRe’s Entity hierarchy, discussed above.
One potential issue with the DAT and the DPAT is that they produce diagrams that are larger than the taxonomy diagrams of Obefore and of Oafter. A DPAT shows all of the areas and partial-areas of the “before partial-area taxonomy” and the “after partial-area taxonomy.” For example, in the DPATs of the SDO and the ERO (Figure 8 and Figures 10–11, respectively), there are many pairs of added/removed areas and partial-areas. One way of simplifying the DAT and DPAT is to define various views that only show certain types of Diff AbN elements. For example, if a curator is only interested in what has changed, then she can hide unmodified areas and unmodified partial-areas. Alternatively, the curator can view only introduced areas and partial-areas, etc.
One potential drawback of the DPAT is that internal changes within diff partial-areas (e.g., changing the subclass hierarchy within an unmodified partial-area) are not identified. For such a case, a structural diff excerpt for the changed classes within the partial-area could be reviewed, thus producing a targeted partial ontology diff that does not overwhelm a user.
In addition to comparing Protégé diff outputs with DPATs, we asked (ST), (SA), and (MH), the curators of OCRe, SDO, and ERO, respectively, and co-authors of the current paper, to comment on how they used structural diff tools during the previously described development phases [23–25]. After correcting the errors found by Ochs et al. [23], (ST) did not use any diff tools to compare the before and after versions due to the small number of relatively simple changes. In general, he uses OWLDiff [18] when there is a specific need to compare the axioms of two ontology versions. When initially designing the SDO (SA) also occasionally used OWL Diff. However, due to the limited benefits he derived from using it, he did not use it to compare the two releases of the SDO reported in previous work [24]. In contrast (SA) found the DPAT very helpful due to the visualization that compactly summarizes changes. In comparison, OWL Diff presents changes in a text-based indented hierarchy, which can be overwhelming in length, making it difficult to find an important change.
During the merge of ERO and VIVO, the ERO development team used an in-house diff tool [25], that integrates spreadsheet-based information, e.g., class equivalences, with Protégé. Their diff tool highlights different classes based on various modeling decisions. They did not use any third-party diff tools, e.g., OWLDiff or Protégé’s Compare Ontologies tool, due to the various needs and levels of experience on the team responsible for the merge. (MH) confirmed that by combining the visualization of the DPAT with an explanation of why the different DPAT elements changed (e.g., as we did for the cell line diff partial-areas), the Diff AbN approach would be helpful when developing and merging ontologies.
A limitation of the DAT and DPAT methodology is they are only applicable to ontologies which assign domains to object properties or use object properties in restrictions. In He et al. [32] it was found that 152/186 ontologies in BioPortal had at least one such object properties while the remaining ontologies had none. Furthermore, some of the 152 ontologies used object properties in a limited fashion. To account for this we will explore new kinds of Diff AbNs in future research (described below). For example, a Diff AbN based on the Tribal Abstraction Network (TAN) [54] could be used to summarize changes in ontologies which have multiple inheritance in their class hierarchies.
5.2 Future Work
This paper presented a method for deriving Diff AbNs based on object properties. We note that several kinds of Diff AbNs can be derived, based on the structural features that are used for the derivation, e.g., data properties can be used instead of object properties. The same general approach for Diff AbN derivation described in this paper can be adapted accordingly. Future research will investigate Diff AbNs based on data properties, equivalence axioms, etc., and their use in uncovering unintended changes.
Ideally, errors should be identified and corrected already during the development process of an ontology. If an ontology curator can see the global impact of an editing operation before she modifies the ontology then certain kinds of errors can be avoided all together. We will investigate the use of Diff AbNs to enable “what if?” analysis in support of ontology development. As an ontology curator is making changes she will be provided with a Diff AbN that reflects the state of the ontology after a given potential editing operation is applied. If the curator determines this Diff AbN exposes an anomaly then the potential editing operation would not be applied to the ontology.
A major part of our AbN research is the development of a comprehensive software tool called the Biomedical Layout Utility for OWL (BLUOWL) for automatically creating and visualizing AbNs for OWL and OBO ontologies. BLUOWL is based on our previously developed BLUSNO tool [55] for SNOMED CT. Currently we are using a prototype of this tool to produce DATs and DPATs. However, a major component that is currently in development implements the visualization of Diff AbNs. The visualization component will provide an interactive environment that allows a user to explore Diff AbNs. Selecting the different Diff AbN elements, e.g., introduced areas or removed areas, will display the list of changes that occurred to the classes summarized by that kind of element. Furthermore, the Diff AbN tool will be able to provide an on-demand explanation of why each diff area/diff partial-area was introduced, modified or removed, by listing the editing operation(s) that affected the diff area/diff partial-area classes (e.g., the removal of an object property or addition of a subclass relationship). The Diff AbN tool will be made available as a standalone application and as a Protégé plugin.
A common ontology design pattern, extensively used in biomedical ontologies, is to import and reuse the content of other ontologies, e.g., a top-level ontology like Basic Formal Ontology (BFO) [12] or a top-domain ontology like the Ontology for General Medical Science (OGMS) [41]. Ontology curators are likely not interested in changes that happened within the imported ontologies. As described in this paper with regards to the ERO, the Diff AbN derivation technique considers all structural changes between two versions of an ontology, including those that occurred to content from imported ontologies. In some situations, this information could be important for detecting errors and inconsistencies in the ontology. Changes in the modeling of the imported ontology could lead to unintended changes to the content added by an ontology curator. However, if an ontology curator is not interested in seeing these changes, she could instead derive a Diff AbN that only captures the changes to her ontology. Such a feature is planned for the Diff AbN tool.
Another visualization issue, which is illustrated by the SDO and ERO DPATs, is the emergence of many removed diff area/introduced diff area pairs and corresponding diff partial-area pairs, summarizing the changes in the object properties for the same set of classes. A planned feature of the Diff AbN tool will identify corresponding diff area/diff partial-area pairs in the DPAT and compare their sets of object properties side-by-side. A similar issue is identifying renamed entities. For example, in the ERO DPAT there are two entity diff partial-areas (one added, one removed). The root classes of these two diff partial-areas are obviously referring to the same entity, though their root classes have different URIs due to using a new version of BFO in the later version of ERO. The Diff AbN tool will detect these kinds of changes and treat them appropriately.
Finally, we will investigate the use of Diff AbNs to compare AbNs of different granularity. We previously compared the granularities of different types of taxonomies for the SDO [24]. A Diff AbN can be used to compare AbNs of different granularities and to determine which is best for QA. Such a Diff AbN will help a curator with identifying which type of AbN does a better job in summarizing an ontology.
6 Conclusions
In this paper we introduced the notion of Diff Abstraction Networks (“Diff AbNs”) for summarizing and visualizing the global structural differences between two ontology releases. We described the Diff Area Taxonomy (DAT) and Diff Partial-area Taxonomy (DPAT) derivation methodologies for summarizing changes related to object properties and their domains. DPATs were derived, using a prototype tool, for the Ontology of Clinical Research (OCRe), Sleep Domain Ontology (SDO), and eagle-i Research Resource Ontology (ERO). For OCRe and SDO, we illustrated how their DPATs reflect changes resulting from a small number of changes due to a previously completed quality assurance review. For ERO, the DPAT compactly summarized thousands of changes that occurred due a merge with the VIVO ontology. The OCRe, SDO, and ERO DPATs were compared to the output of Protégé’s compare ontologies tool and DPATs were shown to compactly summarize important changes within the ontology, many of which were not reflected by the Protégé compare ontologies tool.
Highlights.
Diff Abstraction Networks are introduced to summarize and visualize ontology change.
The Diff Area Taxonomy (DAT) and the Diff Partial-area Taxonomy (DPAT) are introduced.
DPATs are derived for three ontologies and are shown to highlight important changes.
DPATs offer a compact way of viewing many changes due to various editing operations.
Acknowledgments
We thank Natasha Noy and Mark Musen for their early important feedback in specifying Diff AbNs. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA190779.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors have no conflicts of interest. All authors are aware of the submission of this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Briefings in Bioinformatics. 2008;9(1):75–90. doi: 10.1093/bib/bbm059. [DOI] [PubMed] [Google Scholar]
- 2.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- 3.Fensel D. Ontology-based knowledge management. Computer. 2002;35(11):56–9. [Google Scholar]
- 4.Maedche A, Motik B, Stojanovic L, et al. Ontologies for enterprise knowledge management. Intelligent Systems, IEEE. 2003;18(2):26–33. [Google Scholar]
- 5.Cao F, Sun X, Wang X, et al. Ontology-based Knowledge Management for Personalized Adverse Drug Events Detection. Studies in health technology and informatics. 2011;169:699–703. [PubMed] [Google Scholar]
- 6.Kiryakov A. Ontologies for knowledge management. Semantic Web Technologies: trends and research in ontology-based systems. 2006:115–38. [Google Scholar]
- 7.Lussier YA. Ontologies for natural language processing. Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics. 2005 [Google Scholar]
- 8.Estival D, Nowak C, Zschorn A. Towards ontology-based natural language processing. Proceeedings of the Workshop on NLP and XML (NLPXML-2004); 2004. pp. 59–66. [Google Scholar]
- 9.Ovchinnikova E. Integration of world knowledge for natural language understanding. Springer; 2012. [Google Scholar]
- 10.The Gene Ontology Consortium. Gene Ontology annotations and resources. Nucleic acids research. 2013;(41):D530–D5. doi: 10.1093/nar/gks1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodenreider O, Stevens R. Bio-ontologies: current trends and future directions. Briefings in bioinformatics. 2006;7(3):256–74. doi: 10.1093/bib/bbl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grenon P, Smith B, Goldberg L. Biodynamic Ontology: Applying BFO in the Biomedical Domain. In: Pisanelli DM, editor. Ontologies in Medicine. IOS Press; 2004. pp. 20–38. [PubMed] [Google Scholar]
- 13.Noy NF, Musen MA. The PROMPT suite: interactive tools for ontology merging and mapping. International Journal of Human-Computer Studies. 2003;59(6):983–1024. [Google Scholar]
- 14.Noy NF, Crubézy M, Fergerson RW, et al. Protege-2000: an open-source ontology-development and knowledge-acquisition environment. AMIA Annu Symp Proc. 2003:953. [PMC free article] [PubMed] [Google Scholar]
- 15.Sim I, Carini S, Tu S, et al. The human studies database project: federating human studies design data using the ontology of clinical research. AMIA Summits Transl Sci Proc. 2010:51–5. [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt JW, McIlroy MD. An algorithm for differential file comparison. Bell Laboratories; 1976. [Google Scholar]
- 17.Noy NF, Musen M. Promptdiff: A fixed-point algorithm for comparing ontology versions. AAAI/IAAI. 2002;2002:744–50. [Google Scholar]
- 18.Kremen P, Smid M, Kouba Z. OWLDiff: A Practical Tool for Comparison and Merge of OWL ontologies. 22nd International Workshop on Database and Expert Systems Applications; 2011. pp. 229–33. [Google Scholar]
- 19.Jiménez-Ruiz E, Grau BC, Horrocks I, et al. Building Ontologies Collaboratively Using ContentCVS. Description Logics. 2009:447. [Google Scholar]
- 20.Noy NF, Kunnatur S, Klein M, et al. Tracking changes during ontology evolution. The Semantic Web–ISWC. 2004:259–73. [Google Scholar]
- 21.Arabandi S, Ogbuji C, Redline S, et al. Developing a Sleep Domain Ontology. AMIA Clinical Research Informatics Summit. 2010 [Google Scholar]
- 22.Torniai C, Brush MH, Vasilevsky N, et al. Developing an Application Ontology for Biomedical Resource Annotation and Retrieval: Challenges and Lessons Learned. ICBO. 2011;2011:101–8. [Google Scholar]
- 23.Ochs C, Agrawal A, Perl Y, et al. Deriving an Abstraction Network to Support Quality Assurance in OCRe. AMIA Annu Symp Proc. 2012:681–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Ochs C, He Z, Perl Y, et al. Choosing the Granularity of Abstraction Networks for Orientation and Quality Assurance of the Sleep Domain Ontology. Proceedings of the 4th International Conference on Biomedical Ontology; 2013. pp. 84–9. [Google Scholar]
- 25.Torniai C, Essaid S, Lowe B, et al. Finding common ground: integrating the eagle-i and VIVO ontologies. ICBO. 2013;2013:46–9. [Google Scholar]
- 26.Mitchell S, Chen S, Ahmed M, et al. The VIVO ontology: Enabling networking of scientists. ACM WebScience Conference. 2011:14–7. [Google Scholar]
- 27.Motik B, Patel-Schneider PF, Parsia B. OWL 2 Web Ontology Language Structural Specification and Functional Style Syntax. W3C -- World Wide Web Consortium; 2009. [Google Scholar]
- 28.Smith B, Ashburner M, Rosse C, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nature biotechnology. 2007;25(11):1251–5. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonçalves RS, Parsia B, Sattler U. Ecco: A Hybrid Diff Tool for OWL 2 ontologies. OWLED; 2012. [Google Scholar]
- 30.Redmond T, Noy N. Computing the Changes Between Ontologies. Joint Workshop on Knowledge Evolution and Ontology Dynamics; 2011. pp. 1–14. [Google Scholar]
- 31.Halper M, Gu H, Perl Y, et al. Abstraction Networks for Terminologies: Supporting Management of “Big Knowledge”. Artificial intelligence in medicine. 2015 doi: 10.1016/j.artmed.2015.03.005. (accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Z, Ochs C, Agrawal A, et al. A Family-Based Framework for Supporting Quality Assurance of Biomedical Ontologies in BioPortal. Proc AMIA Annu Symp. 2013:581–90. [PMC free article] [PubMed] [Google Scholar]
- 33.Whetzel PL, Noy NF, Sham NH, et al. BioPortal: Enhanced Functionality via New Web services from the National Center for Biomedical Ontology to Access and Use Ontologies in Software Applications. Nucleic Acids Research (NAR) 2011;39(Web Server issue):W541–5. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Z, Ochs C, Soldatova L, et al. Auditing Redundant Import in Reuse of a Top Level Ontology for the Drug Discovery Investigations Ontology. VDOS. 2013 [Google Scholar]
- 35.Da Q, King R, Hopkins A, et al. An ontology for description of drug discovery investigations. J Integrative Bioinformatics. 2010;7(3):126–39. doi: 10.2390/biecoll-jib-2010-126. [DOI] [PubMed] [Google Scholar]
- 36.Zeginis D, Hasnain A, Loutas N, et al. A collaborative methodology for developing a semantic model for interlinking Cancer Chemoprevention linked-data sources. Semantic Web. 2014;5(2):127–42. [Google Scholar]
- 37.Min H, Perl Y, Chen Y, et al. Auditing as part of the terminology design life cycle. J Am Med Inform Assoc. 2006;13(6):676–90. doi: 10.1197/jamia.M2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochs C, Perl Y, Halper M, et al. Gene Ontology Summarization to Support Visualization and Quality Assurance. BICoB. 2015;2015 [Google Scholar]
- 39.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fragoso G, de Coronado S, Haber M, et al. Overview and utilization of the NCI thesaurus. Comp Funct Genomics. 2004;5(8):648–54. doi: 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldfain A. Ontology for General Medical Science (OGMS) 2015 Feb 10; Available from: http://code.google.com/p/ogms/
- 42.Beisswanger E, Schulz S, Stenzhorn H, et al. BioTop: An upper domain ontology for the life sciences. A description of its current structure, contents and interfaces to OBO ontologies. Applied Ontology. 2008;3(4):205–12. [Google Scholar]
- 43.Shearer R, Motik B, Horrocks I. HermiT: a highly-efficient OWL reasoner. OWLED; 2008. [Google Scholar]
- 44.Krafft DB, Cappadona NA, Caruso B, et al. Vivo: Enabling national networking of scientists. Proceedings of the Web Science Conference; 2010. pp. 1310–3. [Google Scholar]
- 45.Noy NF, Klein M. Ontology evolution: Not the same as schema evolution. Knowledge and information systems. 2004;6(4):428–40. [Google Scholar]
- 46.Horridge M, Bechhofer S. The OWL API: A Java API for Working with OWL 2 Ontologies. OWLED. 2009;529:11–21. [Google Scholar]
- 47.Ahmed M, Chen S, Ding Y, et al. Aligning Research Resource and Researcher Representation: The eagle-i and VIVO Use Case. ICBO. 2013;2013:260–2. [Google Scholar]
- 48.Vasilevsky N, Johnson T, Corday K, et al. Research resources: curating the new eagle-i discovery system. Database. 2012;2012:bar067. doi: 10.1093/database/bar067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.connect-isf: The ontology developed in the context of CTSAConnect to represent agents, resources and grants. 2014 Sep 24; Available from: https://code.google.com/p/connect-isf/
- 50.obo-relations: The OBO Relations Ontology. 2014 Sep 18; Available from: https://code.google.com/p/obo-relations/
- 51.Smith CL, Goldsmith CAW, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome biology. 2004;6(1):R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malone J, Brown A, Lister AL, et al. The Software Ontology (SWO): a resource for reproducibility in biomedical data analysis, curation and digital preservation. Journal of Biomedical Semantics. 2014;5(1) doi: 10.1186/2041-1480-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mungall CJ, Torniai C, Gkoutos GV, et al. Uberon, an integrative multi-species anatomy ontology. Genome biology. 2012;13(1):R5. doi: 10.1186/gb-2012-13-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ochs C, Geller J, Perl Y, et al. A Tribal Abstraction Network for SNOMED CT Hierarchies without Attribute Relationships. J Am Med Inform Assoc. 2014 doi: 10.1136/amiajnl-2014-003173. amiajnl-2014-003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geller J, Ochs C, Perl Y, et al. New Abstraction Networks and a New Visualization Tool in Support of Auditing the SNOMED CT Content. AMIA Annu Symp Proc. 2012:237–46. [PMC free article] [PubMed] [Google Scholar]