Abstract
TNFα receptors are constitutively overexpressed in tumor cells, correlating to sustain elevated NFκB and monocyte chemotactic protein-1 (MCP-1/CCL2) expression. The elevation of CCL2 evokes aggressive forms of malignant tumors marked by tumor associated macrophage (TAM) recruitment, cell proliferation, invasion and angiogenesis. Previously, we have shown that the organo-sulfur compound diallyl disulfide (DADS) found in garlic (Allium sativum) attenuates TNFα induced CCL2 production in MDA-MB-231 cells. In the current study, we explored the signaling pathways responsible for DADS suppressive effect on TNFα mediated CCL2 release using PCR Arrays, RT-PCR and western blots. The data in this study show that TNFα initiates a rise in NFκB mRNA, which is not reversed by DADS. However, TNFα induced heightened expression of IKKε and phosphorylated ERK. The expression of these proteins corresponds to increased CCL2 release that can be attenuated by DADS. CCL2 induction by TNFα was also lessened by inhibitors of p38 (SB202190) and MEK (U0126) but not JNK (SP 600125), all of which were suppressed by DADS. In conclusion, the obtained results indicate that DADS down regulates TNFα invoked CCL2 production primarily through reduction of IKKε and phosphorylated-ERK, thereby impairing MAPK/ERK, and NFκB pathway signaling. Future research will be required to evaluate the effects of DADS on the function and expression of TNFα surface receptors.
Keywords: diallyl disulfide, tumor necrosis factor alpha, monocyte chemoattractant protein 1, nuclear factor kappa b, map kinase b
1. INTRODUCTION
Metastatic breast cancer brings together rapid tumor proliferation, detachment and development of secondary tumors with acquired characteristics of the primary tumor. Chemokines such as monocyte chemotactic protein-1 (MCP-1), known as CC chemokine-2 (CCL2) play a critical role in this process. These chemokines recruit monocytes that differentiate into tumor-associated macrophages (TAMs) which subsequently release substances needed for tissue remodeling, angiogenesis and metastasis [1–3]. TAMs can also directly release more TNFα, an inflammatory cytokine [4], furthering the processes of production/release of CCL2 in diverse tumor tissue.[5].
Previously we have shown that diallyl disulfide (DADS), found in garlic (Allium sativum), attenuates TNFα induced CCL2 production in human breast cancer cells. It is likely that TNFα induced CCL2 production occurs through up regulation of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). These three components are concurrently expressed to a greater degree in aggressively advanced tumors marked by TAM recruitment [6–8] and elevated TNF-receptors (TNFRs) on diverse human cancers [9]. Drugs or compounds such as DADS that antagonize these effects are becoming significant therapeutic vehicles. These include infliximab (Remicade), adalimumab (Humira), anti-TNF antibodies [10–11], all of which prevent tumor infiltrating leucocytes [12]. Therefore, the purpose of this study was to determine if the garlic constituent diallyl disulfide, could impact the MDA-MB-231 cells since they are the most studied TNBC to date. In addition, many researchers have used this cell model because it mimics aggressive nature of clinical isolates and have displayed great ability to metastasize in xenograft models. Nakagawa et al. [13] demonstrated the ability of DADS to inhibit tumor growth through its modulation of the apoptotic genes in this cell line. Other studies have used this cell line to study the mechanisms involved in the immune response [14,15]. Moreover, in this study we further explore the mechanism behind the inhibitory effect by DADS on TNFα induced CCL2 release in human breast carcinoma cells, with focus on MAPK/ERK NFκB signaling.
2. MATERIALS AND METHODS
Cell lines, chemicals and reagents: Triple negative human breast tumor (MDA-MB-231) cells were obtained from American Type Culture Collection (Rockville, MD). Dulbecco’s Modified Eagle Medium (DMEM) media, fetal bovine serum (FBS) and penicillin/streptomycin were all obtained from Invitrogen (Carlsbad, CA). Recombinant human TNFα was purchased from RayBiotech (RayBiotech Inc., Norcross, GA, USA). Diallyl disulfide (>80%) was purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.1. Cell culture and Treatment
MDA-MB-231 cells were cultured in 75cm2 or 175cm2 flasks containing DMEM media supplemented with 10% FBS and 1% pen/strep (10,000u/ml penicillin G sodium, 10,000μg/ml streptomycin sulfate). Cells were grown in an environment of 37°C with humidified 95% air and 5% CO2. Control cells received vehicle only (0.01% ethanol). DADS treated cells received 100 μM DADS in vehicle and 40ng of TNFα was given to TNFα-treated and co-treated cells. Cells were incubated for 24 hrs. after treatment. In this study, 100μM was used as the dose for DADS concentration. Although some studies have shown that different concentrations of garlic components exert different responses, such as increasing proliferation and tumor growth, in our laboratory cell viability studies were conducted to determine a working concentration and we recently reported that the 100μM dose was the optimum dose for this cell line [16]. The present study is a continuation of the study performed by Bauer et al. [16]. Moreover, initial studies were done using both 100μM and 400μM with approximately the same effect on CCL2 release. Additionally in our previous studies, we demonstrated an optimum overall response using lower doses of DADS in MCF10A human epithelial cells [17]. Therefore, we used 100μM since it would be less likely to have toxic effects on the normal cells at this dose level.
2.2. Inhibition study
Cultured MDA-MB-231 cells were treated for 24 hours with DADS with and without TNFα treatment at above conditions. Additionally, cells were co-treated with inhibitors of JNK, MEK and p38 at concentrations of 10 μM, 2 μM and 2 μM, respectively. The inhibitors for JNK (SP600125), MEK (U0126) and p38 (SB202190) were purchased from Sigma Aldrich (St. Louis, MO). Cells were detached and the lysate collected.
2.3. ELISA: CCL2 detection
Supernatants from resting and stimulated (24 hrs) MDA-MB-231 cells were collected and centrifuged at 1000× g for 5 min at 4°C. Specific ELISA was performed using MCP-1/CCL2 ELISA kit (Raybiotech, Norcross, GA, USA) following manufacturer’s instructions. Briefly, 100 μl of supernatants from samples and standards were added to 96 well plates pre-coated with capture antibody. After incubation 100μl of prepared biotinylated antibody mixture was added to each well. After 1 hour, mixture was decanted and 100 μl streptavidin solution was placed in each well and incubated. Substrate reagent (100 μl) was then added to each well for 30 min followed by the addition of 50 μl stop solution. Plates were read at 450nm using a UV microplate reader.
2.4. Western Blot: MAPK/ERK pathway and IKKE
Total cell protein concentrations from MDA-MB-231 cells treated with DADS, with and without TNFα co-treatment for 24 hrs, was determined using a modified Bio-Rad “DC” protein assay (Bio-Rad Laboratories, Hercules, CA, USA). A series of concentration standards ranging from 0–20 μg/ml were prepared using IgG. Test samples were prepared by adding 5 μl of a 1/10 dilution of the cell lysate to 795 μl H2O. The standards and samples were mixed with 200 μl Bio-Rad “DC” protein assay dye concentrate and thoroughly mixed by vortexing. Following incubation for 5 minutes at room temperature, the samples were vortexed again and 200 μl of each loaded into a 96-well plate. Protein concentrations were quantified at a wavelength of 595 nm with the Power Wave X 340 microplate reader equipped with KC4 v3.0 PowerReports software (Bio-Tek Instruments, Winooski, VT, USA).
Cell lysates were separated by electrophoresis on 10% SDS-polyacrylamide gels and then transferred to Immobilon-P PVDF membranes. Equal loading was verified by staining with Ponceau S (Sigma-Aldrich Chemical Co, St. Louis, MO). Blots were blocked at 4°C overnight in 5% Carnation Instant Milk in Tris-buffered saline with 0.05% Tween 20 in PBS (PBST) and then incubated overnight at 4°C with mouse anti-human p38 MAPK, ERK and JNK affinity purified antibody (IMGENEX, San Diego, CA). Membranes were washed with PBST and incubated overnight with anti-goat IgG-horseradish peroxidase (Santa Cruz Biotechnology, CA) in PBST overnight at 4°C. Protein loading was monitored in each gel lane by probing the membranes with anti-GAPDH antibodies (R & D Systems, Minneapolis, MN). Immunoblot images were obtained using a Flour-S Max Multimager (Bio-Rad Laboratories, Hercules, CA). Lane density data was acquired with Quantity One Software (Bio-Rad Laboratories, Hercules, CA).
2.5. RT-PCR
MDA-MB-231 cells treated with or without DADS, were subcultured in 6-well plates until confluent. Cells were lysed with 1 ml Trizol reagent. Chloroform (0.2 ml) was added to the lysed samples, tubes were shaken, incubated at 15–30°C for 2–3 min and centrifuged at 10,000 × g for 15 min. at 2–8°C. The aqueous phase was transferred to a fresh tube and the RNA precipitated by mixing 0.5 ml isopropyl alcohol. After incubation the samples were centrifuged, the supernatant was removed and the RNA pellets were washed with 75% ethanol. The samples were mixed before being centrifuged at 7,500 × g for 5 min. at 2–8°C. The RNA pellet was dried and dissolved in RNase-free water and incubated for 10 min. at 55–60°C.
RT reaction
RNA (5 μg/10 μl) was heated for 10 min. then quenched on ice before use. The following components were added to the reaction: 10 μl heat denatured RNA, 3.0 μl 10 x PCR buffer, 2.5 μl 10 mM dNTPs, 6.0 μl 25 mM MgCl2, 1.0 μl random primers, 0.5 μl SuperScript II reverse transcriptase and 17.0 μl water. Samples were allowed to sit for 10 min. at 25°C then incubated for 1 hr. at 42°C. The cDNA was denatured at 95°C and placed on ice. PCR reaction: The following components were mixed in a 0.5 ml PCR tube: 6.0 μl cDNA product, 1.5 μl 10 x PCR buffer, 0.2 μl Taq polymerase, 0.5 μl primer and 10.3 μl water. PCR will be performed with 30 cycles of denaturation: 30 sec. at 95°C; annealing: 45 sec. at 60°C; and extension 60 sec. at 72°C using BioDoc-it System (UVP, Upland CA, USA). cDNA synthesis and Real-Time PCR was performed using First Strand cDNA synthesis kit/SABiosciences RT2 qPCR Master Mix from Qiagen (Gaithersburg, Md., USA) according to manufacturers instructions.
2.6. Statistical Analysis
Statistical analysis on the data was determined by Graph Pad Prism 5.0. All data was expressed as mean ± standard error from at least 3 independent experiments. Differences between mean values were analyzed by a one-way analysis of variance (ANOVA) with Dunnett’s Multiple Comparison test, *p<0.05; **p<0.01.
3. RESULTS
3.1 Effect of DADS on the TNF-mediated release of CCL2 in MDA-MB-231 cells
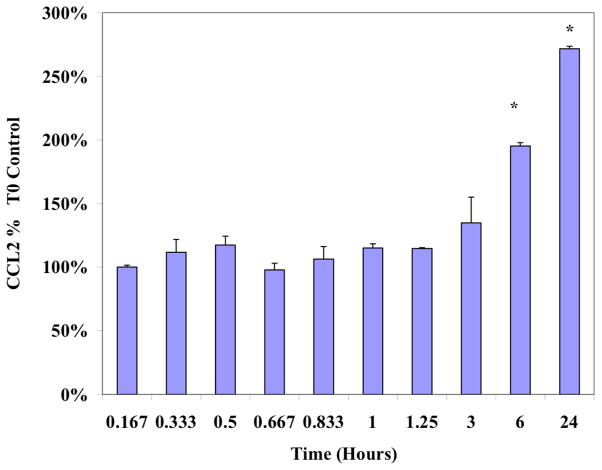
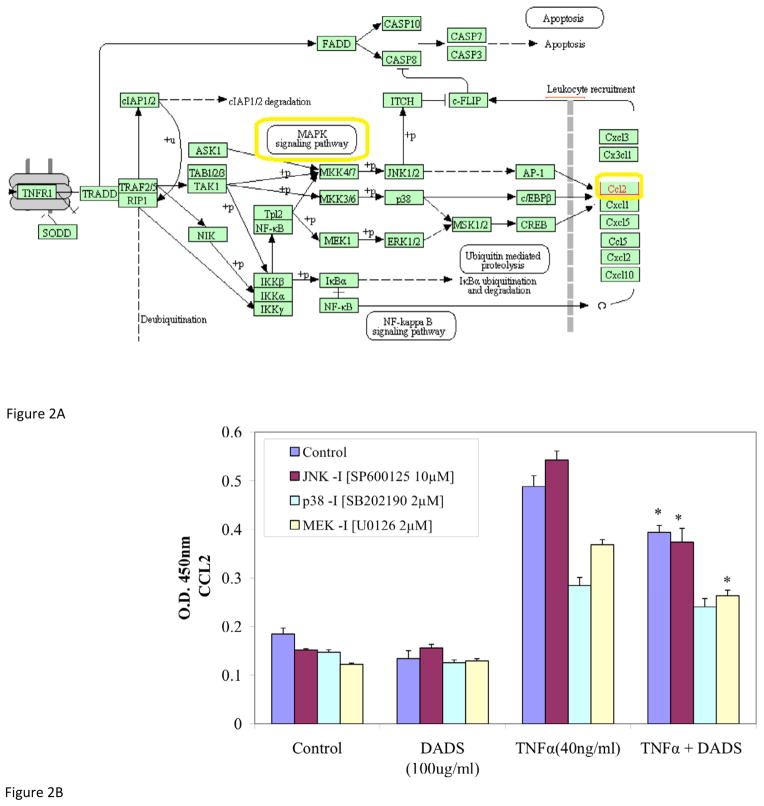
MDA-MB-231 cells were stimulated with TNFα at sub-lethal concentrations (40 ng/ml) and the time-dependent release of CCL2 was monitored. As indicated in Figure 1, the levels of CCL2 remained unchanged for the first 1.25 hours, and began to accumulate after 3 hours with peak levels, approximately 3-fold higher than basal levels, achieved at 24 hours post induction. This time point provides a level of accumulation that is adequate for further analysis (Figure 1). As reported in the literature the induction of TNFα-induced CCL2 is mediated via MAPK signaling pathways (Figure 2A). To evaluate its effect on CCL2 release, cells were treated with DADS in the absence and presence of MAP kinase inhibitors. As shown in Figure 2, neither DADS nor the MAPK inhibitors in combination with DADS, affected CCL2 basal levels in the absence of TNFα-induction. Upon stimulation with TNFα, a significant increase in CCL2 was not prevented by inhibition of JNK (Figure 2). DADS alone reduced the accumulation of CCL2 in TNFα-induced cells, and further enhanced the reduction exerted by p38 and MEK1 inhibitors when co-treated. These data suggest a controlling role for TNFα-induced CCL2 via MAPK involving MEK and P38 signaling, but not JNK.
Figure 1. Time course of the TNFα-dependent accumulation of CCL2 in MDA-MB-231 cells.
MDA-MB-231 cells were exposed to sublethal levels of TNFα (40ng/ml) and CCL2 release was monitored as described in Materials and Methods. The data is presented as % Ctrl at Timezero, displayed as the Mean ± S.E.M., n=4. Differences from Control were determined using a determined by a one-way ANOVA, with a Tukey post hoc test. *p<0.05.
Figure 2.
Figure 2A. Kegg Diagram interconnecting MAPK signaling with CCL2 release. CCL2 release is initiated by TNFα acting on TNFR1 and contributes to leukocyte recruitment. The factors analyzed in this study are highlighted.
Figure 2B. Effect of DADS and MAPK signaling inhibitors on CCL2 release in MDA-MB-231 cells. Additive or synergistic effects of MAPK inhibitors on DADS-treated (100μM), TNFα-treated (40ng/ml) and co-treated MDA-MB-231 cells after 24 hrs. The data are presented as CCL2 release (OD 450nm) and represent the Mean ± S.E.M. n=4. Significance of differences from the control in both groups was determined by T-test. *p<0.05.
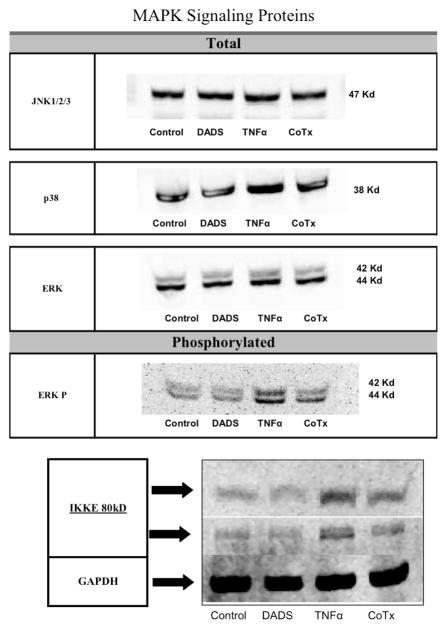
3.2 DADS does not affect the protein levels of MAPK factors, but attenuates the TNFα-dependent phosphorylation of ERK and the induction of IKKε in MDA-MB-231 cells
Based on the ability of DADS to enhance the effect of MAPK inhibitors we evaluated the protein expression levels of the signaling proteins involved in the pathway. Neither DADS, or TNFα, nor their combination considerably altered the protein levels of JNK1/2/3, p38 or ERK (Figure 3). Evaluation of the phosphorylation status of ERK reveals that DADS does not affect its phosphorylation status but significantly attenuates the TNFα-dependent phosphorylation. In addition, DADS also reduced TNFα-dependent induction of IKKε (Figure 3).
Figure 3. Evaluation total or phosphorylated proteins involved with NFκB Signaling Pathway.
DADS-treated (100μM), TNFα-treated (40ng/ml) and co-treated MDA-MB-231 cell lysates were evaluated for the protein levels of JNK1/2/3, p38, ERK and IKKε. Phosphorylation status of ERK was also determined as described in the Materials and methods.
3.3 Evaluation of the effect of DADS on the mRNA profile of NFκB signaling
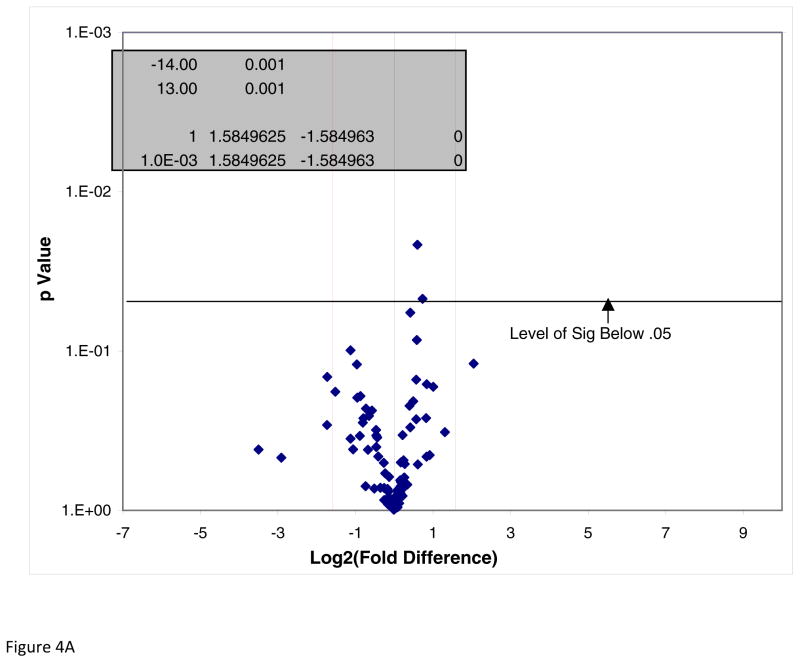
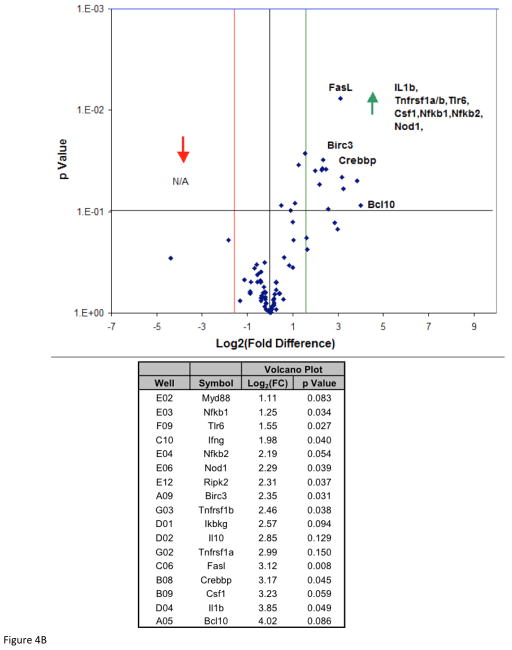
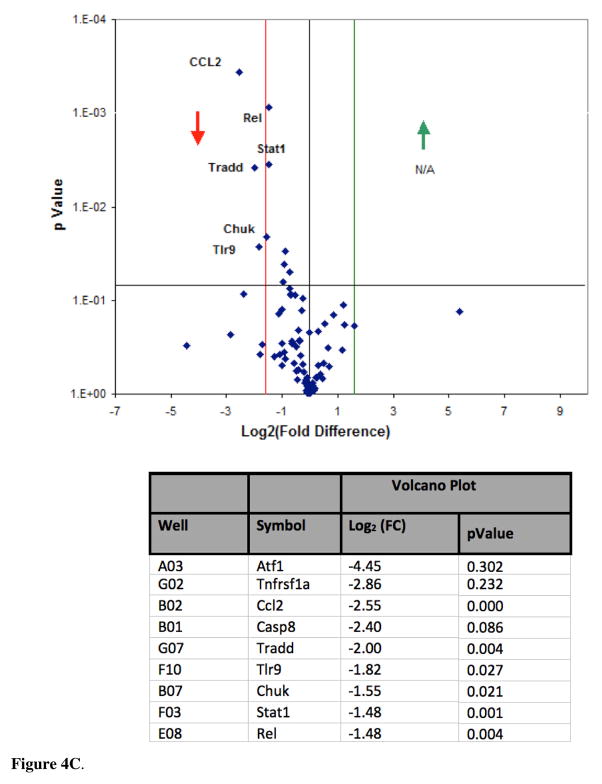
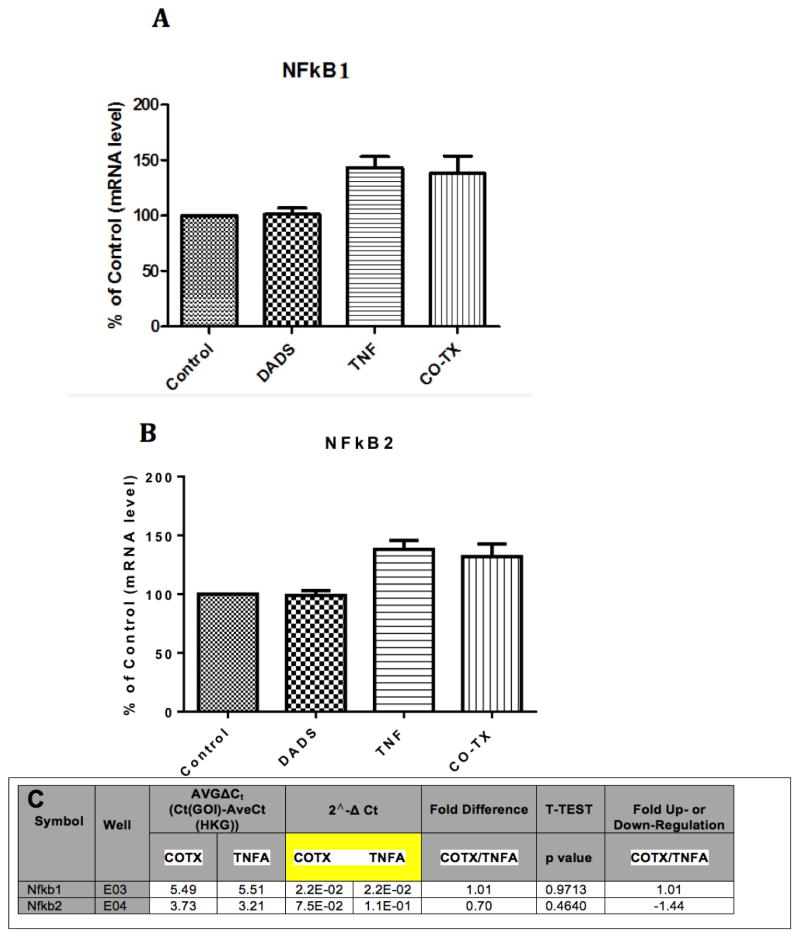
To investigate if DADS inhibition of CCL2 release in the presence of TNFα is mitigated by inhibition of NFκB signaling, we analyzed mRNA profiles of the pathway components using the NFκB Signaling Pathway RT2 Profiler™ PCR Array PAMM-025Z (Qiagen, Gaithersburg, Md., USA) (Table 1). Briefly, the array includes genes involved with Rel, NFκB, and IκB families, NFκB-responsive genes, ligand, receptors, kinases and transcription factors that propagate the signal. The data showed no statistical differences for baseline values between controls vs DADS (Figure 4A) for any gene in the array. The effects of TNFα on mRNA expression showed significant elevation in Bcl10, IL1b, Csf1, Crebbp, Fas, Tnfrsf1a, IL-10, Ikbkγ and Tnfrsf1b (Figure 4B). Genes were entered into bio-informatic analysis (Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7) to identify major systems affected (Table 2), where classifications provided by DAVID enrichment scores are presented for those averaging less than p<0.01 [18, 19]. There were significant differences found between Control vs TNFα-treated groups in the upward direction showing statistical probabilities for: regulation of cytokine biosynthesis, positive regulator of NFκB signaling, cell death and Nod-Like receptor signaling. Effect of TNFα vs TNFα/DADS shows DADS downregulation of Ccl2, Casp8 and Tradd at equal to or greater than 2-fold with p<0.05 (Figure 4C). We further investigated the effect of DADS on the TNFα NFκB expression pattern using RT-PCR. As shown in Figure 5, there were no changes in the mRNA expression for NFκB1 corroborating the results of the mRNA profile analysis.
Table 1.
Gene Table.
NFκB Signaling Pathway RT2 Profiler™ PCR Array PAMM – 025Z plate layout. Well position, gene identifiers, official gene symbol and gene description.
| Position | Unigene | GeneBank | Symbol | Description |
|---|---|---|---|---|
| A01 | Mm.301626 | NM_007428 | Agt | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| A02 | Mm.6645 | NM_009652 | Akt1 | Thymoma viral proto-oncogene 1 |
| A03 | Mm.676 | NM_007497 | Atf1 | Activating transcription factor 1 |
| A04 | Mm.209903 | NM_009715 | Atf2 | Activating transcription factor 2 |
| A05 | Mm.239141 | NM_009740 | Bcl10 | B-cell leukemia/lymphoma 10 |
| A06 | Mm.479217 | NM_009742 | Bcl2a1a | B-cell leukemia/lymphoma 2 related protein A1a |
| A07 | Mm.238213 | NM_009743 | Bcl2l1 | Bcl2-like 1 |
| A08 | Mm.439658 | NM_033601 | Bcl3 | B-cell leukemia/lymphoma 3 |
| A09 | Mm.2026 | NM_007464 | Birc3 | Baculoviral IAP repeat-containing 3 |
| A10 | Mm.17629 | NM_130859 | Card10 | Caspase recruitment domain family, member 10 |
| A11 | Mm.46187 | NM_175362 | Card11 | Caspase recruitment domain family, member 11 |
| A12 | Mm.1051 | NM_009807 | Casp1 | Caspase 1 |
| B01 | Mm.336851 | NM_009812 | Casp8 | Caspase 8 |
| B02 | Mm.290320 | NM_011333 | Ccl2 | Chemokine (C-C motif) ligand 2 |
| B03 | Mm.284248 | NM_013653 | Ccl5 | Chemokine (C-C motif) ligand 5 |
| B04 | Mm.367714 | NM_001033126 | Cd27 | CD27 antigen |
| B05 | Mm.271833 | NM_011611 | Cd40 | CD40 antigen |
| B06 | Mm.486313 | NM_009805 | Cflar | CASP8 and FADD-like apoptosis regulator |
| B07 | Mm.3996 | NM_007700 | Chuk | Conserved helix-loop-helix ubiquitous kinase |
| B08 | Mm.392384 | NM_001025432 | Crebbp | CREB binding protein |
| B09 | Mm.795 | NM_007778 | Csf1 | Colony stimulating factor 1 (macrophage) |
| B10 | Mm.4922 | NM_009969 | Csf2 | Colony stimulating factor 2 (granulocyte-macrophage) |
| B11 | Mm.1238 | NM_009971 | Csf3 | Colony stimulating factor 3 (granulocyte) |
| B12 | Mm.420648 | NM_007912 | Egfr | Epidermal growth factor receptor |
| C01 | Mm.181959 | NM_007913 | Egr1 | Early growth response 1 |
| C02 | Mm.378990 | NM_011163 | Eif2ak2 | Eukaryotic translation initiation factor 2-alpha kinase 2 |
| C03 | Mm.490895 | NM_007922 | Elk1 | ELK1, member of ETS oncogene family |
| C04 | Mm.24816 | NM_010169 | F2r | Coagulation factor II (thrombin) receptor |
| C05 | Mm.5126 | NM_010175 | Fadd | Fas (TNFRSF6)-associated via death domain |
| C06 | Mm.3355 | NM_010177 | Fasl | Fas ligand (TNF superfamily, member 6) |
| C07 | Mm.246513 | NM_010234 | Fos | FBJ osteosarcoma oncogene |
| C08 | Mm.276389 | NM_010442 | Hmox1 | Heme oxygenase (decycling) 1 |
| C09 | Mm.435508 | NM_010493 | Icam1 | Intercellular adhesion molecule 1 |
| C10 | Mm.240327 | NM_008337 | Ifng | Interferon gamma |
| C11 | Mm.277886 | NM_010546 | Ikbkb | Inhibitor of kappaB kinase beta |
| C12 | Mm.386783 | NM_019777 | Ikbke | Inhibitor of kappaB kinase epsilon |
| D01 | Mm.12967 | NM_010547 | Ikbkg | Inhibitor of kappaB kinase gamma |
| D02 | Mm.874 | NM_010548 | Il10 | Interleukin 10 |
| D03 | Mm.15534 | NM_010554 | Il1a | Interleukin 1 alpha |
| D04 | Mm.222830 | NM_008361 | Il1b | Interleukin 1 beta |
| D05 | Mm.896 | NM_008362 | Il1r1 | Interleukin 1 receptor, type I |
| D06 | Mm.38241 | NM_008363 | Irak1 | Interleukin-1 receptor-associated kinase 1 |
| D07 | Mm.152142 | NM_172161 | Irak2 | Interleukin-1 receptor-associated kinase 2 |
| D08 | Mm.105218 | NM_008390 | Irf1 | Interferon regulatory factor 1 |
| D09 | Mm.275071 | NM_010591 | Jun | Jun oncogene |
| D10 | Mm.87787 | NM_010735 | Lta | Lymphotoxin A |
| D11 | Mm.3122 | NM_010736 | Ltbr | Lymphotoxin B receptor |
| D12 | Mm.15918 | NM_011945 | Map3k1 | Mitogen-activated protein kinase kinase kinase 1 |
| E01 | Mm.8385 | NM_011952 | Mapk3 | Mitogen-activated protein kinase 3 |
| E02 | Mm.213003 | NM_010851 | Myd88 | Myeloid differentiation primary response gene 88 |
| E03 | Mm.256765 | NM_008689 | Nfkb1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, p105 |
| E04 | Mm.102365 | NM_019408 | Nfkb2 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2, p49/p100 |
| E05 | Mm.170515 | NM_010907 | Nfkbia | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| E06 | Mm.28498 | NM_172729 | Nod1 | Nucleotide-binding oligomerization domain containing 1 |
| E07 | Mm.184163 | NM_029780 | Raf1 | V-raf-leukemia viral oncogene 1 |
| E08 | Mm.4869 | NM_009044 | Rel | Reticuloendotheliosis oncogene |
| E09 | Mm.249966 | NM_009045 | Rela | V-rel reticuloendotheliosis viral oncogene homolog A (avian) |
| E10 | Mm.1741 | NM_009046 | Relb | Avian reticuloendotheliosis viral (v-rel) oncogene related B |
| E11 | Mm.374799 | NM_009068 | Ripk1 | Receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| E12 | Mm.112765 | NM_138952 | Ripk2 | Receptor (TNFRSF)-interacting serine-threonine kinase 2 |
| F01 | Mm.272675 | NM_015747 | Slc20a1 | Solute carrier family 20, member 1 |
| F02 | Mm.7320 | NM_016769 | Smad3 | MAD homolog 3 (Drosophila) |
| F03 | Mm.487336 | NM_009283 | Stat1 | Signal transducer and activator of transcription 1 |
| F04 | Mm.34580 | NM_019786 | Tbk1 | TANK-binding kinase 1 |
| F05 | Mm.273024 | NM_030682 | Tlr1 | Toll-like receptor 1 |
| F06 | Mm.87596 | NM_011905 | Tlr2 | Toll-like receptor 2 |
| F07 | Mm.33874 | NM_126166 | Tlr3 | Toll-like receptor 3 |
| F08 | Mm.38049 | NM_021297 | Tlr4 | Toll-like receptor 4 |
| F09 | Mm.42146 | NM_011604 | Tlr6 | Toll-like receptor 6 |
| F10 | Mm.44889 | NM_031178 | Tlr9 | Toll-like receptor 9 |
| F11 | Mm.1293 | NM_013693 | Tnf | Tumor necrosis factor |
| F12 | Mm.116683 | NM_009397 | Tnfaip3 | Tumor necrosis factor, alpha-induced protein 3 |
| G01 | Mm.193430 | NM_020275 | Tnfrsf10b | Tumor necrosis factor receptor superfamily, member 10b |
| G02 | Mm.474976 | NM_011609 | Tnfrsf1a | Tumor necrosis factor receptor superfamily, member 1a |
| G03 | Mm.235328 | NM_011610 | Tnfrsf1b | Tumor necrosis factor receptor superfamily, member 1b |
| G04 | Mm.1062 | NM_009425 | Tnfsf10 | Tumor necrosis factor (ligand) superfamily, member 10 |
| G05 | Mm.483369 | NM_019418 | Tnfsf14 | Tumor necrosis factor (ligand) superfamily, member 14 |
| G06 | Mm.103551 | NM_023764 | Tollip | Toll interacting protein |
| G07 | Mm.264255 | NM_001033161 | Tradd | TNFRSF1A-associated via death domain |
| G08 | Mm.3399 | NM_009422 | Traf2 | Tnf receptor-associated factor 2 |
| G09 | Mm.27431 | NM_011632 | Traf3 | Tnf receptor-associated factor 3 |
| G10 | Mm.389227 | NM_011633 | Traf5 | Tnf receptor-associated factor 5 |
| G11 | Mm.292729 | NM_009424 | Traf6 | Tnf receptor-associated factor 6 |
| G12 | Mm.8038 | NM_009539 | Zap70 | Zeta-chain (TCR) associated protein kinase |
| H01 | Mm.391967 | NM_007393 | Actb | Actin, beta |
| H02 | Mm.163 | NM_009735 | B2m | Beta-2 microglobulin |
| H03 | Mm.304088 | NM_008084 | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase |
| H04 | Mm.3317 | NM_010368 | Gusb | Glucuronidase, beta |
| H05 | Mm.2180 | NM_008302 | Hsp90ab1 | Heat shock protein 90 alpha (cytosolic), class B member 1 |
| H06 | N/A | SA_00106 | MGDC | Mouse Genomic DNA Contamination |
| H07 | N/A | SA_00104 | RTC | Reverse Transcription Control |
| H08 | N/A | SA_00104 | RTC | Reverse Transcription Control |
| H09 | N/A | SA_00104 | RTC | Reverse Transcription Control |
| H10 | N/A | SA_00103 | PPC | Positive PCR Control |
| H11 | N/A | SA_00103 | PPC | Positive PCR Control |
| H12 | N/A | SA_00103 | PPC | Positive PCR Control |
Figure 4.
Figure 4A. NFκB Signaling Pathway RT2 Profiler™ PCR Array of DADS vs Control. Effect of DADS vs control in MDA-MB-231 cells, displayed on a volcano plot showing significance, fold change and direction. There were no significant differences found between these groups at p<0.05.
Figure 4B. NFκB Signaling Pathway RT2 Profiler™ PCR Array of TNFα vs control. Analysis of TNFα vs control displayed on a volcano plot showing significance, fold change and direction. Transcriptome upward directional shifts (A), with significance and Log2 (Fold Change) listed along official gene symbols (B).
Figure 4C. NFκB Signaling Pathway RT2 Profiler™ PCR Array of TNFα vs TNFα/DADS. Effect of TNFα vs TNFα/DADS on gene expression: displayed on a volcano plot showing significance, fold change and direction. There were significant differences found between these groups using the PCR Array PAMM – 025Z in the downward direction (A), with significance and Log2 (Fold Change) listed along official gene symbols (B).
Table 2.
Function Categories Affected by TNF-alpha vs Control
Effect of TNFα vs Controls on targeted pathways using DAVID functional annotation statistical program. Gene-annotation enrichment analysis, functional annotation clustering on statistically differentially expressed genes. Statistical DAVID genes were analyzed for annotation cluster enrichment scores. The data represent the score, number of genes, shift direction and p-values (Fisher Exact/EASE Score).
| Annotation Cluster 1 | Enrichment Score: 7.62 | Count | P_Value | Benjamini | |
|---|---|---|---|---|---|
| GOTERM_BP_FAT | regulation of cytokine biosynthetic process | RT | 6 | 2.50E-10 | 1.40E-08 |
| Annotation Cluster 2 | Enrichment Score: 7.02 | Count | P_Value | Benjamini | |
| GOTERM_BP_FAT | positive regulation of protein kinase cascade | RT | 7 | 5.10E-12 | 8.5E-10 |
| GOTERM_BP_FAT | regulation of interleukin-6 production | RT | 6 | 6.5E-12 | 8.10E-10 |
| GOTERM_BP_FAT | positive regulation of I-kappaB kinase/NF-kappaB cascade | RT | 6 | 7.7E-12 | 7.70E-10 |
| GOTERM_BP_FAT | regulation of stress-activated protein kinase signaling pathway | RT | 5 | 1.40E-08 | 4.20E-07 |
| Annotation Cluster 3 | Enrichment Score: 4.08 | Count | P_Value | Benjamini | |
| GOTERM_BP_FAT | regulation of chemokine product | RT | 4 | 3.30E-08 | 8.60E-07 |
Figure 5. Effect of DADs on TNFα NF-Kappa B expression pattern using RT-PCR.
The data represent the Mean ± S.E.M. n=3. There were no statistical differences found between the Control and TNFα controls ± DADS for NF-Kappa B1 (A) or NF-KappaB2 (B), also corroborating PCR expression arrays for both subtypes (C).
DISCUSSION
CCL2 is a cancer promoting chemokine with capacity to enhance malignant cell migration, proliferation and invasive properties [20]. This enhancement occurs by mobilization of monocytes, macrophages and other inflammatory components to infiltrate the tumor area [21] enabling differentiation into TAMs [22]. The potent effects of CCL2 on metastatic invasion are likely due to it association with the elevation of matrix metalloproteinases (MMPs) (e.g. MMP-1, MMP-9) [23], prooncogenic substances such as TNF-α, vascular endothelial growth factor (VEGF)-A, TGFβ1 and IL-8 which collectively assist differentiation of human monocytes into TAMS. In a self-perpetuating cycle, CCL2 release is indirectly controlled by TNFα via TNF-receptors (TNFRs), with both CCL2 and TNFα being highly co-expressed in many human cancers [9,24]. Overactive TNFα receptor signaling is associated with coordination of tumor angiogenesis and metastasis [5] necessitating the development of therapeutic anti-cancer drugs. These drugs were designed to sequester TNFα [25] or block TNFRs [26 27] such as infliximab (Remicade), adalimumab (Humira), all of which downregulate TAMS [10] or other tumor infiltrating leucocytes in the tumor microenvironment [12]. Likewise, drugs or agents that can suppress CCL2-CCR2 signaling, can block monocyte recruitment and inhibit metastasis in vivo [28].
DADS, one of the major organo-sulfur compounds in garlic, is becoming recognized as a potential cancer chemopreventive compound. DADS is effective against growth of diverse cancer cell types such as HT-29 [24] HL-60 [30] cultured human colon tumor cells (HCT-15) skin (SK MEL-2) and lung (A549) [31]. Preliminary studies in our lab have indicated that DADS can attenuate CCL2 release in TNFα stimulated human breast carcinoma cells. DADS has recently been shown to reduce migration and invasion of human colon cancer in part mediated by NF-κB, ERK1/2, JNK1/2 and p38 signaling. [32] In this study, we explore signaling involved with DADS ability to down-regulate CCL2 release in TNFα-stimulated MDA-MB-231 cells.
In tumor cells, elevated NFκB signaling is triggered by TNFα, corresponding to a rise in CCL2 and TAM recruitment, cell proliferation, invasiveness and angiogenesis.[6–8]. TNF-α activation of NFκB requires its translocation from the cytoplasm to the nucleus to function. The location of NFκB is controlled by IκBs, which binds NFκB and prevents nuclear uptake. Further downstream, IκBs are themselves regulated by phosphorylation which can trigger ubiquitin-dependent degradation. The phosphorylation of IκB by IκB kinase (IKK) occurs on IKKbeta, itself a component of IKK complexes housing regulatory subunits IKKα, IKKγ and NEMO. [33]. Phosphorylation enables the recognition by E3RS (IκB/β-TrCP) to E3 ubiquitin ligase, leading to degradation, and thereby breaking controlling elements for IκB, enabling rapid NFκB translocation to the nucleus to turn on proinflammatory molecules [34]. The data in this study suggest that TNFα initiated a rise in NFκB1/2 gene expression (confirmed by PCR Array PAMM – 025Z and RT-PCR), both sustained in the presence or absence of DADS. However, DADs reduced protein expression of IKKε, which could negatively control NFκB activation signaling, and account for loss of CCL2 protein expression.
IKKi/IKKε plays an important role in carrying out TNFα signaling, via acting as a serine-threonine kinase [33]. It is capable of phosphorylating NFκB subunit RelA (also known as p65) correlating to NFκB activation [35], a rise in CCAAA/enhancer-binding protein (C/EBPδ) [36] and phosphorylation/rapid degradation of inhibitors of NFκB. Subsequent dissociation of the inhibitor/NFκB complex allows free NFκB translocation to the nucleus and initiates gene transcription. The ability of DADS to downregulate IKKε could in effect hamper TNFα induced IKKε-mediated NFκB activation [37]. This is an otherwise strong correlate to many human cancers, including, breast, ovarian, prostate, glial, [38, 39], esophageal, [40] and aggressive metastasis, tumor survival, [41] and poor clinical prognosis in diverse cancers [42]. Further, the correlation of IKKε with cell proliferation and transformation, has given rise to its being classified as oncogene [43]. Silencing or inhibition of IKKε results in inhibition of cell growth, proliferation, invasion, [44] clonogenicity, migration [45] and overcoming its contributory resistance to tamoxifen [46] in breast cancer, as well as cisplatin in ovarian tumors.[42] The identification of novel molecules that can inhibit IKKε is currently underway as a means to inflammatory processes associated with cancer progression.[47] Moreover, if DADS can reduce IKKε, this could also prevent events downstream to IKKε over expression such as activation of p52 NF-κB dimers [48], [49] estrogen receptor ERα activation, upregulation of cyclin D1 and chemotherapy resistance in breast cancer cells in particular to tamoxifen [50]. In the current study we were focused on IKKi/IKKε because it plays an important role in TNFα signaling. The presented data show a correlation between IKKε and cell proliferation and transformation as well as many different cancers. The data also show the involvement of IKKi/IKKε in tumor survival and aggressive metastasis. We are reporting that IKKε expression is reduced in this model, which is not isolated to this model but is important since this model has been known to be highly aggressive and has fewer treatment options. In the study we have not examined signaling molecules in other TNBC cell lines but we are planning to do so in future studies.
The data presented in this study suggest DADS can down-regulate IKK and CCL2 but the mechanism for this is unclear. It is possible that DADS could be down regulating the TNFα receptor complex, which would correlate to subcellular localization of NFκB, and its influence on induction of CCL2. The effect of DADS in this study, did not appear to involve mRNA of NFκB, but possibly reduced TNFRSF1A gene and adaptor protein tumor necrosis factor receptor (TNFR-associated death domain, TRADD), which are well known to “activate” via altering subcellular localization of NFκB. [51–53] Future research will be required to evaluate if the effects of DADS on CCL2 occur due to upstream events including TNFR down-regulation or potential involvement of AKT, which directly leads to up regulation of IKKε protein expression in MDA-MB-231 cells [54].
Down stream TNFα-triggered multiple signaling pathways that lead to expressed and secreted RANTES and CCL2 [55]. These pathways are believed to involve TNFR1 association with Jak2, c-Src, which could lead to CCL2 release through one or more of activating p38 MAPK, JNK, and Akt or activation of NFκB [56]. The data from this study show no change in the total proteins for ERK, P38, MEK and JNK, in the presence or absence of TNFα and DADS, however TNFα induction of CCL2 is mediated through signaling which involves MAPK phosphorylation signaling, which were further reduced in the presence of DADS. These findings suggest that level of control of DADS in CCL2 reduction is either or both occurring at the TNFα receptor or phosphorylation of ERK and P38, the former being confirmed by the data.
In summary, the findings of this study contribute to the body of work describing garlic as a chemopreventive agent, carrying diverse properties which range from carcinogen detoxification to cell-cycle arrest/apoptosis and a reduced expression of monocyte-chemoattractant protein [57]. DADS or any compound that can suppress TAM recruitment is considered an effective therapeutic approach in treatment of human cancers [58–59].
Highlights.
DADS/TNFα decreased CCL2, Casp8, and Tradd gene expression in a TNBC cell line
DADS/TNFα reduced IKKε protein expression in a TNBC cell line.
DADS/TNFα reduced phosphorylated ERK protein expression in a TNBC cell line.
Acknowledgments
This project was supported by the National Center for Research Resources NIH NCRR RCMI program (G12RR 03020) and the National Institute on Minority Health and Health Disparities, NIH (8G12MD007582-28 and 1P20 MD006738-01)
Footnotes
This manuscript version is made available under the CC BY-NC-ND 4.0 license.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zlotnik A. Chemokines in neoplastic progression. Seminars in cancer biology. 2004;14:181–5. doi: 10.1016/j.semcancer.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka T, Bai Z, Srinoulprasert Y, Yang BG, Hayasaka H, Miyasaka M. Chemokines in tumor progression and metastasis. Cancer science. 2005;96:317–22. doi: 10.1111/j.1349-7006.2005.00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annual review of immunology. 2000;18:217–42. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 4.Jaattela M. Biologic activities and mechanisms of action of tumor necrosis factor-alpha/cachectin. Laboratory investigation; a journal of technical methods and pathology. 1991;64:724–42. [PubMed] [Google Scholar]
- 5.Balkwill F. TNF-alpha in promotion and progression of cancer. Cancer metastasis reviews. 2006;25:409–16. doi: 10.1007/s10555-006-9005-3. [DOI] [PubMed] [Google Scholar]
- 6.Kehlen A, Haegele M, Menge K, Gans K, Immel UD, Hoang-Vu C, et al. Role of glutaminyl cyclases in thyroid carcinomas. Endocrine-related cancer. 2013;20:79–90. doi: 10.1530/ERC-12-0053. [DOI] [PubMed] [Google Scholar]
- 7.Hopewell EL, Zhao W, Fulp WJ, Bronk CC, Lopez AS, Massengill M, et al. Lung tumor NF-kappaB signaling promotes T cell-mediated immune surveillance. The Journal of clinical investigation. 2013;123:2509–22. doi: 10.1172/JCI67250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lerebours F, Vacher S, Andrieu C, Espie M, Marty M, Lidereau R, et al. NF-kappa B genes have a major role in inflammatory breast cancer. BMC cancer. 2008;8:41. doi: 10.1186/1471-2407-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrzycka B, Terlikowski SJ, Garbowicz M, Niklinska W, Bernaczyk PS, Niklinski J, et al. Tumor necrosis factor-alpha and its receptors in epithelial ovarian cancer. Folia histochemica et cytobiologica/Polish Academy of Sciences, Polish Histochemical and Cytochemical Society. 2009;47:609–13. doi: 10.2478/v10042-008-0117-1. [DOI] [PubMed] [Google Scholar]
- 10.Sethi G, Sung B, Kunnumakkara AB, Aggarwal BB. Targeting TNF for Treatment of Cancer and Autoimmunity. Advances in experimental medicine and biology. 2009;647:37–51. doi: 10.1007/978-0-387-89520-8_3. [DOI] [PubMed] [Google Scholar]
- 11.Zhou P, Qiu J, L’Italien L, Gu D, Hodges D, Chao CC, et al. Mature B cells are critical to T-cell-mediated tumor immunity induced by an agonist anti-GITR monoclonal antibody. J Immunother. 2010;33:789–97. doi: 10.1097/CJI.0b013e3181ee6ba9. [DOI] [PubMed] [Google Scholar]
- 12.Schaer DA, Cohen AD, Wolchok JD. Anti-GITR antibodies--potential clinical applications for tumor immunotherapy. Curr Opin Investig Drugs. 2010;11:1378–86. [PubMed] [Google Scholar]
- 13.Nakagawa H, Tsuta K, Kiuchi K, Senzaki H, Tanaka K, Hioki K, Tsubura A. Growth inhibitory effects of diallyl disulfide on human breast cancer cell lines. Carcinogenesis. 2001 Jun;22(6):891–7. doi: 10.1093/carcin/22.6.891. [DOI] [PubMed] [Google Scholar]
- 14.Fang WB, Jokar I, Zou A, Lambert D, Dendukuri P, Cheng N. CCL2/CCR2 Chemokine signaling coordinates survival and motility of breast cancer cells through smad3 protein and p42/44 mitogen activated protein kinase (MAPK)-dependent mechanisms. The Journal of Biological Chemistry. 2012 Oct;287(43):36593–36608. doi: 10.1074/jbc.M112.365999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon AH, O’Keefe RJ, Schwartz EM, Rosier RN, Puzas JE. Nuclear Factor κB-dependent mechanisms in breast cancer cells regulate tumor burden and osteolysis in bone. Cancer Res. 2005;65:3209–3217. doi: 10.1158/0008-5472.CAN-04-4017. [DOI] [PubMed] [Google Scholar]
- 16.Bauer D, Mazzio E, Soliman KF, Taka E, Oriaku E, Womble T. Darling-Reed S. Diallyl disulfide inhibits TNFα-induced CCL2 release by MDA-MB-231 cells. Anticancer Res. 2014 Jun;34(6):2763–70. [PMC free article] [PubMed] [Google Scholar]
- 17.Nkrumah-Elie YM1, Reuben JS, Hudson AM, Taka E, Badisa R, Ardley T, Israel B, Sadrud-Din SY, Oriaku ET, Darling-Reed SF. The attenuation of early benzo(a)pyrene-induced carcinogenic insults by diallyl disulfide (DADS) in MCF-10A cells. Nutr Cancer. 2012;64(7):1112–21. doi: 10.1080/01635581.2012.712738. Epub 2012 Sep 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–75. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craddock JA, Lu A, Bear A, Pule M, Brenner MK, Rooney CM, et al. Enhanced tumor trafficking of GD2 chimeric antigen receptor T cells by expression of the chemokine receptor CCR2b. J Immunother. 2010;33:780–8. doi: 10.1097/CJI.0b013e3181ee6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry JA, Thamm DH, Eickhoff J, Avery AC, Dow SW. Increased monocyte chemotactic protein-1 concentration and monocyte count independently associate with a poor prognosis in dogs with lymphoma. Veterinary and comparative oncology. 2011;9:55–64. doi: 10.1111/j.1476-5829.2010.00235.x. [DOI] [PubMed] [Google Scholar]
- 22.Mishra P, Banerjee D, Ben-Baruch A. Chemokines at the crossroads of tumor-fibroblast interactions that promote malignancy. Journal of leukocyte biology. 2011;89:31–9. doi: 10.1189/jlb.0310182. [DOI] [PubMed] [Google Scholar]
- 23.Hatfield KJ, Reikvam H, Bruserud O. The crosstalk between the matrix metalloprotease system and the chemokine network in acute myeloid leukemia. Current medicinal chemistry. 2010;17:4448–61. doi: 10.2174/092986710794183033. [DOI] [PubMed] [Google Scholar]
- 24.Cendan JC, Topping DL, Pruitt J, Snowdy S, Copeland EM, 3rd, Lind DS. Inflammatory mediators stimulate arginine transport and arginine-derived nitric oxide production in a murine breast cancer cell line. The Journal of surgical research. 1996;60:284–8. doi: 10.1006/jsre.1996.0044. [DOI] [PubMed] [Google Scholar]
- 25.Moran AE, Kovacsovics-Bankowski M, Weinberg AD. The TNFRs OX40, 4-1BB, and CD40 as targets for cancer immunotherapy. Current opinion in immunology. 2013;25:230–7. doi: 10.1016/j.coi.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daniel D, Wilson NS. Tumor necrosis factor: renaissance as a cancer therapeutic? Current cancer drug targets. 2008;8:124–31. doi: 10.2174/156800908783769346. [DOI] [PubMed] [Google Scholar]
- 27.Mahmood Z, Shukla Y. Death receptors: targets for cancer therapy. Experimental cell research. 2010;316:887–99. doi: 10.1016/j.yexcr.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–5. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YS, Xie N, Su Q, Su J, Huang C, Liao QJ. Diallyl disulfide inhibits the proliferation of HT-29 human colon cancer cells by inducing differentially expressed genes. Molecular medicine reports. 2011;4:553–9. doi: 10.3892/mmr.2011.453. [DOI] [PubMed] [Google Scholar]
- 30.Yi L, Ji XX, Lin M, Tan H, Tang Y, Wen L, et al. Diallyl disulfide induces apoptosis in human leukemia HL-60 cells through activation of JNK mediated by reactive oxygen. Die Pharmazie. 2010;65:693–8. [PubMed] [Google Scholar]
- 31.Sundaram SG, Milner JA. Diallyl disulfide induces apoptosis of human colon tumor cells. Carcinogenesis. 1996;17:669–73. doi: 10.1093/carcin/17.4.669. [DOI] [PubMed] [Google Scholar]
- 32.Lai KC, Hsu SC, Kuo CL, Yang JS, Ma CY, Lu HF, et al. Diallyl sulfide, diallyl disulfide, and diallyl trisulfide inhibit migration and invasion in human colon cancer colo 205 cells through the inhibition of matrix metalloproteinase-2, -7, and -9 expressions. Environmental toxicology. 2011 doi: 10.1002/tox.20737. [DOI] [PubMed] [Google Scholar]
- 33.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Science’s STKE: signal transduction knowledge environment. 2006;2006:re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 34.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annual review of immunology. 2000;18:621–63. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Lu X, Zhu L, Shen Y, Chengedza S, Feng H, et al. IKK epsilon kinase is crucial for viral G protein-coupled receptor tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:11139–44. doi: 10.1073/pnas.1219829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang N, Ahmed S, Haqqi TM. Genomic structure and functional characterization of the promoter region of human IkappaB kinase-related kinase IKKi/IKKvarepsilon gene. Gene. 2005;353:118–33. doi: 10.1016/j.gene.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou AY, Shen RR, Kim E, Lock YJ, Xu M, Chen ZJ, et al. IKKepsilon-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell reports. 2013;3:724–33. doi: 10.1016/j.celrep.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peant B, Forest V, Trudeau V, Latour M, Mes-Masson AM, Saad F. IkappaB-Kinase-epsilon (IKKepsilon/IKKi/IkappaBKepsilon) expression and localization in prostate cancer tissues. The Prostate. 2011;71:1131–8. doi: 10.1002/pros.21329. [DOI] [PubMed] [Google Scholar]
- 39.Cheng A, Guo J, Henderson-Jackson E, Kim D, Malafa M, Coppola D. IkappaB Kinase epsilon expression in pancreatic ductal adenocarcinoma. American journal of clinical pathology. 2011;136:60–6. doi: 10.1309/AJCP2JJGYNIUAS2V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang MR, Kim MS, Kim SS, Ahn CH, Yoo NJ, Lee SH. NF-kappaB signalling proteins p50/p105, p52/p100, RelA, and IKKepsilon are over-expressed in oesophageal squamous cell carcinomas. Pathology. 2009;41:622–5. doi: 10.3109/00313020903257756. [DOI] [PubMed] [Google Scholar]
- 41.Baldwin AS. Regulation of cell death and autophagy by IKK and NF-kappaB: critical mechanisms in immune function and cancer. Immunological reviews. 2012;246:327–45. doi: 10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- 42.Guo JP, Shu SK, He L, Lee YC, Kruk PA, Grenman S, et al. Deregulation of IKBKE is associated with tumor progression, poor prognosis, and cisplatin resistance in ovarian cancer. The American journal of pathology. 2009;175:324–33. doi: 10.2353/ajpath.2009.080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhelst K, Verstrepen L, Carpentier I, Beyaert R. IkappaB kinase epsilon (IKKepsilon): a therapeutic target in inflammation and cancer. Biochemical pharmacology. 2013;85:873–80. doi: 10.1016/j.bcp.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Chen L, Zhang A, Wang G, Han L, Yu K, et al. Silencing of IKKepsilon using siRNA inhibits proliferation and invasion of glioma cells in vitro and in vivo. International journal of oncology. 2012;41:169–78. doi: 10.3892/ijo.2012.1452. [DOI] [PubMed] [Google Scholar]
- 45.Qin B, Cheng K. Silencing of the IKKepsilon gene by siRNA inhibits invasiveness and growth of breast cancer cells. Breast cancer research: BCR. 2010;12:R74. doi: 10.1186/bcr2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grandvaux N. The IKKepsilon kinase in breast cancer: from oncogenesis to treatment resistance. Medecine sciences: M/S. 2011;27:619–25. doi: 10.1051/medsci/2011276014. [DOI] [PubMed] [Google Scholar]
- 47.Hutti JE, Porter MA, Cheely AW, Cantley LC, Wang X, Kireev D, et al. Development of a high-throughput assay for identifying inhibitors of TBK1 and IKKepsilon. PloS one. 2012;7:e41494. doi: 10.1371/journal.pone.0041494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wietek C, Cleaver CS, Ludbrook V, Wilde J, White J, Bell DJ, et al. IkappaB kinase epsilon interacts with p52 and promotes transactivation via p65. The Journal of biological chemistry. 2006;281:34973–81. doi: 10.1074/jbc.M607018200. [DOI] [PubMed] [Google Scholar]
- 49.Li Q, Sun H, Zou J, Ge C, Yu K, Cao Y, et al. Increased Expression of Estrogen Receptor alpha-36 by Breast Cancer Oncogene IKKepsilon Promotes Growth of ER-Negative Breast Cancer Cells. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2013;31:833–41. doi: 10.1159/000350101. [DOI] [PubMed] [Google Scholar]
- 50.Guo JP, Shu SK, Esposito NN, Coppola D, Koomen JM, Cheng JQ. IKKepsilon phosphorylation of estrogen receptor alpha Ser-167 and contribution to tamoxifen resistance in breast cancer. The Journal of biological chemistry. 2010;285:3676–84. doi: 10.1074/jbc.M109.078212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Guan YJ, Zhang Z, Yu C, Ma L, Hu W, Xu L, et al. Phospho-SXXE/D motif mediated TNF receptor 1-TRADD death domain complex formation for T cell activation and migration. J Immunol. 2011;187:1289–97. doi: 10.4049/jimmunol.1003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng L, Bidere N, Staudt D, Cubre A, Orenstein J, Chan FK, et al. Competitive control of independent programs of tumor necrosis factor receptor-induced cell death by TRADD and RIP1. Molecular and cellular biology. 2006;26:3505–13. doi: 10.1128/MCB.26.9.3505-3513.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pobezinskaya YL, Liu Z. The role of TRADD in death receptor signaling. Cell Cycle. 2012;11:871–6. doi: 10.4161/cc.11.5.19300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnamurthy S, Basu A. Regulation of IKKepsilon Expression by Akt2 Isoform. Genes & cancer. 2011;2:1044–50. doi: 10.1177/1947601912444604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westlund KN, Zhang L, Ma F, Oz HS. Chronic inflammation and pain in a tumor necrosis factor receptor (TNFR) (p55/p75−/−) dual deficient murine model. Translational research: the journal of laboratory and clinical medicine. 2012;160:84–94. doi: 10.1016/j.trsl.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pincheira R, Castro AF, Ozes ON, Idumalla PS, Donner DB. Type 1 TNF receptor forms a complex with and uses Jak2 and c-Src to selectively engage signaling pathways that regulate transcription factor activity. J Immunol. 2008;181:1288–98. doi: 10.4049/jimmunol.181.2.1288. [DOI] [PubMed] [Google Scholar]
- 57.Tsubura A, Lai YC, Kuwata M, Uehara N, Yoshizawa K. Anticancer effects of garlic and garlic-derived compounds for breast cancer control. Anti-cancer agents in medicinal chemistry. 2011;11:249–53. doi: 10.2174/187152011795347441. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Patel L, Pienta KJ. Targeting chemokine (C-C motif) ligand 2 (CCL2) as an example of translation of cancer molecular biology to the clinic. Progress in molecular biology and translational science. 2010;95:31–53. doi: 10.1016/B978-0-12-385071-3.00003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rafei M, Galipeau J. A CCL2-based fusokine as a novel biopharmaceutical for the treatment of CCR2-driven autoimmune diseases. Critical reviews in immunology. 2010;30:449–61. doi: 10.1615/critrevimmunol.v30.i5.40. [DOI] [PubMed] [Google Scholar]