Abstract
The generation of regulatory T (Treg) cells is driven by Foxp3 and is responsible for dampening inflammation and reducing autoimmunity. In this study the epigenetic regulation of iTreg cells was examined and identified a H3K4 histone methyltransferase, SMYD3, which regulates expression of Foxp3 by a TGFβ1/Smad3 dependent mechanism. Using ChIP assays, SMYD3 depletion led to reduction in H3K4me3 in the promoter region and CNS-1 of the foxp3 locus. SMYD3 abrogation affected iTreg cell formation while allowing dysregulated IL-17 production. In a mouse model of respiratory syncytial virus infection (RSV), a model where iTreg cells play a critical role in regulating lung pathogenesis, SMYD3−/− mice demonstrated exacerbation of RSV-induced disease related to enhanced proinflammatory responses and worsened pathogenesis within the lung. Our data highlight a novel activation role for the TGFβ-inducible SMYD3 in regulating iTreg cell formation leading to increased severity of virus-related disease.
INTRODUCTION
Epigenetic regulation of gene activation is organization of loci into transcriptionally active or silent states altering the accessibility of transcription factors and polymerases to gene promoters and enhancers 1, 2. Histone modifications that regulate chromatin accessibility include methylation, acetylation, ubiquitination, phosphorylation, etc, and determine the transcriptional status of the gene loci by exposing or sequestering the promoter region 3. Methylation of lysines on histone H3 for the regulation of chromatin accessibility, especially H3K4 trimethylation, is associated with transcriptional activation. This activation mark is offset by methylation of H3K9 and H3K27, associated with transcriptional silencing of the gene. The modifications rely on both methyltransferases that add and demethylases that remove methyl groups from specific lysines allowing plasticity to gene activation 4. Thus, the specific regulation of genes by chromatin modifications is likely both gene and cell specific.
The SET and MYND Domain (SMYD) are a family of SET histone methyltransferases involved in chromatin regulation and gene transcription 5. SMYD3 was previously identified as an H3K4me3 histone methyltransferase (HMTase) that may be a proto-oncogene based upon its expression in numerous cancers and due to cellular function observed in overexpression studies of normal cells or in silencing studies in tumors 6–8. SMYD3 is a regulator of MMP9 modifying H3K4me3 marks on the MMP9 promoter and affecting tumor invasiveness 9. The function and regulation of SMYD3 in non-transformed cells or its regulation in immune cells has not been examined.
The differentiation of mature T cells into different phenotypes is controlled by multiple cytokines and related transcription factors that allow the immune system to “fine tune” responses to pathogen insult 10, 11. An important T cell phenotype is the Foxp3-expressing regulatory T (Treg) cell that can affect the other T helper phenotypes and their accompanying responses 12. The central determinant of Treg development is Foxp3 expression, a transcription factor that is constitutively expressed in thymus-derived naturally occurring Treg (nTreg) cells and upregulated in inducible Treg (iTreg) cells 13, 14. Also important in the generation of iTreg cells is the activation of TGFβ/Smad3 signaling pathway 15, which correlates with the alteration of a conserved non-coding DNA sequence (CNS1) element at the foxp3 locus and regulates Foxp3 expression in iTreg cells 16–18. The present studies expand our knowledge of epigenetic regulation during the development of Treg cells 10.
In the present study SMYD3 was identified as a TGFβ/Smad3 associated primary epigenetic mediator of Foxp3 in iTreg cells, while also regulates IL-17 production. In vitro silencing or CD4 specific genetic deficiency of TGFβ-inducible SMYD3 reduces iTreg cell development and in vivo leads to exacerbated virus-induced lung pathology associated with dysregulated proinflammatory cytokine production. Overall these data highlight a novel activation role for SMYD3 methyltransferase in the regulation of Foxp3 expression, generation of iTreg cells, and modulation of proinflammatory cytokine production.
RESULTS
TGFβ induces SMYD3, a H3K4 methyltransferase, in iTreg differentiating cells
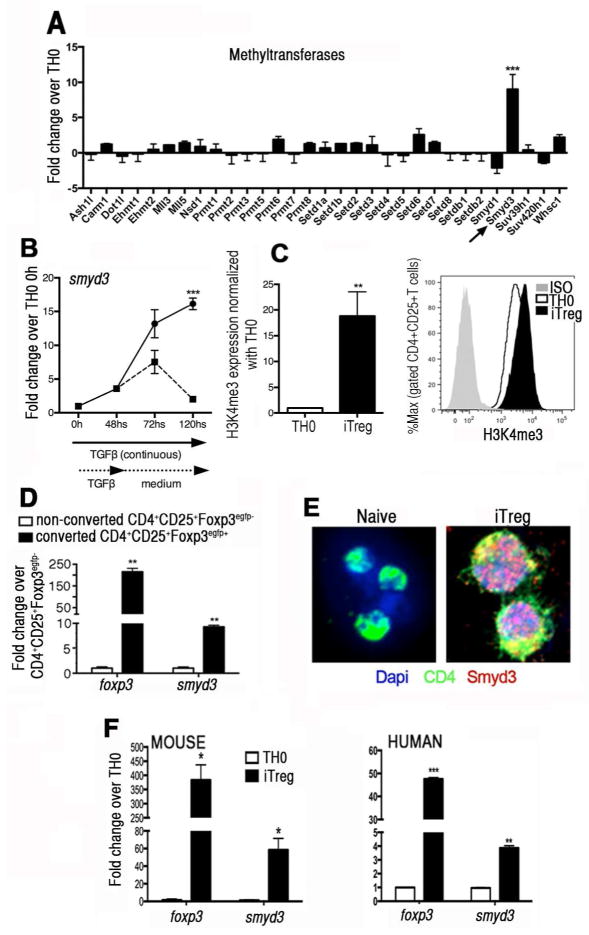
The present studies focused upon examining the overall epigenetic regulation in iTreg cells by initiating an unbiased examination of epigenetic enzymes using a gene subset array during iTreg cell development. After 48 hours of iTreg skewing conditions, mRNA analysis of chromatin remodeling enzymes was performed. The data in Fig. 1A depict the methyltransferases analyzed and show the only gene that was significantly upregulated during the iTreg skewing process was SMYD3 (H3K4 methyltransferase). Subsequent studies using q-RT-PCR analysis of SMYD3 mRNA levels in naïve CD4+T cells under iTreg skewing conditions in vitro showed a continuous increase in the gene expression levels over a period of 120 hours (Fig. 1B). Also, the sustained expression of smyd3 required the continuous presence of TGFβ in culture as expression levels diminished once TGFβ was removed (Fig. 1B). When analyzing the SMYD3 chromatin modifying mark H3K4me3 after 3 days under skewing conditions, results showed that iTreg cells had a significant increase in H3K4me3 expression compared to TH0 cells (Fig. 1C). By stimulating naïve CD4+T cells with each component of the iTreg cell differentiation individually, our data demonstrated that TGFβ is a primary inducer of SMYD3 (Supplementary Fig. 1A). Moreover, using naïve CD4+T cells derived from mice (Supplementary Fig. 1B) and from human PBMCs (Supplementary Fig. 1C) TGFβ-induced SMYD3 protein level was dose dependent as indicated by Western blot and by q-RT-PCR. Next, to verify whether SMYD3 was upregulated in all cells exposed to TGFβ or only in converted Foxp3+iTreg cells, naïve T cells were isolated from Foxp3egfp mice and skewed toward iTreg cells. Converted iTregs (CD4+CD25+Foxp3egfp+) and non-converted effector T cells (CD4+CD25+Foxp3egfp−) were FACS sorted. As shown in Fig. 1D, smyd3 was overexpressed in Foxp3egfp+ converted iTreg cells but not in non-converted Foxp3egfp− effector T cells, indicating that SMYD3 is correlated with the expression of Foxp3.
Figure 1. TGFβ1 regulates the expression of SMYD3 in inducible Treg cells.
(A) Naïve T cells were cultured under TH0 or skewed towards iTregs. After 48 hours the levels of methyltransferases were each assessed by PCR arrays. Data are representative of 3 independent experiments. (B) SMYD3 expression was assessed by q-RT-PCR in naïve T cells skewed under iTreg conditions over 120 hours. At 48 hours some wells had TGFβ1 withdrawn from the culture, mRNA extracted, and gene expression of SMYD3 assessed. (C) Bar graph shows expression of H3K4me3 in iTreg cells normalized with TH0 and histogram show H3K4me3 expression in gated CD4+CD25+T cells in iTreg and TH0 cells at 3 days of differentiation. (D) Naïve CD4+T cells from Foxp3egfp mice were skewed towards iTreg cells. Converted CD4+CD25+Foxp3egfp+ iTregs and CD4+CD25+Foxp3egfp− effector cells were FACS sorted from 5-day cultures and mRNA was extracted with Foxp3 and SMYD3 expression was then analyzed by q-RT-PCR. (E) Naïve CD4+T cells and iTreg cells were stained with anti-CD4 and anti-SMYD3 and evaluated by confocal microscopy. (F) Naïve CD4+T cells were isolated from mouse spleens and human PBMCs, activated under TH0 or iTreg skewing condition for 5 days, mRNA extracted, and gene expression of Foxp3 and SMYD3 assessed. Data are means ± SD of triplicate samples and are representative of at least two independent experiments. *P<0.05, **P<0.01, ***P<0.001.
To verify the effect of TGFβ for SMYD3 protein induction in iTreg cells and its co-localization in the nucleus, both naïve CD4+T cells and 4-day skewed iTreg cells were assessed using a SMYD3 specific antibody and confocal microscopy. The cells under iTreg skewing conditions, but not naïve T cells, had SMYD3 protein expressed in the nucleus (Fig. 1E). Next, to further confirm the correlation of SMYD3 with Foxp3 expression, using murine and human naïve CD4+T cells skewed towards iTreg cells in vitro it was verified that smyd3 expression paralleled the expression of foxp3. Both mRNA levels of foxp3 and smyd3 were upregulated in iTreg-skewing cells in mouse and human cells (Fig. 1F). Also, Foxp3 and SMYD3 protein levels were checked in both TH0 and iTreg cells and showed higher SMYD3 and Foxp3 protein levels in iTreg cells (Supplementary Fig. 1D). Thus, these results suggest that the epigenetic enzyme SMYD3 is specifically induced by TGFβ and coordinated with Foxp3 expression in iTregs cells.
SMYD3 correlates with Foxp3 expression only in iTreg but not nTreg cells
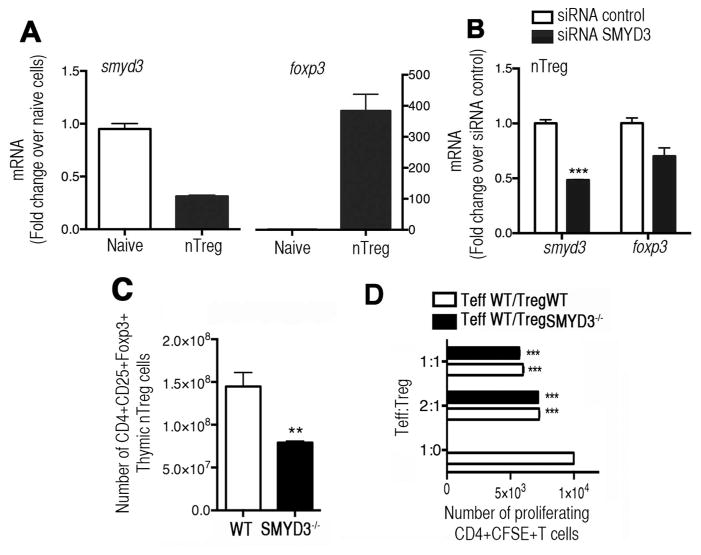
We also characterized the expression of SMYD3 in nTreg cells by isolating thymic nTregs (CD4+CD25+Foxp3egfp+) or naïve (CD4+CD25−Foxp3egfp−) T cells from Foxp3egfp mice. While Foxp3 expression was high, the expression level of SMYD3 in thymic derived nTreg cells was low compared to naïve cells (Fig. 2A), suggesting that SMYD3 correlates with Foxp3 only in iTreg cells but not in nTreg cells. To determine whether SMYD3 would affect Foxp3 expression in nTreg cells, thymic nTreg (CD4+CD25+Foxp3egfp+) cells from Foxp3egfp mice were isolated and transfected with control or SMYD3 siRNA. The results show that SMYD3 knockdown did not affect Foxp3 expression in nTreg cells compared to control siRNA (Fig. 2B).
Figure 2. TGFβ-inducible SMYD3 correlates with Foxp3 expression in inducible but not in natural Treg cells.
(A) CD4+CD25+Foxp3egfp+ nTregs and CD4+CD25−Foxp3egfp− T cells were isolated from thymus of Foxp3egfp mice. Smyd3 and foxp3 mRNA expression was analyzed by q-RT-PCR. (B) nTreg cells were isolated as in A and transfected with SMYD3 or control siRNA. After 24 hours smyd3 and foxp3 expression was analyzed. (C) Flow cytometric analysis of CD4+CD25+Foxp3+nTreg cells in thymus of SMYD3−/− and WT mice. (D) Treg suppression assay comparing suppressive function of nTreg cells derived from WT and SMYD3−/− mice co-cultured with CFSE-labeled WT naïve CD4+T cells stimulated with anti-CD3 and anti-CD28. Data are representative of 3 independent experiments and show means ± SEM of 4 replicates. *P<0.05, **P<0.01, ***P<0.001.
To further determine the possible role of SMYD3 in the regulation of Foxp3 in nTreg cells in vivo we conditionally deleted SMYD3 in CD4+T cells by crossing novel SMYD3fl/fl mice generated by our lab, with CD4Cre mice. Therefore, resultant mice (hereafter SMYD3−/−) are SMYD3 deficient in all mature CD4+ T cells, as well as in CD8+ T cells and CD4+CD8+ T cells. All animals were viable, healthy, and fertile. SMYD3−/− mice had reduced number of thymic CD4+CD25+Foxp3+ nTreg cells in the SMYD3−/− mice compared to WT (Fig. 2C). However, despite the reduced numbers of nTreg cells all SMYD3−/− mice were born at normal Mendelian ratios and displayed neither phenotypic alteration nor autoimmunity with no signs of weight loss, lymphadenopathy or splenomegaly, further suggesting that nTreg cells may not be substantially regulated by SMYD3 when using the CD4-Cre targeted mice. Furthermore, histological sections of key tissues from SMYD3−/− showed no pathological alterations at age 8 weeks or 6 months (Supplementary Fig. 3A). To assess whether SMYD3 was regulating the suppressive function of nTreg cells, a suppression assay was performed using WT and SMYD3−/− FACS-sorted CD4+CD25HI cells (Foxp3+nTreg cells; >95% purity) isolated from naïve mice. Our data show that both WT and SMYD3−/− derived nTreg cells were able to suppress proliferation of CFSE-labeled CD4+T effector cells when cultured at Teff:Treg ratio of 2:1 or 1:1, suggesting that the Foxp3+ nTreg cells generated by the SMYD3−/− mice have no defect in their suppressive function (Fig. 2D). Thus, our results indicate that SMYD3 is not required for Foxp3 expression in nTreg cells and do not alter their suppressive function.
The induction of SMYD3 in iTreg cells is mediated by a Smad-dependent mechanism
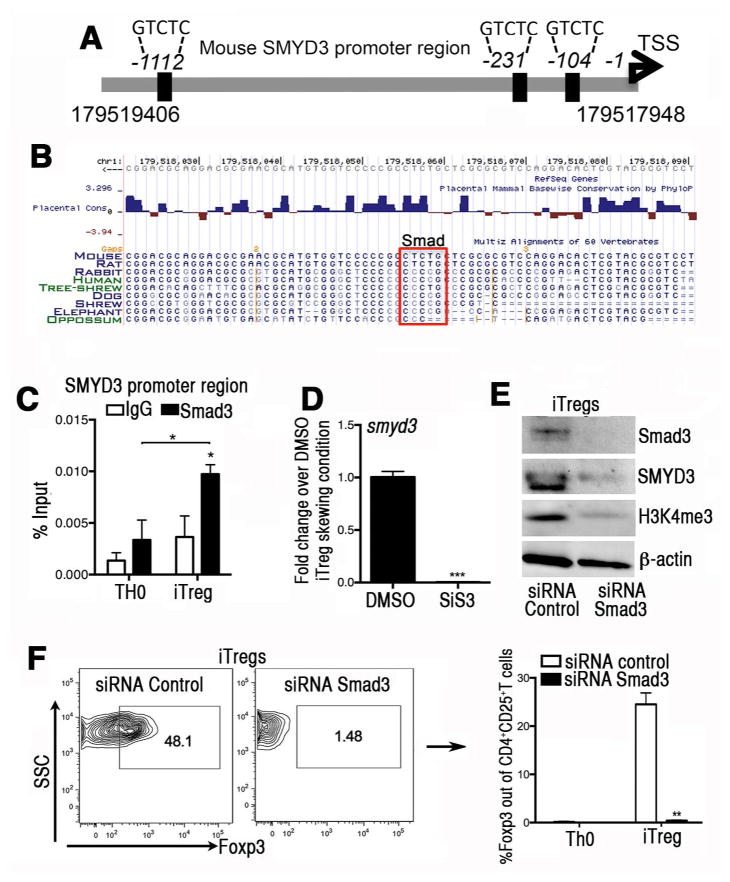
Since our data in Fig. 1 indicated that TGFβ was a primary inducer of SMYD3, we interrogated the SMYD3 promoter gene for Smad binding element (SBE) (5′-GTCTG-3′) 19. Three SBEs were identified within 1.1kb of the SMYD3 promoter region (Fig. 3A) with one SBE that is evolutionary conserved within the minimal SMYD3 promoter (-104) (Fig. 3B). Using Chip assay, iTreg cells showed enhanced Smad3 at the SMYD3 promoter region when compared to TH0 cells (Fig. 3C). When SiS3, a Smad3 inhibitor, was used in iTreg skewing cell cultures, it inhibited the expression of SMYD3 (Fig. 3D). Compared to a control siRNA, Smad3 siRNA transfected iTreg cells displayed significant decreases in SMYD3 as well as its chromatin mark H3K4me3 by the Western blot (Fig. 3E). The downregulation of SMYD3 was accompanied by decreased Foxp3 protein using flow cytometry (Fig. 3F), in agreement with previous studies reporting the role of TGFβ/Smad3 signaling pathway for iTreg conversion 15. The data show that SMYD3 expression and H3K4 trimethylation are reduced in the absence of Smad3 signaling.
Figure 3. The induction of SMYD3 in iTregs is mediated in a Smad3-dependent manner.
(A) Schematic of the murine SMYD3 promoter region (accession # NM_027188) highlighting potential putative Smad3 binding sites at −104bp, −231 bp, and −1112 bp upstream of the SMYD3 transcription starting site. (B) A Smad3 binding site is evolutionary conserved within the minimal SMYD3 promoter (104bp). Conserved Smad3 binding site on SMYD3 promoter extracted from USCS Genome browser on Mouse (GRCm38/mm10) Assembly. (C) Cells were cultured in complete RPMI under TH0 or iTreg cell skewing conditions for 18 hours. Chromatin immunoprecipitation (ChIP)-quantitative PCR was performed in the SMYD3 promoter site using Smad3 antibody or species-matched IgG as control. Data show four replicates and are representative of three independent experiments (*P<0.05). (D) Naïve CD4+T cells were isolated and stimulated under iTreg differentiation conditions in the presence of the Smad3 inhibitor SiS3 (10 μm) or DMSO for 4 days and assessed for SMYD3. (E) Western blots of Smad3, SMYD3, H3K4me3 and β-actin from Smad3 or control siRNA transfected T cells under iTregs skewing conditions. (F) Foxp3 protein levels in naïve CD4+T cells transfected with Smad3 or control siRNA under iTreg skewing condition. Data are means ±SD and are representative of three independent experiments. *P<0.05, **P<0.01, ***P<0.001.
SMYD3 depletion inhibits Foxp3 expression during iTreg cell development
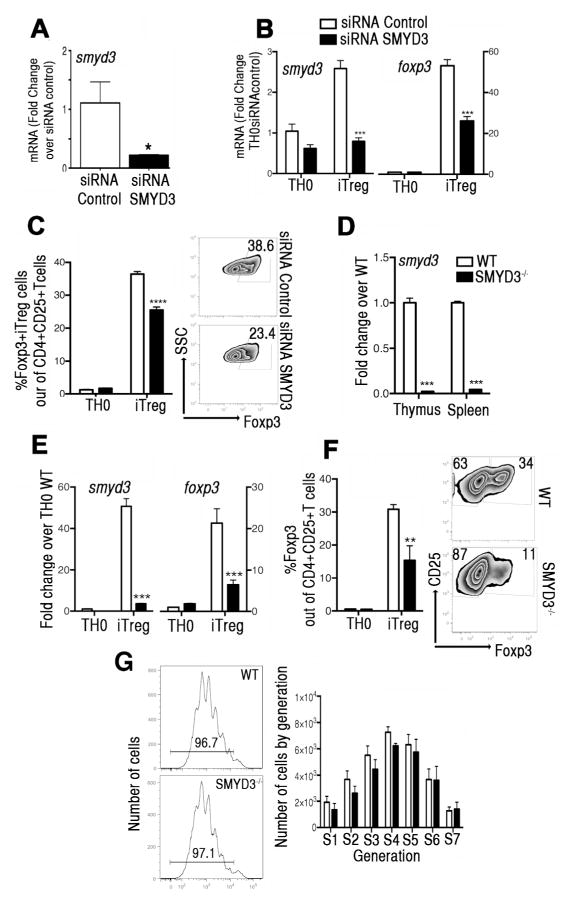
In order to determine whether SMYD3 was involved in iTreg development, a siRNA knockdown approach was used. Compared to control siRNA, SMYD3 siRNA significantly reduced the smyd3 mRNA expression in splenic naive CD4+T cells 24hs after transfection (Fig. 4A). Examination of the SMYD3 knockdown effect in iTreg cell conversion, assessed by smyd3 and foxp3, demonstrated that in the SMYD3 siRNA transfected naïve CD4+T cells there was an abrogation of iTreg cell development, by q-RT-PCR (Fig. 4B) and flow cytometry (Fig. 4C), suggesting that SMYD3 regulates iTreg cell conversion. Importantly, inhibition of SMYD3 had no effect on apoptosis (Supplementary Fig. 2A) or the proliferative capacity of TH0 or iTreg cells (Supplementary Fig. 2B), suggesting that the absence of SMYD3 did not alter cell survival or proliferation. The effect of SMYD3 on Foxp3 stability was investigated by transfecting iTreg cells differentiated in vitro with SMYD3 or control siRNA. Silencing SMYD3 in differentiated iTregs resulted in decreased foxp3 expression when compared to control siRNA (Supplementary Fig. 2C), suggesting that SMYD3 has a role in the stability of Foxp3 in iTreg cells.
Figure 4. SMYD3 depletion inhibits Foxp3 expression during Treg cell development.
(A) Naïve CD4+T cells, transfected with SMYD3 or control siRNA, were assessed for smyd3 expression. (B) After 5 days, expression levels of smyd3 and foxp3 were assessed in transfected cells under TH0 or iTreg skewing conditions. (C) Bar graphs and zebra plots showing Foxp3 expression in gated CD4+CD25+ T cells. Data are presented as the mean of four wells and are representative of four independent experiments. (D) Expression levels of smyd3 mRNA in CD4+T cells derived from thymus and spleens of WT and SMYD3−/− mice. (E) Expression levels of smyd3 and foxp3 mRNA in naïve CD4+T cells from WT (white) and SMYD3−/− (black) differentiated under TH0 and iTreg skewing conditions for 3 days. (F) Bar graphs of CD4+CD25+T cells from WT (white) and SMYD3−/− mice (black) and zebra plots showing Foxp3 expression in gated CD4+CD25+T cells skewed as in E. (G) CFSE-labeled naïve CD4+T cells derived from WT and SMYD3−/− mice under iTreg skewing conditions were assessed by flow cytometry. Histogram plots show proliferation and bar graph shows number of proliferating cells by generation at day 4. Data are representative of 4 independent experiments performed with n=3 per group and show means ± SEM of 4 replicates. *P<0.05, **P<0.01, ***P<0.001.
To examine physiological relevance of SMYD3 in iTreg cells in vivo we used SMYD3fl/fl mice. Q-RT-PCR of smyd3 in T cells derived from thymus and spleens of SMYD3−/− compared to WT mice showed that the expression of smyd3 was inhibited in SMYD3−/− mice (Fig. 4D). Importantly, the frequency and numbers of naïve and activated/memory CD4+ and CD8+T cells in SMYD3−/− were equivalent to WT mice (Supplementary Fig. 3B). To characterize the effect of SMYD3 depletion in iTreg cell conversion, naïve CD4+T cells were isolated from WT and SMYD3−/− mice and differentiated into iTreg cells in vitro. SMYD3−/− derived and skewed iTreg cells had a significant reduction in the expression of smyd3 and foxp3 compared to those from WT mice (Fig. 4E). This latter finding was verified using flow cytometry of 5 day-skewed cell cultures demonstrating that SMYD3−/− mice generated decreased numbers of CD4+CD25+Foxp3+ iTreg cells compared to WT mice (Fig. 4F). Importantly, SMYD3 deletion did not modify proliferative competence of naïve CD4+T cells derived from SMYD3−/− mice stimulated with anti-CD3/CD28 (Fig. 4G). These results offer genetic evidence of the functional relevance of SMYD3 in the development of TGFβ-inducible iTreg cells.
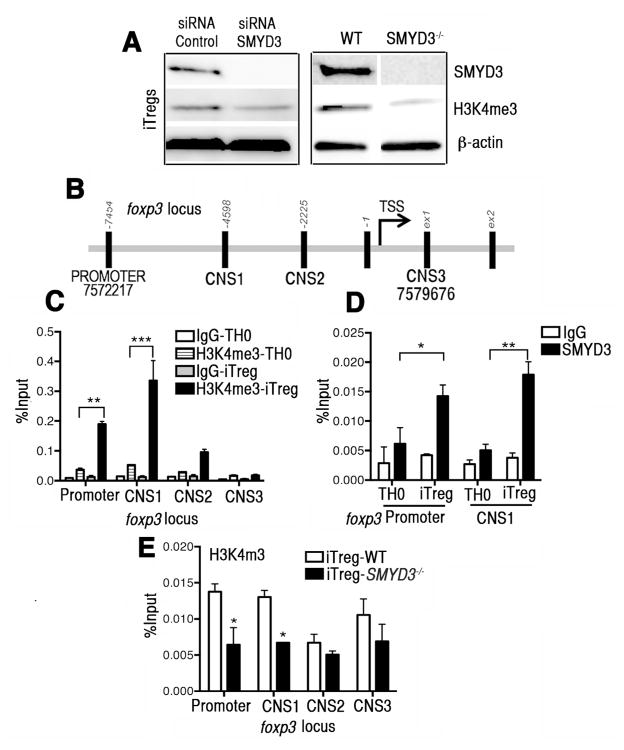
H3K4 trimethylation chromatin immunoprecipitation (ChIP) of Foxp3 in iTreg cells is partially regulated by SMYD3
To further characterize SMYD3 function, we investigated whether SMYD3 depletion affects the H3K4 trimethylation in iTreg cells. Western blot analysis of naive CD4+T cells transfected with SMYD3 siRNA or control siRNA, and of naive CD4+T cells derived from SMYD3−/− or WT mice under iTreg skewing conditions show that by either knocking down or genetically depleting SMYD3, the expression of H3K4me3 in iTreg-skewed cells was significantly reduced (Fig. 5A). Transcription of the foxp3 gene is regulated by the promoter region but also by conserved non-coding DNA sequence (CNS) elements. CNS-1, -2 and -3 encode essential information defining composition and stability of the Treg population 16. Using ChIP analysis of H3K4 trimethylation, we specifically examined the promoter site and CNS 1, 2 and 3 of the foxp3 locus in cells from WT mice (Fig. 5B). Compared to TH0 cells, it was found that iTreg cells have enhanced H3K4 trimethylation at the promoter site and CNS-1 (Fig. 5C). To verify that SMYD3 was the methyltransferase responsible for the modification in H3K4 in the promoter region and CNS-1 of foxp3, we performed a ChIP assay of SMYD3. The results showed significant enhancement of SMYD3 in these regions in iTreg cells (Fig. 5D). Finally, to confirm that SMYD3 was the enzyme responsible for modifying H3K4me3 at the promoter and CNS-1, SMYD3−/− and WT derived iTreg-skewed cells were examined. The results showed significant reduction in the H3K4 trimethylation levels at the promoter and CNS-1 sites, but not CNS-2 and -3, of foxp3 locus from SMYD3−/− mice (Fig. 5E). Although our results indicate that SMYD3 is not the sole factor regulating Foxp3, the TGFβ-inducible SMYD3 enhances to the trimethylated state of H3K4 in specific regions of the foxp3 locus, namely the promoter and the CNS-1 element, thus facilitating gene transcription.
Figure 5. SMYD3 is partially responsible for H3K4 trimethylation at the foxp3 locus in iTreg cells.
(A) Western blots of SMYD3, H3K4me3 and β-actin from lysates of control or SMYD3 siRNA transfected WT naïve CD4+T cells and from WT and SMYD3−/− derived naïve CD4+T cells, 4 days under iTreg differentiating conditions. (B) Schematic of the murine foxp3 locus highlighting the regions investigated. ChIP assays were performed in the foxp3 promoter site, CNS -1, -2, and -3 of the foxp3 locus using H3K4me3 antibody or IgG as control (C), and in the foxp3 promoter site and CNS-1 using SMYD3 antibody or IgG as control (D). (E) ChIP assays were performed in the foxp3 locus using H3K4me3 antibody or IgG as control or skewed T cells. Data show means ± SEM of four replicate wells and are representative of three independent experiments. *P<0.05, ** P< 0.01, ***P<0.001.
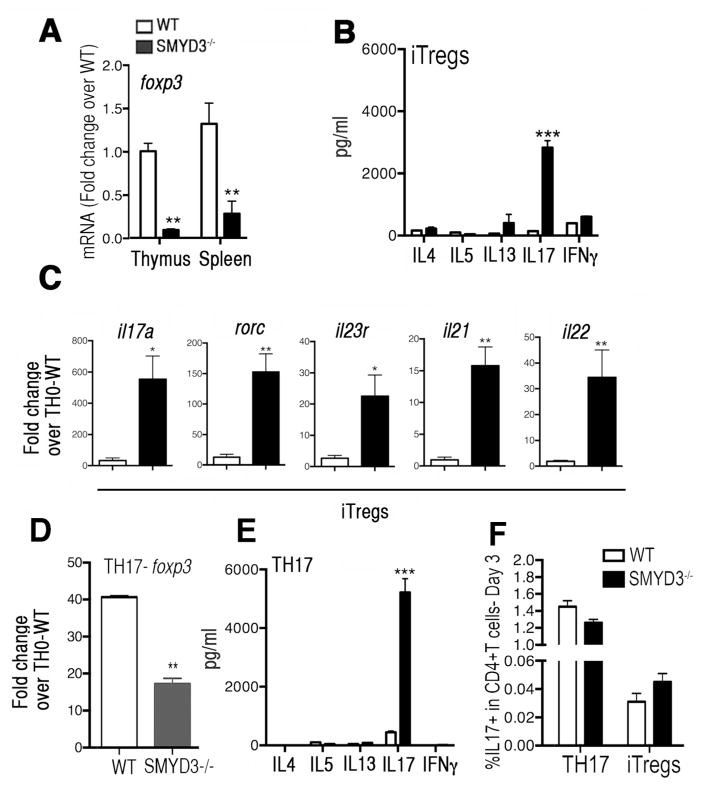
SMYD3 depleted CD4+T cells are poised to produce IL-17
Given that the overall T cell proportions between SMYD3−/− and WT mice were similar (Supplementary Fig. 3B), we explored the phenotypic consequences of SMYD3 ablation and Foxp3 inhibition in iTreg developing cells. CD4+T cells were isolated from WT and SMYD3−/− mice and show that the baseline levels of Foxp3 expression was significantly decreased in CD4+T cells derived from SMYD3−/− mice (Fig. 6A). Moreover, because Foxp3 protein prevents iTreg cell deviation to a effector cell lineage20, we determined if iTreg skewed SMYD3−/− CD4+T cells would exhibit differences in their production of proinflammatory cytokines. Indeed, the production of IL-17 was enhanced in the supernatants derived from iTreg skewed SMYD3−/− T cell cultures compared to WT, suggesting that SMYD3 depletion and Foxp3 inhibition may preferentially permit IL-17 production by CD4+T cells (Fig. 6B). To determine the transcriptional level of other TH17-associated genes, mRNA from SMYD3−/− and WT iTreg skewed cells was assessed. Our results show that besides il17a, SMYD3 abrogation also led to enhancement of rorc, il23r, il21, and il22 transcripts in iTreg skewing cells (Fig. 6C). Furthermore, since TH17 skewed cultures also utilize TGFβ and up-regulate foxp3 gene expression our studies examined those cultures. Our data demonstrate that even in TH17 cultures SMYD3 appears to regulate foxp3 expression (Fig. 6D) and the generation of IL-17 associated phenotype (Fig. 6E). To investigate whether SMYD3 was impacting the TH17/iTreg lineage decision or influencing cell function WT and SMYD3−/− naive CD4+T cells were differentiated into iTregs and TH17 and intracellular IL-17 was measured by flow cytometry to access whether the increase in IL-17 production observed in the SMYD3-deficient cells was due to increased IL-17 production/cell and/or an increase in the frequency of IL-17-producing cells. Our results show similar frequency of IL-17 producing cells in either iTregs or TH17 cell cultures, suggesting that the increased IL-17 production observed in the supernatants of SMYD3-deficient cells represents increased IL-17/cell as opposed to more cells differentiating. Collectively, these data suggest that SMYD3 is important for the regulation of Foxp3 leading to the subsequent regulation of IL-17 production by CD4+T cells.
Figure 6. SMYD3 depleted CD4+T cells are poised to produce the proinflammatory cytokine IL-17.
(A) Expression levels of foxp3 mRNA in CD4+T cells derived from thymus and spleens of WT and SMYD3−/− mice. (B) Naïve CD4+T cells derived from WT and SMYD3−/− were skewed into iTreg for 3 days and analyzed for cytokines. (C) WT and SMYD3−/− derived iTreg cells were examined for il17a, rorc, il23r, il21 and il22 mRNA by q-RT-PCR. Naïve CD4+T cells derived from WT and SMYD3−/− were skewed into TH17 for 3 days and analyzed for foxp3 mRNA expression (D) and cytokine production (E). (F) IL-17 expression was accessed by flow cytometry in WT and SMYD3−/− differentiated TH17 and iTreg cells. Data show means ± SEM of triplicate wells and are representative of three independent experiments performed with n=3 per group. *P<0.05, **P<0.01, ***P<0.001.
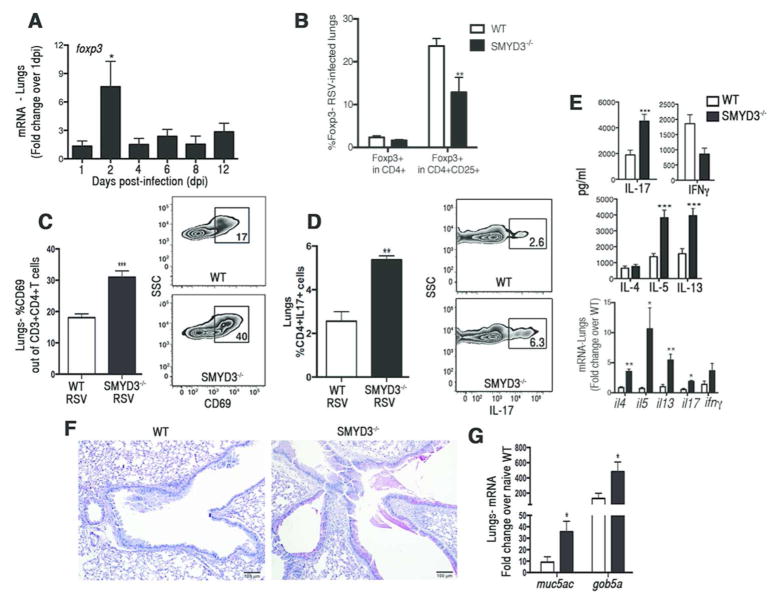
SMYD3 deficiency leads to exacerbation of RSV-induced pathology
Given that Treg cells contribute to the control of immune responses against respiratory pathogens, and previous studies related IL-17 with the exacerbation of pulmonary tissue damage and mucus production 21, 22, we examined the effect of SMYD3 deficiency in lung pathology using a mouse model of respiratory syncytial virus (RSV) infection. RSV infected animals showed up-regulation of foxp3 expression at 2 days post-infection (dpi) (Fig. 7A), as previously described 23. SMYD3−/− and WT mice infected with RSV showed that the frequency of CD4+CD25+Foxp3+ Treg cells found in SMYD3−/− mice was significantly lower compared to WT mice even though the total number of Foxp3+ CD4 T cells were not reduced (Fig. 7B). Infiltration of lymphocytic populations in the lungs showed that activated CD4+CD69+T cells (Fig. 7C) and percentage of TH17 cells (CD4+IL17+) (Fig. 7D) infiltrating the lungs of SMYD3−/− mice was significantly higher compared to WT mice. Supernatants from single cell suspensions of lung-associated LNs isolated from RSV-infected SMYD3−/− mice re-stimulated ex-vivo with RSV showed enhanced production of IL-17, as well as IL-5 and IL-13 (Fig. 7E), cytokines known to contribute to lung pathogenesis and airway mucus production during RSV infection 21, 24, 25. Cytokine expression studied in lung homogenates of RSV-infected SMYD3−/− mice also showed enhanced cytokine responses similar to the restimulated lymph nodes (Fig. 7E). Due to the effect of SMYD3 ablation in the expression of TH2-associated cytokines in the lungs of RSV infected mice additional experiments performed in vitro explored the effect of SMYD3 in TH2 differentiation. Results showed that SMYD3 depletion leads to enhancement in the production of IL-4 by TH2 differentiating cells (data not shown), suggesting that this molecule may also regulate TH2-associated cytokine production. Histopathological analysis of PAS-stained lung sections showed enhanced goblet cells hyperplasia and increased mucus secretion in the large airways of SMYD3−/− mice compared to WT (Fig. 7F) as well as increased expression of the mucus-associated genes muc5ac and gob5a (Fig. 7G). Thus, during virus-induced respiratory infection, SMYD3 deficiency led to decreased Treg cell infiltration and dysregulated immune responses with increased IL-5, IL-13 and IL-17 production that exacerbated pathology in the lungs.
Figure 7. SMYD3 is required to mount a protective immune response against RSV-infection.
(A) C57BL/6 wild type mice were infected RSV and lungs were assessed for foxp3 expression at 1, 2, 4, 6, 8, and 12 (n=3) days post-infection (dpi). WT (n=4) and SMYD3−/− (n=4) were infected with RSV and assessed at 8 dpi. Percentage of Foxp3+ in gated CD3+CD4+CD25+T cells (B), percentage of activated CD69+ in gated CD3+CD4+T cells (C), and percentage of CD4+IL17+ TH17 cells in gated CD3+T cells (D). (E) Lungs from RSV-infected WT and SMYD3−/− mice were homogenized, mRNA was extracted and expression of cytokine was assessed by q-RT-PCR. (F) PAS staining of lung sections of WT (left panel) and SMYD3−/− mice (right panel) infected with RSV at 8 dpi. (G) muc5ac and gob5a mRNA in the lungs of RSV-infected WT and SMYD3−/− mice. Data show means ± SEM of triplicate wells and are representative of three independent experiments. *P<0.05, **P<0.01, ***P<0.001.
DISCUSSION
The regulation of the immune response by Treg cells has been examined in nearly every disease response scenario and found to be central in controlling the outcome of immunity and disease severity while demonstrating a tumor-promoting role in cancer 12, 14, 26, 27. The central determinant of Treg cell development is Foxp3 expression that is constitutively expressed in thymus-derived nTreg cells and transcriptionally regulated by NFAT, and AP1 in iTreg cells 13, 28–30. Also important in the generation of iTreg cells is the accompanying activation of the TGFβ/Smad3 signaling pathway 15 that correlates with alteration of the CNS-1 element at the foxp3 locus, and that has a functional effect on Foxp3 expression 16–18. It has been reported that Treg cells can lose their Foxp3 expression in an inflammatory environment and acquire the ability to produce proinflammatory cytokines 20, 31. Elucidating distinct mechanisms that impact Foxp3 expression and defining pathways that can contribute to the stability of Treg cell lineage remain topics of ongoing research. The differentiation of T cells is linked to epigenetic changes in chromatin structure 31. Previous studies examining T cell regulation have primarily defined key components that regulate H3K4 (an activating mark) and H3K27 (an inhibitory mark) trimethylation 32–34. However, the specific epigenetic enzymes that control gene expression in iTreg cells have not been elucidated. In the present studies a focus on H3K4 regulation was pursued due to the identification of SMYD3 expression early during the iTreg skewing process. The maintained expression of SMYD3 was dependent upon sustained stimulation with TGFβ that corresponded to the expression of Foxp3 and SMYD3 when the cytokine stimulus was removed.
Importantly, lower expression of SMYD3 was observed in freshly isolated nTreg cells and along with data from siRNA knockdown that indicate no alteration in Foxp3 suggest that SMYD3 is not critical in nTreg cells. These data also correlate with previous studies indicating greater stability of nTreg cell commitment to the Foxp3+ phenotype, while iTreg cells may have greater plasticity to be redirected toward other phenotypes and necessitate the presence of TGFβ for maintenance of SMYD3 and stability of Foxp3 31, 35, 36. Recent studies indicate that nTreg cells are more potent in vitro in T cell proliferation assays compared to iTreg cells, and further, that nTreg cells were more stable in vivo transfer studies allowing protection from inflammatory bowel disease (IBD) in Rag2−/− mice 37. The latter study further demonstrated that nTreg cells also had a characteristic CpG hypomethylation pattern that began in the thymus upon T cell receptor ligation by self-ligands attributing to their greater stability. Interestingly, a study conducted with CNS elements and their role in the foxp3 gene showed that CNS-2 controls the maintenance of Foxp3 expression in mature Treg cells 16. In the present study no alteration in the CNS2 region of the foxp3 promoter was observed when SMYD3 was depleted. Instead, the regulation of Foxp3 by SMYD3 appears to be associated with the foxp3 promoter and the TGFβ/Smad3 response element CNS1, sites described to be important for iTreg cell conversion16. Importantly, although Foxp3 was significantly inhibited, its expression was not completely abolished upon SMYD3 ablation suggesting that epigenetic changes mediated by SMYD3 enhance foxp3 transcription in iTregs but is not the only element that regulates foxp3. This latter observation is consistent with the function of chromatin modifying enzymes that regulate gene accessibility and allow enhanced access to transcription factors such as Smad3.
The data from the present study also outline the requirement of Smad3 activation for the expression and function of SMYD3 and consequent Foxp3 regulation, clearly implicating SMYD3 as necessary for iTreg development linked to TGFβ activation. We confirmed the importance of TGFβ-mediated signaling for SMYD3 expression using both Smad3 inhibition and knockdown. TGFβ induces Foxp3 expression in T cells and causes TH- to iTreg-cell conversion and subsequent expansion through Smad signaling 38. While these data do not examine other critical gene targets of SMYD3 in Treg cell development, it does confirm that Foxp3 is a transcriptional target of SMYD3. The data show that SMYD3 ablation inhibits H3K4me3 in the CNS-1 and promoter region of the foxp3 locus. CNS-1 has been identified as a TGFβ-sensitive element that contains binding sites for NFAT and Smad3 16, 18. Our results suggest that SMYD3 is recruited to CNS-1, trimethylates H3K4, making the foxp3 locus poised for transcription. The foxp3 promoter region containing NFAT-AP1 response elements, was also affected upon SMYD3 ablation, suggesting that TGFβ/SMYD3 induced chromatin remodeling of CNS-1 might affect the accessibility of the upstream foxp3 promoter. Other epigenetic modifications have been associated with repression of Foxp3 in conventional T cells that is at least partially dependent upon PIAS1, a SUMO E3 ligase, that binds to the foxp3 promoter and recruits DNA methyltransferases that keep the foxp3 promoter in a methylated and silent state (heterochromatin) 39. Removal of PIAS1 in knockout experiments verified its function for regulation of Foxp3 and generation of nTreg cells that are derived in the thymus 39. We have not examined whether reduced PIAS1 expression or function is coordinated with the expression of SMYD3.
Abrogation of Foxp3 or its diminished expression in iTreg cells leads to decreased suppressive capability and to the acquisition of proinflammatory properties including production of cytokines including IL-4, IL-17, and IFN-γ 20. In this study SMYD3 inhibition and Foxp3 downregulation in CD4+T cells led to increased production of IL-17. An intimate link between Treg and TH17 cell programs has been identified previously 40, 41. The increased production of IL17 by SMYD3−/− CD4+T cells under iTreg skewing conditions was also demonstrated by the expression of the TH17-associated genes il17a, rorc, il23r, il21 and il22. Foxp3 expression reduces TH17 differentiation through inhibition of RORγt function 41. Given that several transcription factors have been described to be associated with the transcription regulation of Foxp342, T-bet and Gata3 were also investigated and not affected by SMYD3 depletion (data not shown). Together, these results suggest that the loss of Foxp3 by SMYD3 depleted CD4+T cells reverse Foxp3-mediated inhibition of RORγt, thereby promoting IL-17 production.
iTreg cells are crucial players in immune regulation of the lungs during pulmonary disease 22, 23, 43, 44. During RSV infection, Treg cells suppress antigen-specific T effector cell responses, contribute to the control of viral replication, and minimize tissue damage 22, 44. Accordingly, RSV-infected SMYD3 deficient mice had lower levels of Treg cells in the lungs and LNs accompanied by increased T cell responses with greater lung and airway inflammation. Our results also demonstrated that in addition to decreased Treg cells, RSV-infected SMYD3 deficient mice fail to control the T cell cytokine (IL-5, IL-13, and IL-17) and RSV-associated pathogenic responses, such as airway goblet cell hyperplasia. IL-17 can increase mucus production with consequent exacerbation of tissue damage. These data in virus infection demonstrate that without proper iTreg cell development in SMYD3−/− mice a dysregulated immune environment develops.
In summary, our studies demonstrate a novel role for SMYD3 as a TGFβ/SMAD3-inducible epigenetic enzyme that plays a central role for enhancing Foxp3 expression during immune responses and regulating proinflammatory IL-17 production. Enhancement of lung pathology during pulmonary infection triggered by RSV in mice deficient in SMYD3 lends further support to this notion. Our observations suggest that TGFβ signaling is required for SMYD3 activation, subsequent H3K4 methylation in the foxp3 loci, and maintenance of Foxp3 expression levels in iTreg cells. Overall, these data highlight SMYD3 as a potential target for manipulating Foxp3 and iTreg cell response during infection, autoimmunity and cancer.
METHODS
1. Mice
Female wild type (WT) C57BL/6 mice (6-10 weeks old) were purchased from Taconic Company. Breeding pairs of Foxp3 eGFP reporter mice (Foxp3eGFP, B6.Cg-Foxp3TM2Tch/J) were purchased from The Jackson Laboratory and bred in-house. CD4-Cre mice were obtained from Taconic. SMYD3tm1a (KOMP) Wtsi mice in a C57BL/6 background were derived from KOMP Repository at UC Davis. The allele contains loxP and FRT sites so Cre and FLP deletion were used to create a SMYD3 conditional knockout resulting in a null allele in CD4+ cells (SMYD3fl/flCD4Cre+). All mice were housed in the University Laboratory Animal Facility at the University of Michigan Medical School. All animal experiments were conducted according to animal protocols approved by The Animal Use Committee at the University of Michigan.
2. Extracellular and intracellular staining for flow cytometry
Cells were stained using antibodies against surface markers CD3 (145-2C11), CD4 (RM4-5) and CD25 (7D4) (all from eBioscience or BD Biosciences), permeabilized, fixed and labeled for intracellular Foxp3 (FJK-16s), IL-4 (11B11), IFNγ (XMG1.2) and IL-17 (eBio17B7). Samples were acquired on LSRII flow cytometer with use of FACSDiva software (BD Biosciences). Data were analyzed using FlowJo software (TreeStar).
3. In vitro T cell conversion
Naïve CD4+ T cells were enriched from spleens of WT, SMYD3−/− or Foxp3eGFP mice as described 18. Cells stimulated for 48 hours to 5 days using plate-bound anti-CD3 (2.5 μg/ml; eBioscience), soluble anti-CD28 (3 μg/ml; eBioscience) plus TGFβ (2 to 20 ng/ml) and rIL-2 (10 ng/ml; all from R&D Systems). Human naïve CD4+T cells were isolated as above and converted into iTreg cells using 5 ng/ml of TGFβ. For T helper (H) cell differentiation, naïve CD4+T cells were skewed towards TH17 using IL-6 (1 ng/ml) and TGFβ (2 ng/ml) plus anti-IFNγ (10 μg/ml), anti-IL12 (10 μg/ml), anti-IL4 (10 μg/ml), and anti-CD28 (3 μg/ml).
4. Quantitative RT-PCR
Gene expression levels were determined as described 45. For PCRarrays, RNA was converted to cDNA and analyzed on a mouse epigenetic chromatin modification enzymes PCR array platform (PAMM-085, SA Biosciences) following the manufacturer’s protocol. PCR arrays results were analyzed using the RT2 Profiler PCR Array Data Analysis (SA Biosciences).
5. Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as described 3 using anti-Smad3 Ab (C67H9; Millipore), anti-SMYD3 Ab (ab16027; Abcam), anti-H3K4me3 Ab (ab8580; Abcam) and normal rabbit immunoglobulin G (IgG) as control. Regions of the Foxp3 promoter, Foxp3 CNS1, CNS2, CNS3 were amplified by SYBER Green qPCR (Applied Biosciences) and quantified in triplicates or quadruplicates with the percent of input method. The following primers were used: SMYD3 (forward 5′ccccgttgcactcagacccc3′ and reverse 5′ggctgtccagaccagaagctca3′) Foxp3 promoter (forward 5′gggcactcagcacaaacatgatg3′ and reverse 5′gaggcttccttctgcccaaac3′), Foxp3 CNS1 (forward 5′gttttgtgttttaagtcttttgcacttg3′ and reverse 5′cagtaaatggaaaaaatgaagccata3′), Foxp3 CNS2 (forward 5′gttgccgatgaagcccaat3′ and reverse 5′atctgggccctgttgtcaca3′), and Foxp3 CNS3 (forward 5′aatgaatgagacacagaactattaagatga3′ and reverse 5′cagacggtgccaccatgac3′).
6. Cytokine Assay
Cytokines from supernatants of in vitro culture assays were measured using BioPlex System and Suspension Array Technology (Bio-Rad).
7. Western Blot
Western blots were performed using antibodies anti-SMYD3 (Abcam), anti-H3K4me3 (Millipore), anti-SMAD3 (Abcam), anti-Foxp3 (eBioscience) or β-actin (Sigma) as control primary antibody, the membrane was counterstained with horseradish peroxidase-conjugated rabbit or mouse IgG antibody and visualized with enhanced chemiluminescence detection reagents (GE Healthcare).
8. Short-interfering RNA (siRNA) assay
Naïve CD4+T cells were transfected with 2uM of Smad3-specific, SMYD3-specific or nontargeting control siRNAs (Dharmacon) using mouse T cell nucleofector® kit (Lonza Walkersville). Transfected cells were kept in mouse T cells nucleofector medium for 18 to 24 hours at 37°C, 5%CO2, harvested, and stimulated under TH0 or Treg cell skewing conditions.
9. Suppression assay
Splenic CD4+CD25HInTreg cells derived form naïve WT and SMYD3−/− mice were stained as before and FACS sorted. Splenic naïve CD4+T cells were isolated from WT mice, labeled with 2.5 μM CFSE (Invitrogen) and co-cultured with nTregs derived from WT or SMYD3−/− in complete medium with plate-bound anti-CD3 mAb (2.5 μg/ml) and soluble anti-CD28 (3 μg/ml) (eBiocience) for 3 days. Suppression was analyzed by flow cytometry.
10. Cell proliferation assay
Naïve CD4+T cells were labeled with CFSE and incubated under iTreg skewing conditions for 3 days. Proliferation was analyzed by flow cytometry.
11. Confocal Analysis
Cell suspension from naïve and iTreg skewing cells stained as described 45 using mouse anti-CD4 (BD-Pharmigen), rabbit anti-Smyd3 (Abcam), goat anti-rabbit Alexa Fluor 568, and anti-mouse Alexa Fluor 488 (Invitrogen). The stained tissue sections were analyzed using a Nikon A1 confocal microscope system (Nikon Instruments).
12. RSV infection and primary cell isolation from infected mice
The RSV A strain (line 19) was derived from a clinical isolate at the University of Michigan and propagated in HEp-2 cells (ATCC). Line 19 elicits disease in mice comparable to severe RSV infection in humans, including significant airway hyperresponsiveness and mucus overproduction 46. LN cells were cultured (5×105/well in a 96 well plate) and re-stimulated with RSV (MOI = 0.5) for 48 hours for RNA and cytokine measurements.
13. Histology
Organs were perfused with 4% formaldehyde for fixation and embedded in paraffin using standard histological techniques. Lung sections were stained with periodic acid-Schiff (PAS) to detect mucus production. All other tissues were stained with hematoxilin and eosin. Tissue sections were evaluated using light microscopy by an independent blinded observer.
14. Statistical Analysis
Data were analyzed using GraphPad prism software Version 6 (GraphPad). Measures between two groups were performed with a Student’s t-test (two-tailed). Groups of three or more were analyzed by one-way ANOVA followed by with Tukey’s post-testing. P < 0.05 was considered significant. * P < 0.05, ** P < 0.01, *** P < 0.001.
Supplementary Material
Acknowledgments
The authors thank Susan Morris, Andrew Rasky, Ron Allen, Ana L. Coelho, Amrita D. Joshi, Lisa Riggs Johnson, and Pamela Lincoln for technical assistance; Robin Kunkel for art work; and Dr. Judith Connett for critical reading of the manuscript. This work was supported by the NIH grants: AI073876 (NWL) and T32 AI007413 (DDAN).
Footnotes
DISCLOSURES
There is no conflict of interest to declare.
References
- 1.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Ehrensberger AH, Svejstrup JQ. Reprogramming chromatin. Crit Rev Biochem Mol Biol. 2012;47:464–482. doi: 10.3109/10409238.2012.697125. [DOI] [PubMed] [Google Scholar]
- 3.Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood. 2008;111:1797–1804. doi: 10.1182/blood-2007-08-106443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nature reviews Immunology. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 5.Ruden DM, Lu X. Hsp90 affecting chromatin remodeling might explain transgenerational epigenetic inheritance in Drosophila. Curr Genomics. 2008;9:500–508. doi: 10.2174/138920208786241207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- 7.Tsuge M, Hamamoto R, Silva FP, Ohnishi Y, Chayama K, Kamatani N, et al. A variable number of tandem repeats polymorphism in an E2F-1 binding element in the 5′ flanking region of SMYD3 is a risk factor for human cancers. Nat Genet. 2005;37:1104–1107. doi: 10.1038/ng1638. [DOI] [PubMed] [Google Scholar]
- 8.Frank B, Hemminki K, Wappenschmidt B, Klaes R, Meindl A, Schmutzler RK, et al. Variable number of tandem repeats polymorphism in the SMYD3 promoter region and the risk of familial breast cancer. Int J Cancer. 2006;118:2917–2918. doi: 10.1002/ijc.21696. [DOI] [PubMed] [Google Scholar]
- 9.Cock-Rada AM, Medjkane S, Janski N, Yousfi N, Perichon M, Chaussepied M, et al. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res. 2012;72:810–820. doi: 10.1158/0008-5472.CAN-11-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locksley RM. Nine lives: plasticity among T helper cell subsets. J Exp Med. 2009;206:1643–1646. doi: 10.1084/jem.20091442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Shakya A, Guo X, Zhang H, Tantin D, Jensen PE, et al. Constitutive nuclear localization of NFAT in Foxp3+ regulatory T cells independent of calcineurin activity. J Immunol. 2012;188:4268–4277. doi: 10.4049/jimmunol.1102376. [DOI] [PubMed] [Google Scholar]
- 14.Shevach EM, DiPaolo RA, Andersson J, Zhao DM, Stephens GL, Thornton AM. The lifestyle of naturally occurring CD4+ CD25+ Foxp3+ regulatory T cells. Immunol Rev. 2006;212:60–73. doi: 10.1111/j.0105-2896.2006.00415.x. [DOI] [PubMed] [Google Scholar]
- 15.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nature immunology. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 18.van der Veeken J, Arvey A, Rudensky A. Transcriptional Control of Regulatory T-Cell Differentiation. Cold Spring Harbor symposia on quantitative biology. 2013;78:215–222. doi: 10.1101/sqb.2013.78.020289. [DOI] [PubMed] [Google Scholar]
- 19.Kitani A, Fuss I, Nakamura K, Kumaki F, Usui T, Strober W. Transforming growth factor (TGF)-beta1-producing regulatory T cells induce Smad-mediated interleukin 10 secretion that facilitates coordinated immunoregulatory activity and amelioration of TGF-beta1-mediated fibrosis. J Exp Med. 2003;198:1179–1188. doi: 10.1084/jem.20030917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nature immunology. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee S, Lindell DM, Berlin AA, Morris SB, Shanley TP, Hershenson MB, et al. IL-17-induced pulmonary pathogenesis during respiratory viral infection and exacerbation of allergic disease. The American journal of pathology. 2011;179:248–258. doi: 10.1016/j.ajpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lund JM, Hsing L, Pham TT, Rudensky AY. Coordination of early protective immunity to viral infection by regulatory T cells. Science. 2008;320:1220–1224. doi: 10.1126/science.1155209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fulton RB, Meyerholz DK, Varga SM. Foxp3+ CD4 regulatory T cells limit pulmonary immunopathology by modulating the CD8 T cell response during respiratory syncytial virus infection. J Immunol. 2010;185:2382–2392. doi: 10.4049/jimmunol.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotz MT, Peebles RS., Jr Mechanisms of respiratory syncytial virus modulation of airway immune responses. Current allergy and asthma reports. 2012;12:380–387. doi: 10.1007/s11882-012-0278-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durant LR, Makris S, Voorburg CM, Loebbermann J, Johansson C, Openshaw PJ. Regulatory T cells prevent Th2 immune responses and pulmonary eosinophilia during respiratory syncytial virus infection in mice. Journal of virology. 2013;87:10946–10954. doi: 10.1128/JVI.01295-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziegler SF, Buckner JH. Influence of FOXP3 on CD4+CD25+ regulatory T cells. Expert Rev Clin Immunol. 2006;2:639–647. doi: 10.1586/1744666X.2.4.639. [DOI] [PubMed] [Google Scholar]
- 27.Bos PD, Plitas G, Rudra D, Lee SY, Rudensky AY. Transient regulatory T cell ablation deters oncogene-driven breast cancer and enhances radiotherapy. J Exp Med. 2013;210:2435–2466. doi: 10.1084/jem.20130762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 29.Lee SM, Gao B, Fang D. FoxP3 maintains Treg unresponsiveness by selectively inhibiting the promoter DNA-binding activity of AP-1. Blood. 2008;111:3599–3606. doi: 10.1182/blood-2007-09-115014. [DOI] [PubMed] [Google Scholar]
- 30.Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Bailey-Bucktrout S, Jeker LT, Bluestone JA. Plasticity of CD4(+) FoxP3(+) T cells. Curr Opin Immunol. 2009;21:281–285. doi: 10.1016/j.coi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114:3727–3735. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, et al. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24:611–622. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X, Kong N, Wang J, Fan H, Zou H, Horwitz D, et al. Cutting edge: all-trans retinoic acid sustains the stability and function of natural regulatory T cells in an inflammatory milieu. J Immunol. 2010;185:2675–2679. doi: 10.4049/jimmunol.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murai M, Krause P, Cheroutre H, Kronenberg M. Regulatory T-cell stability and plasticity in mucosal and systemic immune systems. Mucosal Immunol. 2010;3:443–449. doi: 10.1038/mi.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, Neurath MF. Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25− T cells through Foxp3 induction and down-regulation of Smad7. J Immunol. 2004;172:5149–5153. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- 39.Liu B, Tahk S, Yee KM, Fan G, Shuai K. The ligase PIAS1 restricts natural regulatory T cell differentiation by epigenetic repression. Science. 2010;330:521–525. doi: 10.1126/science.1193787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 41.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nature immunology. 2012;13:1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 44.Loebbermann J, Thornton H, Durant L, Sparwasser T, Webster KE, Sprent J, et al. Regulatory T cells expressing granzyme B play a critical role in controlling lung inflammation during acute viral infection. Mucosal Immunol. 2012;5:161–172. doi: 10.1038/mi.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kittan NA, Allen RM, Dhaliwal A, Cavassani KA, Schaller M, Gallagher KA, et al. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PloS one. 2013;8:e78045. doi: 10.1371/journal.pone.0078045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huber S, Gagliani N, Esplugues E, O’Connor W, Jr, Huber FJ, Chaudhry A, et al. Th17 cells express interleukin-10 receptor and are controlled by Foxp3(−) and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 2011;34:554–565. doi: 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.