Abstract
Background
RNA-directed regulation of epigenetic processes has recently emerged as an important feature of mammalian differentiation and development. Perturbation of this regulatory system in the brain may contribute to the development of neuropsychiatric disorders.
Methods
RNA sequencing (RNA-seq) was used to identify changes in the experience-dependent expression of long non-coding RNAs (lncRNAs) within the medial prefrontal cortex (mPFC) of adult mice. Transcripts were validated by real-time quantitative PCR and a candidate lncRNA, Gomafu, was selected for further investigation. The functional role of this schizophrenia-related lncRNA was explored in vivo by antisense oligonucleotide-mediated gene knockdown in the mPFC, followed by behavioral training and assessment of fear-related anxiety. LncRNA-directed epigenetic regulation of gene expression was investigated by chromatin and RNA immunoprecipitation assays.
Results
RNA-seq analysis revealed changes in the expression of a significant number of genes related to neural plasticity and stress, as well as the dynamic regulation of lncRNAs. In particular, we detected a significant down-regulation of Gomafu lncRNA. Our results revealed that Gomafu plays a role in mediating anxiety-like behavior, and suggest that this may occur through an interaction with a key member of the polycomb repressive complex 1, BMI1, which regulates the expression of the schizophrenia-related gene beta crystallin (Crybb1). We also demonstrated a novel role for Crybb1 in mediating fear-induced anxiety-like behavior.
Conclusion
Experience-dependent expression of lncRNAs plays an important role in the epigenetic regulation of adaptive behavior, and the perturbation of Gomafu may be related to anxiety and the development of neuropsychiatric disorders.
Keywords: non-coding RNA, Gomafu, anxiety, epigenetics, behavior, Crybb1
Introduction
Once considered vestiges of our evolutionary history associated with “junk” DNA, non-coding transcriptional activation, and post-transcriptional gene silencing (1-5). Advances in RNA sequencing (RNA-seq) technology have resulted in the discovery of a large number of long ncRNAs (lncRNAs) (6-11), many of which show features of functionality (12-14). There are at least 50,000 genes specifying lncRNAs scattered throughout the human genome, with many expressed in a highly cell type- and developmental stage-specific manner (15-20). Moreover, a significant number of brain-enriched or brain-specific lncRNAs are found adjacent to genes encoding transcriptional regulators and key drivers of neural development (21), including those involved in the regulation of stem cell pluripotency, neuronal differentiation, and synaptogenesis (16, 18, 22-24).
In agreement with these findings, lncRNAs have been implicated in neurodevelopmental disorders such as Rett syndrome (25), autism (26, 27), schizophrenia (SZ) (28, 29) and Fragile X syndrome (30). Screening of lncRNA activity has also shown links between lncRNA expression and drug abuse (31, 32), suicidal behavior (33) and potentially anxiety disorders. For instance, it has been reported that experimental knockdown BC1 leads to increased anxiety-like behavior in mice (34, 35).
Recent evidence indicates that the expression of lncRNAs can be altered in an activity-dependent manner (29, 36). LncRNAs have been found to be co-expressed with activity-dependent genes such as C-fos, Arc, Nr4a2 and Bdnf, suggesting a coordinated network of coding and non-coding gene expression associated with neuronal plasticity (29, 36, 37).
Despite these correlative links, little is known about the expression and function of brain lncRNAs, nor the mechanisms by which these transcripts influence protein-coding gene expression within the context of neuropsychiatric disease. Here we combined high-throughput RNA-seq with molecular and behavioral approaches to identify changes in the expression of lncRNAs. We also determined whether these changes contribute to the epigenetic regulation of gene expression underlying the development of anxiety disorders.
Materials and Methods
Animals
Naïve 9-week-old C57BL/6 male mice were housed individually in sections of divided cages, with free access to food and water under a 12h light/dark cycle in a humidity- and temperature-controlled vivarium. Behavioral tests were conducted during the light cycle, and all procedures were performed with approval from the Animal Ethics Committee of The University of Queensland.
Fear conditioning
For tissue collection and sequencing library preparation, 3 groups (n=8 per group) of mice were used: a naïve, age-matched, home-cage control group, a fear-conditioned group that received six pairings of a 2 min 80 dB white noise conditioned stimulus (CS) that co-terminated with a 1 s foot-shock at 0.7 mA as the unconditioned stimulus (US, inter-trial interval of 2 min), and a context only group that was exposed to the CS but not the US. Animals were sacrificed 90 min post-training, followed by medial prefrontal cortex (mPFC) dissection and nuclear-enriched RNA extraction. To determine the effect of Gomafu knockdown on fear conditioning, animals were infused with 600nM of a Gomafu antisense oligonucleotide (ASO), directly into the mPFC, whereas the control group received an infusion of a scrambled control. 3h after infusion, mice were trained using a mild fear acquisition protocol in order to avoid a non-specific response (3 CS-US pairing with foot shocks of 0.4mA), followed by a 2CS test for memory recall in the same context 24h later.
Measures of anxiety
Anxiety tests for the knockdown and control groups were performed 3h after ASO infusion, and 2 weeks after lentiviral infusion respectively, and involved the use of mild stressors (38). This included a 10min exposure to a 27x27x20.3cm open field chamber with 200 lux of light intensity, which was followed by an interval of 30min. Mice were then introduced for 10min into an elevated plus maze (under bright light at 900 lux in the open arms and 200 lux in the closed arms), and the time spent in the open arms was recorded. Following a second interval of 10min mice were once more placed in the open field for 10min, at which point time in the center was automatically determined in seconds, while ambulatory time, distance traveled and self-grooming time were calculated as a ratio between zone results.
Stereotaxic surgery and cannula implantation
Mice were anesthetized with ketamine (100 mg/kg) and xylazine (10mg/kg). Surgeries were performed under stereotaxic guidance and cannulas were implanted bilaterally within the prelimbic region of the PFC (PLPFC) at +1.8mm anterioposterior and −2mm dorsoventral to Bregma. Mice were allowed to recover for a minimum of 7 days before experiments. Animals were transcardially perfused post-training with cold phosphate buffered saline (PBS) (Lonza, Walkersville, MD, USA), followed by 4% paraformaldehyde in PBS. Brains were dissected out, sectioned and the localization of the ASO and shRNA infusion was determined by fluorescence microscopy.
Nuclear enrichment, RNA extraction and reverse transcription
mPFC derived from fear-conditioned mice was homogenized in nuclear buffer (detailed in Supplement 1) followed by RNA extraction using the RNAeasy Mini kit (Qiagen, Hilden, Germany). Reverse transcription and cDNA synthesis was performed following the Quantitech Reverse Transcription kit protocol (Qiagen, Hilden, Germany), with the exception of RNA eluted from RNA immunoprecipitation (RIP) assays, which was reverse transcribed using Super Script III First Strand Synthesis (Invitrogen, Carslbad, CA, USA). The assay was validated both in vitro and in vivo (Fig S6 A-B in Supplement 1).
RNA sequencing
1μg of nuclear-enriched RNA, with a minimum RNA Integrity (RIN) value of 8, from 6 samples per treatment was used to build a total RNA library using the Illumina TruSeq RNASample Preparation v2 protocol. Each animal was individually indexed, representing a single library per mouse, which was further multiplexed in 2 pools of 9 animals before loading into the Illumina flow cell. Samples with low read alignment were excluded, resulting in 5 animals for Naïve and Context groups and 4 mice for FC. The quality of the RNA library was verified on an Agilent DNA 1000 chip and run on the Bioanalyzer, while quantification was determined by qPCR, after which the samples were sequenced using the Illumina HiSeq v3 platform.
Bioinformatic analysis
Mapping of reads to the mouse genome (mm10) was performed using the Bowtie2 and TopHat 2.0.6 programs (39). Cufflinks 2.0.2 (BETA) algorithms were then implemented for assembly of RNA sequencing reads into transcripts and analysis of differential levels of transcript expression among treatment groups as previously described (40, 41). For the purpose of this investigation we used a cutoff of p<0.03.
Gene knockdown
ASOs targeting Gomafu lncRNA were designed using the IDT Antisense design software targeting the splicing regulatory regions known as exonic splicing enhancers, as well as sequences with higher G and C content and low potential for RNA secondary structure formation. Primer sequences are detailed in Table S4 (Supplement 1). ShRNAs (Genecoepoa) targeting Crybb1 mRNA were packaged in-house using a 3rd generation lentiviral packaging system.
Primary neurons cell culture and electroporation
Cortical neuronal cells were dissociated from C57BL/6 mouse embryos at embryonic day 16 and plated onto poly-L-ornithine hydrochloride-coated plates. Cultures were grown in Neurobasal medium (GIBCO Life Technologies, NY, USA) containing 5% fetal bovine serum, 1% penicillin/streptomycin, 1% GlutaMAX, and 2% B27 supplement (GIBCO Life Technologies, NY, USA), and were maintained at 37°C with 5% CO2. Electroporation of ASOs was performed using the Nucleofector transfection system (Lonza, Basel, Switzerland) according to the manufacturer’s instructions. KCl treatments were performed at 50mM concentration.
Technical validation of RNA sequencing gene target selection
Gene expression was measured using the SYBR Green quantitative real-time PCR (qRT-PCR) detection method on a RotorGene 3000 (Qiagen, Hilden, Germany). The 2^-AACt method was applied to estimate differential levels of gene expression. ANOVA and Holm-Sidak’s multiple comparison tests were applied to establish gene expression differences among groups. Validation was performed with the original RNA used for RNA-seq.
Chromatin immunoprecipitation (ChIP)
Dissociated cortical neurons were subjected to electroporation with 200nM ASO for Gomafu knockdown followed by BMI1 pull-downs 3h post-transfection. For in vivo studies, tissues were dissected out and homogenized 1.5h after training. Tissue and cells were processed as previously described (42). Chromatin was immunoprecipitated using 4 μg ChIP-grade antibodies specific to BMI1 (1T21, Abcam, Cambridge, England, United Kingdom), SUZ12 (ab12073, Abcam, Cambridge, England, United Kingdom), and RING1B (D22F2, Cell Signaling, Danvers, Massachusetts, USA), H2AK119 (D2764, Cell Signaling, Danvers, Massachusetts, USA), mouse IgG (103533, Active Motif, Carslbad, CA, USA) and rabbit IgG (2295402, Millipore, Billerica, Massachusetts, USA). DNA-protein interactions were analyzed by qRT-PCR using primers specific to the binding motifs of the proteins being investigated.
RNA immunoprecipitation (RIP)
Naïve tissues were dissected out and homogenized followed by fixation and cross-linking. Immunoprecipitation with 4 μg of the antibody of interest was performed as previously described (22), and RNA was extracted using the Trizol method (Ambion, Carslbad, CA, USA), followed by DNase treatment using the TURBO DNA-free Ambion kit (Ambion, Carslbad, CA, USA).
Results
Long non-coding RNAs are dynamically expressed in the mPFC in response to behavioral experience
Fear-conditioned mice exhibited a robust increase in fear-related behavior at the end of training compared to context-only exposed mice (Fig. S1A in Supplement 1). Nuclear RNA was extracted from the mPFC of mice sacrificed 90 min after behavioral training, and transcriptome-wide profiles were obtained by RNA-seq. In total, we detected more than 400 loci that exhibited differential expression of protein-coding and non-coding RNAs (Fig. 1A). A complete list of differentially expressed transcripts identified by RNA-seq is provided in Table S1 in Supplement 2.
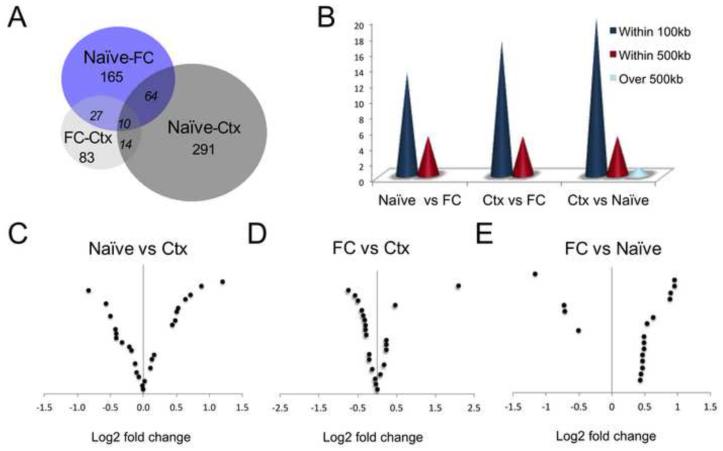
Figure 1. Transcriptome-wide analysis of nuclear RNA expression in the mouse prefrontal cortex reveals dynamic regulation of coding genes as well as lncRNAs.
(A) BioVenn graphic of loci detected by nuclear-enriched RNA-Seq with p-value (p) <0.03 corresponding to each pairwise comparison for naïve, context exposure (Ctx) and fear conditioning (FC) groups. (B) Distribution analysis of lncRNA loci revealing close proximity to coding genes. (C, D, E) Volcano plot showing log2 fold change magnitude of lncRNAs with p-value <0.03 in each pairwise analysis. Up-regulation of genes in context (D-E) and in naïve (F) animals is shown to the right side of the 0 in each figure.
Intergenic lncRNAs from 53 loci were identified as being significantly altered in either context-exposed or fear-conditioned mice, with the majority of these transcripts being found within 100kb of the nearest protein-coding gene, consistent with a potential cis-regulatory function (21, 43) (Fig. 1B-E; Table S2 in Supplement 1). RNA-seq analysis also revealed a cluster of modulated protein-coding genes, some of which have been implicated in behavioral regulation, in mediating the stress response, and in neuropsychiatric disorders (Table S3 in Supplement 1).
Experience-dependent alterations in lncRNA expression within the mPFC
In order to validate the RNA-seq findings, candidates were selected for downstream analyses. We first examined the expression of two protein-coding genes, Bdnf (exon IV) and Homer1, which are involved in neural plasticity and memory (Table S3 in Supplement 1; Fig. 2A and B). Bdnf has been reviewed as the master regulator of neuronal circuitries driving learning and synaptic plasticity (44). This gene has a complex structure that allows its differential epigenetic response to stimuli (45, 46); however, Bdnf exon IV has particularly been shown to participate in memory processes within the mammalian PFC (47). There was a significant increase in Bdnf exon IV and Homer1 mRNA expression in context-exposed mice, confirming previous studies demonstrating the activity-dependent nature of these genes in models of experience-dependent plasticity.
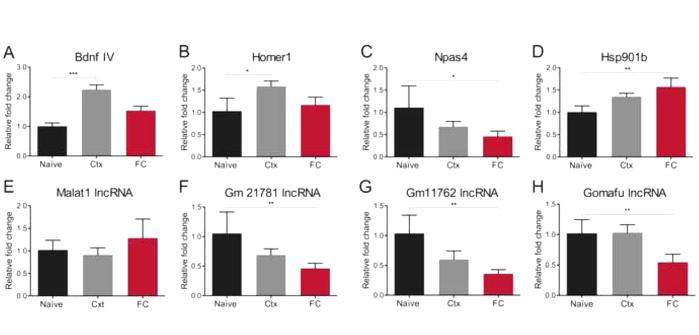
Figure 2. RT-qPCR validation of plasticity-related genes and non-coding RNA transcripts.
(A-D) Analyses of protein coding genes confirm up-regulation of Bdnf exon IV mRNA (naïve vs Ctx ***p<0.001, Kruskal Wallis test statistics 16.58), up-regulation of Homer1 (naïve vs Ctx, *p<0.05, ANOVA, F(2, 20)=5.119), down-regulation of Npas4 (naïve vs FC, *p<0.05, ANOVA, F (2, 20)=5.5), and up-regulation of Hsp901b (naïve vs FC, **p<0.01, Kruskal Wallis test statistics 9.459). (E-G) Analyses of lncRNAs demonstrate a non-significant slight up-regulation of Malat1, while Gm21781 lincRNA and Gm11762 lncRNA were down-regulated following fear conditioning relative to naïve mice (ANOVA, **p<0.01, F (2, 20) =7.308 and ANOVA, **p<0.01, F(2, 20)=5.892 respectively). (H) Specific down-regulation of Gomafu lncRNA in fear-conditioned mice (ANOVA, **p<0.01, F (2, 20)=5.953 naïve vs FC and Ctx vs FC). n=8 per group.
We further observed a significant decrease in the expression of the activity-dependent immediate early gene Npas4 (Fig. 2C). Although Npas4 expression is related to fear learning, a down-regulation of this gene is correlated with both exposure to stress and impairments in the formation of fear memory (48-51). Indeed, stressors can also lead to fear incubation or enhanced fear-potentiated startle, which may be related to anxiety rather than learning per se (52-54). In the experiments described here, we used a strong fear conditioning protocol (6CS-US pairings of a 0.7mA foot shock), which we expected could lead to an increase in stress reactivity and anxiety. In this paradigm, fear-conditioned mice showed high levels of freezing, not only during CS onset but also during the inter-trial interval (Fig. S1 A and B in Supplement 1).
Concordantly, we also observed an increase in the expression of the stress- and anxiety-related gene, Hsp901b, following fear conditioning (Fig. 2D). Although there was no effect of fear conditioning on the expression of the synaptogenesis-related lncRNA Malat1 (Fig. 2E), two intergenic lncRNAs of unknown function exhibited a significant, but non-specific, decrease in expression following behavioral training (Fig. 2F and G). The first of these, Gm21781, is upstream of a gene that encodes a DNA binding protein known as zinc finger and BTB domain containing 2 (Zbtb2), which has been shown to be a potent epigenetic regulator (55), and the second, Gm11762, is antisense to neuronal pentraxin 1 (Nptx1), which has been reported to enhance synaptogenesis and glutamate signalling through clustering of AMPA receptors (56). Finally, we observed a significant down-regulation of the neuropsychiatric disease-related lncRNA Gomafu in the mPFC, which occurred following fear conditioning but not after context exposure (Fig. 2H). Importantly, the down-regulation of Gomafu in fear-conditioned animals was also observed in a targeted RNA-seq experiment performed in these mice (Fig. S2A in Supplement 1) using a recently developed highly sensitive RNA capture protocol (57), thereby confirming the previously reported activity-dependent nature of the expression of this lncRNA (29). Together these data suggest that experience-induced regulation of lncRNAs may play a role in regulating neuronal plasticity and cognitive processes in the adult brain.
ASO-mediated knockdown of Gomafu within the mPFC induces anxiety-like behavior
In order to determine whether Gomafu regulates adaptive behavior, we employed an ASO-mediated knockdown approach. The efficacy of an ASO designed to specifically target Gomafu (Gomafu KD) was first tested in cultured primary cortical neurons at different time points (Fig. S3A and B in Supplement 1). The preliminary screening indicated an approximately 50% knockdown at a concentration of 1μM, when measured 3h post-administration (Fig. 3A), which decreased further by 6h post-infusion (Fig. S3A and B in Supplement 1).
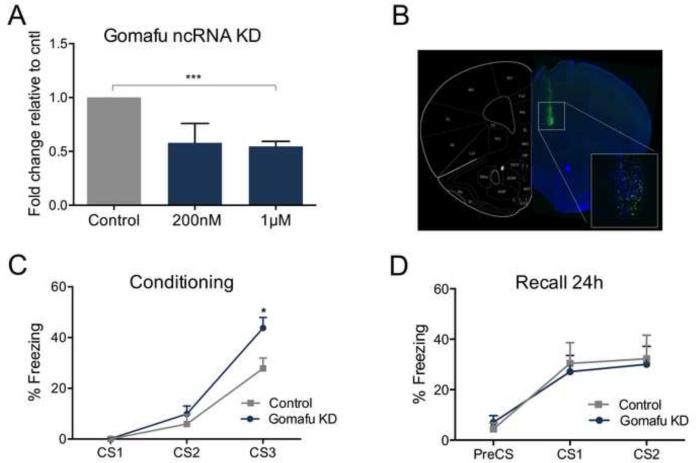
Figure 3. Knockdown of Gomafu lncRNA in the PLPFC enhances fear response during behavioral training.
(A) Knockdown level by Gomafu ASO was assessed in cortical neurons 3h post-transfection (***p<0.001, t=11.59 df=4, n=3 per group) relative to control (cntl). (B) Coronal section of mouse brain infused with 6-Fam fluorescein-labeled Gomafu ASO (600nM) and co-stained with DAPI (blue). (C) Fear acquisition profile of animals trained at 3CS-US, 3h post infusion (CS3: *p<0.05, t=2.672 df=12, n=6 in cntl and n=8 in ASO-induced Gomafu KD). (D) Recall test performed in mice 24h after the initial fear conditioning.
ASO-mediated Gomafu KD also produced a high rate of neuronal transfection when infused directly into the PLPFC in vivo (Fig. 3B) prior to fear conditioning, and was preliminarily verified by gene expression assay compared to a saline and a scrambled control group (Fig. S4A and B in Supplement 1). Following a mild fear-conditioning protocol, mice infused with ASO-mediated Gomafu KD exhibited a moderate but significant enhancement in freezing behavior (Fig. 3C). However, 24h later these mice did not demonstrate a difference in fear recall of the previous conditioning episode, when compared to the control group, indicating there was no effect of Gomafu on long-term memory per se (Fig. 3D).
Given that stress-induced anxiety has been shown to enhance fear responding and interfere with memory retrieval events (58-61), we also examined the effect of Gomafu KD on anxiety-like behavior in an independent cohort of mice. Infusion of ASO-mediated Gomafu KD into the PLPFC led to a decrease in the amount of time spent in the center of an open field (Fig. 4A and E), which was accompanied by a significant increase in distance traveled (Fig. 4B), ambulatory time (Fig. 4C), and stereotypic grooming behavior (Fig. 4D), all of which are characteristic features of increased anxiety in mice (62). These data suggest that a reduction in the expression of the lncRNA Gomafu in the mPFC promotes behaviors that have been implicated in the development of anxiety disorders.
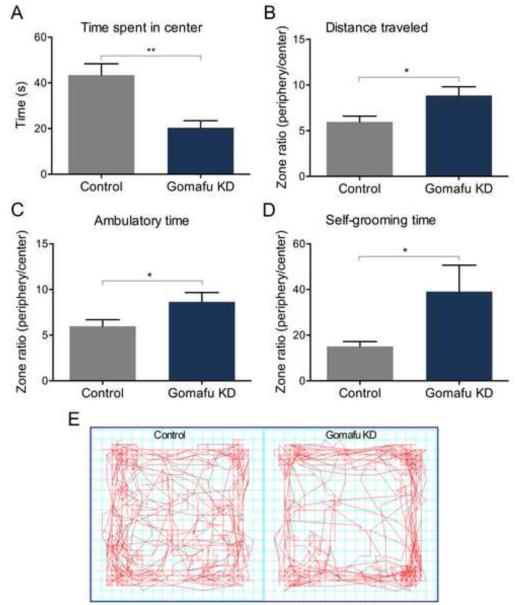
Figure 4. Knockdown of Gomafu lncRNA induces anxiety-like behavior.
(A and E) A mild anxiety test performed on Gomafu knockdown animals in an activity monitoring chamber demonstrated that significantly less time was spent in the center of the field (**p<0.01, t=3.922 df=16, n=9 per group). (B, C, D) Gomafu knockdown mice also exibited a significant tendency to travel longer distances outside the center of the field, as well as having an increased ambulatory and stereotypic grooming time compared to control animals (*p<0.05, t=2.523 df=16, n=9 per group; *p<0.05, t=2.158 df=16, n=9 per group and *p<0.05, Mann-Witney U 16, n=9 per group respectively).
A dual role for the neuropsychiatric-disease related lncRNA Gomafu
LncRNAs have been shown to exert dual control over gene expression through both cis- and trans-mediated mechanisms (63-65). As indicated above, the majority of lncRNAs that were differentially expressed in an experience-dependent manner were encoded within 100kb of the nearest protein-coding gene, suggesting that these experience-dependent lncRNAs may control gene expression via a cis-mediated mechanism. Given that the activity-dependent lncRNA Gomafu was down-regulated in the mPFC following behavioral training, and that it has previously been shown to regulate alternative splicing associated with neuropsychiatric disease through a trans-mediated mechanism (29), we examined whether this lncRNA also regulates gene expression associated with fear-induced anxiety.
Crystallin clusters are molecular chaperone proteins belonging to the heat shock family of genes, and are known to play a major role in maintaining cellular homeostasis in response to stress (66-68). While the alpha-beta cluster has been associated with neurodegenerative diseases (69, 70), a beta family member, Crybb1, has been reported to exert a major effect over these crystallin clusters by acting as an intermolecular chaperone that regulates their correct folding and misfolding (71). Furthermore, Crybb1 has previously been associated with SZ and stress (72-75). In contrast to Gomafu, fear conditioning led to a significant and specific increase in the level of Crybb1 mRNA (Fig. 5A), with no change being observed with its close homolog, Cryba4, which is located on the same strand as Gomafu (Fig. S2 in Supplement 1).
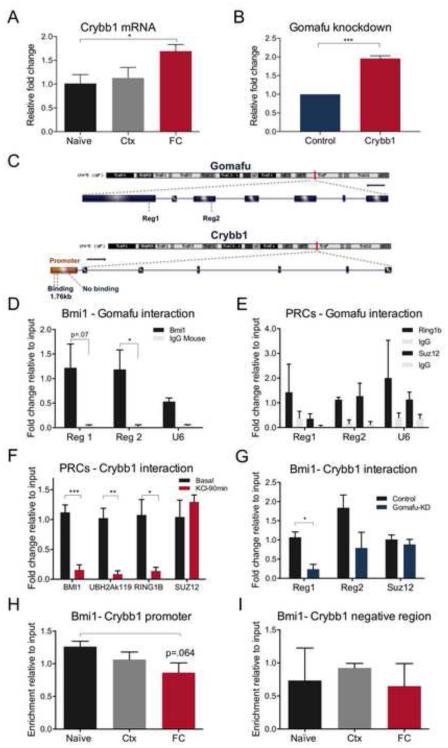
Figure 5. Interaction of Gomafu with the PRC1 complex in the Crybb1 promoter region leads to transcriptional repression.
(A). In vivo RT-qPCR analysis of Crybb1 in fear conditioned (FC) and context (Ctx) mice showed a significant up-regulation in response to the fear stimulus relative to naïve animals (ANOVA *p<0.05, Kruskal-Wallis statistic 6.455, n=8 per group). (B) Significant in vitro up-regulation of Crybb1 resulted from ASO-mediated Gomafu KD at 1μM concentration in cortical neurons (***p<0.001, t=12.89 df=4, n=3 per group). (C) Schematic diagram showing Gomafu lncRNA and Crybb1 genomic structure and loci on chromosome 5, including the promoter region of Crybb1 containing the BMI1 consensus sequence. (D) In vivo RIP assay from mPFC tissue revealed a significant binding of BMI1 on Gomafu lncRNA relative to IgG in exonic region 2 (Reg 2, *p<0.05, t=2.916 df=4, n=3 per group) and a marked binding in the consecutive primer for exonic region 1 (Reg 1, p=0.07, t=2.434 df=4, n=3 per group), as opposed to the negative locus on U6 RNA. (E) RIP analysis for the same regions containing RING1B and SUZ12 binding sites showed no significant enrichment compared to the negative control, U6 (n=3 per group). (F) In vitro ChIP analysis under basal conditions and KCl stimulation exposed a significant dysregulation of BMI1 at the Crybb1 promoter region, approximately 1.76kb from TSS (region 1) in KCl-treated cells (***p<0.001, t=6.369 df=6, n=4 per group) which was followed by down-regulation of the RING1B and UBH2Ak119 repressive marks (**p<0.01, t= 3.511 df=4, n=3 per group and **p<0.01, t=5.426 df=4, n=3 per group respectively). No significant changes were seen in the same region for SUZ12. (G) In vitro ChIP analysis revealed a significant decrease in BMI1 binding at 1.76kb from the Crybb1 TSS, using two consecutive primers, Reg1 and Reg2, following ASO-mediated Gomafu KD in cortical neurons (*p<0.05, t=4.427 df=4, n=3 per group and at 1.35kb from TSS p=0.118, t=1.986 df=4, n=3 per group), whereas no changes were observed in a negative control containing the SUZ12 binding site. (H) In vivo ChIP analysis from mPFC tissue of trained mice showed BMI1 binding at approximately 1.76kb from the Crybb1 TSS, which markedly decreased after fear conditioning (ANOVA, p=0.116, F (2,6)=3.143, n=3 per group; t-test between naïve and fear conditioned mice, p=0.064, n=3 per group). (I) ChIP analysis in a negative region about 1kb from the Crybb1 TSS did not show similar regulation of BMI1 on Crybb1 (ANOVA, p=0.755, F (2, 6)=0.2943, n=3 per group).
In order to test the hypothesis that Gomafu exerts its influence on proximal gene expression via a cis-mediated mechanism, we employed a model of primary cortical neurons in vitro. As expected, infusion of ASO-mediated Gomafu KD in primary cortical neurons led to a significant increase in Crybb1 mRNA expression (Fig. 5B). Next, considering that members of the polycomb repressive complex (PRC) have been shown to interact with lncRNAs to affect gene expression (22, 76-80), we explored the relationship between Gomafu and major components of PRC1 and PRC2, namely BMI1 and RING1B and SUZ12 respectively. The binding motifs for these molecules have been identified (81, 82) and were present within 2kb of the transcription start site (TSS) of Crybb1, and within the gene body of Gomafu (Fig. 5C). RNA immunoprecipitation using an antibody specific to BMI1 revealed an interaction with Gomafu lncRNA (Fig. 5D). In contrast, there was no significant interaction between Gomafu and the PRC2 components SUZ12 and RING1B (Fig. 5E). In addition, no interaction was observed between a U6 negative control and these proteins (Fig. 5E), further supporting the presence of a specific interaction between Gomafu and BMI1.
We further verified through in vitro ChIP assay an activity-dependent binding of the PRC1 proteins BMI1 and RING1B accompanied by the ubiquitin histone 2A lysine 119 repressive mark within the Crybb1 promoter, which was released upon KCl-induced depolarization (Fig. 5F). Although binding of the PRC2 component SUZ12 was observed, the interaction was unresponsive to stimulation (Fig. 5F), confirming the specific activity-dependent recruitment of PRC1 to the Crybb1 promoter.
To test whether Gomafu is required for the interaction between BMI1 and Crybb1, we performed a Gomafu knockdown experiment in cortical neurons, followed by ChIP analysis. After transfection with an ASO targeting Gomafu KD, there was a significant decrease in BMI1 occupancy at the Crybb1 promoter (Fig. 5G). However, importantly, there was no effect of the knockdown on SUZ12 occupancy (Fig. 5G). This finding is consistent with the idea that, under basal conditions, BMI1 is maintained at the Crybb1 promoter through a direct interaction with Gomafu, which serves to repress the transcriptional activity of Crybb1.
To investigate the interaction between BMI1 and Crybb1 in vivo, similar ChIP analyses were performed which revealed BMI1 occupancy at the proximal promoter of Crybb1 in mPFC tissue obtained from naïve animals. Although not significant, a marked decrease was observed in fear-conditioned compared to naïve animals (Fig. 5H). Relative to input, there was little evidence of BMI1 occupancy in a region of the Crybb1 promoter distal to the BMI1 binding motif (Fig. 5I). These results suggest that, upon neuronal stimulation or behavioral training, a decrease in Gomafu releases PRC1 from the Crybb1 promoter, leading to an activity-dependent increase in Crybb1 mRNA expression.
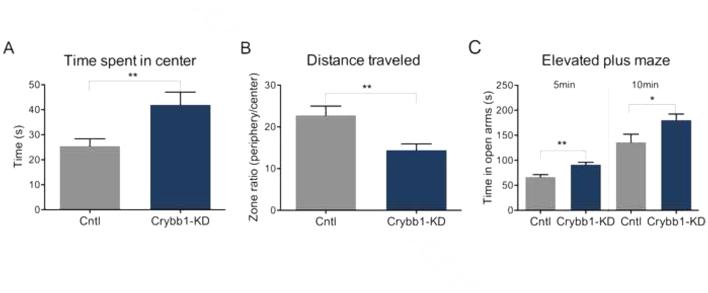
A novel function for Crybb1 in the adult brain. To test the hypothesis that fear-related anxiety in mice is linked to Crybb1 expression, we examined the behavioral effect of Crybb1 knockdown (Crybb1 KD) (Fig. S5 in Supplement 1). Consistent with our Gomafu findings, knockdown of Crybb1 decreased anxiety-like behaviour in the activity-monitoring chamber (Fig. 6A and 6B). Interestingly, in the pre-training test used to induce stress in these subjects, mice treated with Crybb1 KD spent significantly more time in the open arms, also supporting the idea that decreased Crybb1 led to a decrease in anxiety (Fig. 6C). We therefore show that Crybb1 directed knockdown within the mPFC has a role in stress-associated responses in mice.
Figure 6. Knockdown of Crybb1 mRNA expression decreases anxiety in mice.
(A-B) An anxiety test demonstrated that Crybb1 knockdown animals (Crybb1-KD) spent significantly more time within the center of the activity-monitoring chamber (*p<0.05, t= 2.559 df=12, cntl n=6, Crybb1-KD n=8) while travelling significantly shorter distances (**p<0.01, t= 3.263 df=12, cntl n=6, Crybb1-KD n=8). C) Crybb1 knockdown mice also spent significantly more time within the open arms of the elevated plus maze (*p<0.05, t= 2.269 df=12, cntl n=6, Crybb1-KD n=8), an effect that was more pronounced within the first 5 min of this trial (**p<0.01, t= 3.634 df=12, cntl n=6, Crybb1-KD n=8).
Discussion
The present findings reveal widespread experience-dependent activation of lncRNAs in the mPFC, including differential expression of a significant number of non-overlapping intergenic lncRNAs. In particular, the expression of the SZ-associated lncRNA, Gomafu, was decreased in the mPFC following fear conditioning, and ASO-mediated knockdown of this lncRNA promoted stress reactivity and anxiety-like behavior with no effect on long-term memory.
Fear is an evolutionary programed response for self-preservation and survival, but it can also induce pathological behaviors fueled by the persistent memory of adverse events (83). The PFC is a major site mediating stress-associated disorders and can elicit an anxiety response in the fear-conditioning paradigm (84-86). Fear conditioning has further been demonstrated to induce greater anxiety-like behaviors in ‘high fear learning mouse lines’, establishing a direct genetic link between fear conditioning and anxiety (87). Concordantly, the PFC is also vulnerable to stress inducing SZ (88), and has been implicated as a likely neurological site for stress and anxiety behavioral responses following fear-related learning (85, 86, 89).
Consistent with our findings, Gomafu has been reported to show evidence of dysregulation in post mortem cortical tissue of patients who had suffered SZ (29), which is commonly associated with anxiety disorders (90-96). Gomafu is expressed in specific neurons, localized in sub-nuclear speckle-like structures (97), and has been found to be associated with the splicing factors SRSF1 and QK1 (29), the latter of which has been linked with SZ (98-100). Moreover Gomafu knockdown and overexpression affects expression and alternative splicing of genes associated with SZ, such as Erbb4 and Disc1 (29).
Here we have identified a potential pathway of Gomafu regulatory function through recruitment of the PRC1 complex to the site of gene transcription. This suggests a dual function of Gomafu on distal and local genes that guide epigenetic modulators in abnormal behaviors. It is well established that splicing is regulated by epigenetic factors (101, 102), possibly via the 4-dimensional organization of transcription-splicing complexes (103), which may themselves be built on RNA scaffolds (104). Although the mechanistic basis of the interaction of Gomafu with epigenetic processes has yet to be resolved, these results add to the emerging evidence that epigenetic function of lncRNAs may play a role in behavior and neurological diseases (105, 106).
We also showed that the expression of the Crybb1 gene was increased in the mPFC following fear conditioning and after Gomafu knockdown. Dysregulation of Crybb1 has been associated with SZ and autism (72) and both Gomafu and Crybb1 reside in the conserved human locus 22q12.1 (29), which has been related to SZ and generalized anxiety disorders (73-75, 107). Although it has been linked to the development of cataracts (68, 108, 109), the specific function of Crybb1 in the adult brain has not been determined. Here, in conjunction with lncRNA activity, we have discovered a novel role for Crybb1 in mediating fear and stress-associated responses, with its repression appearing to reduce anxiety-like behavior in mice.
We suggest that Gomafu, in addition to its role in regulating alternative splicing in trans (29), acts in cis to direct the epigenetic regulation of Crybb1. Under basal conditions, Gomafu maintains the PRC1 at the promoter of Crybb1, which serves to repress its expression. In response to neuronal activation or fear conditioning, the activity-dependent down-regulation of Gomafu, and its subsequent dissociation from BMI1, relieves its repressive control over the Crybb1 promoter, leading to increased Crybb1 gene expression (Fig. 7). Bmi1 has been reported to act as a major regulator of the cell stress response (110-113), and to sustain stem cell self-renewal in both the peripheral and central nervous systems (114-118). It also plays a key role in the defensive response to oxidative stress by affecting neuronal survival through repression of p53 (119, 120).
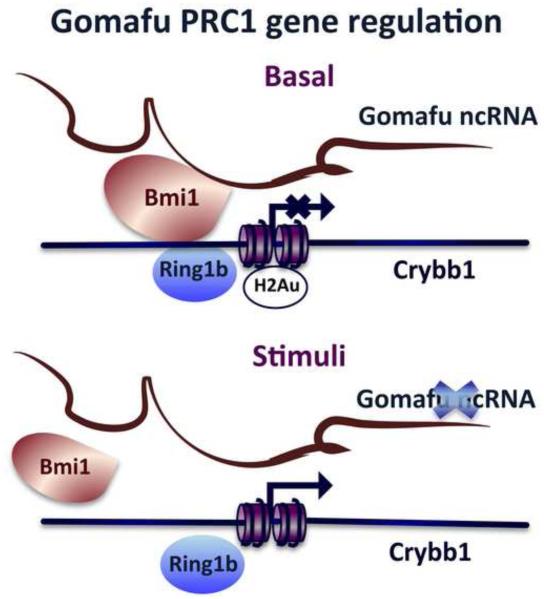
Figure 7. Proposed working model depicting activity-dependent regulation of Gomafu lncRNA in the mouse prefrontal cortex.
Under basal conditions, Gomafu lncRNA associates with the PRC1 (BMI1) complex to mediate in cis repression of Crybb1, potentially through the histone ubiquitination mark. In response to an environmental stimulus, such as that triggered by the fear-conditioning paradigm, Gomafu lncRNA is significantly down-regulated, rapidly releasing BMI1 from the Crybb1 promoter and allowing its transcription, resulting in the up-regulation of Crybb1.
These results imply a complex relationship between lncRNA activity and transcriptional regulation in the adult brain. The dual function of Gomafu is reminiscent of previous work demonstrating that the lncRNA Malat1 can both function in trans, by interacting with hPSF protein (121) and serine/arginine splicing factors to regulate gene expression and cell cycle fate, respectively (63, 64), and in cis to regulate the expression of adjacent genes (65).
In summary, we have shown that the down-regulation of Gomafu lncRNA drives anxiety-like behavior. We further propose a novel role for lncRNAs within the mPFC in directing the epigenetic regulation of gene expression associated with adaptive behavior. Together with its role in governing SZ-related alternative splicing (29), in trans, Gomafu also may function in cis to control gene expression and complex behavior. These findings suggest that further investigation into lncRNAs as molecular links between epigenetic mechanisms, and the development of neuropsychiatric disorders is warranted.
Supplementary Material
Acknowledgements
The authors thank Ms Rowan Tweedale for helpful editing of the manuscript, Ms Vikki Marshall for performing RNA-Seq, Miss Marie-Jeanne Kempen and Gabriela Minervini for their assistance with behavioral data revision. The authors acknowledge grant and fellowship support from the National Health & Medical Research Council of Australia (APP1042051-TWB; Australia Fellowship 631688 - JSM) and the Australian Research Council (DP1096148 - TWB). PAS is supported by a postgraduate scholarship from the Australian Government and the Queensland Brain Institute, The University of Queensland. TWB is supported by a grant from the NIMH (1R21MH103812)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 2.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 3.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 4.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Current Opinion in Genetics & Development. 2010;20:142–148. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nature Structural & Molecular Biology. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 6.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 7.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 8.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 9.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Research. 2011;39:D146–151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R, et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Research. 2012;40:D210–215. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattick JS. RNA regulation: a new genetics? Nature Reviews Genetics. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 13.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genetics. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neuroscience. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, et al. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biology. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin M, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng D, et al. RNA-Seq of human neurons derived from iPS cells reveals candidate long non-coding RNAs involved in neurogenesis and neuropsychiatric disorders. PLoS ONE. 2011;6:e23356. doi: 10.1371/journal.pone.0023356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv J, Liu H, Huang Z, Su J, He H, Xiu Y, et al. Long non-coding RNA identification over mouse brain development by integrative modeling of chromatin and genomic features. Nucleic Acids Research. 2013;41:10044–10061. doi: 10.1093/nar/gkt818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipovich L, Tarca AL, Cai J, Jia H, Chugani HT, Sterner KN, et al. Developmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcripts. Cerebral Cortex. 2014;24:1451–1459. doi: 10.1093/cercor/bhs414. [DOI] [PubMed] [Google Scholar]
- 21.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genetics. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. The Embo Journal. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Molecular Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 24.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. The Embo Journal. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petazzi P, Sandoval J, Szczesna K, Jorge OC, Roa L, Sayols S, et al. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biology. 2013;10:1197–1203. doi: 10.4161/rna.24286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams JM, Beck TF, Pearson DM, Proud MB, Cheung SW, Scott DA. A 1q42 deletion involving DISC1, DISC2, and TSNAX in an autism spectrum disorder. American Journal of Medical Genetics Part A. 2009;149A:1758–1762. doi: 10.1002/ajmg.a.32941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. Journal of Molecular Neuroscience: MN. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Human Molecular Genetics. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 29.Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Molecular Psychiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- 30.Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Human Genetics. 2014;133:59–67. doi: 10.1007/s00439-013-1356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. Journal of Neurochemistry. 2011;116:459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bu Q, Hu Z, Chen F, Zhu R, Deng Y, Shao X, et al. Transcriptome analysis of long non-coding RNAs of the nucleus accumbens in cocaine-conditioned mice. Journal of Neurochemistry. 2012;123:790–799. doi: 10.1111/jnc.12006. [DOI] [PubMed] [Google Scholar]
- 33.Punzi G, Ursini G, Shin JH, Kleinman JE, Hyde TM, Weinberger DR. Increased expression of MARCKS in post-mortem brain of violent suicide completers is related to transcription of a long, noncoding, antisense RNA. Molecular Psychiatry. 2014 doi: 10.1038/mp.2014.41. [DOI] [PubMed] [Google Scholar]
- 34.Lewejohann L, Skryabin BV, Sachser N, Prehn C, Heiduschka P, Thanos S, et al. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behavioural Brain Research. 2004;154:273–289. doi: 10.1016/j.bbr.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Centonze D, Rossi S, Napoli I, Mercaldo V, Lacoux C, Ferrari F, et al. The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. The Journal of Neuroscience: the official journal of the Society for Neuroscience. 2007;27:8885–8892. doi: 10.1523/JNEUROSCI.0548-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipovich L, Dachet F, Cai J, Bagla S, Balan K, Jia H, et al. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics. 2012;192:1133–1148. doi: 10.1534/genetics.112.145128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramos A. Animal models of anxiety: do I need multiple tests? Trends in Pharmacological Sciences. 2008;29:493–498. doi: 10.1016/j.tips.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carey MF, Peterson CL, Smale ST. Chromatin immunoprecipitation (ChIP) Cold Spring Harbor Protocols. 2009;2009 doi: 10.1101/pdb.prot5279. pdb prot5279. [DOI] [PubMed] [Google Scholar]
- 43.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Bekinschtein P, Cammarota M, Medina JH. BDNF and memory processing. Neuropharmacology. 2014;76:677–683. doi: 10.1016/j.neuropharm.2013.04.024. Pt C. [DOI] [PubMed] [Google Scholar]
- 45.Boulle F, van den Hove DL, Jakob SB, Rutten BP, Hamon M, van Os J, et al. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Molecular Psychiatry. 2012;17:584–596. doi: 10.1038/mp.2011.107. [DOI] [PubMed] [Google Scholar]
- 46.Ratnu VS, Wei W, Bredy TW. Activation-induced cytidine deaminase regulates activity-dependent BDNF expression in post-mitotic cortical neurons. The European Journal of Neuroscience. 2014;40:3032–9. doi: 10.1111/ejn.12678. [DOI] [PubMed] [Google Scholar]
- 47.Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learning & Memory. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furukawa-Hibi Y, Yun J, Nagai T, Yamada K. Transcriptional suppression of the neuronal PAS domain 4 (Npas4) gene by stress via the binding of agonist-bound glucocorticoid receptor to its promoter. Journal of Neurochemistry. 2012;123:866–875. doi: 10.1111/jnc.12034. [DOI] [PubMed] [Google Scholar]
- 49.Yun J, Koike H, Ibi D, Toth E, Mizoguchi H, Nitta A, et al. Chronic restraint stress impairs neurogenesis and hippocampus-dependent fear memory in mice: possible involvement of a brain-specific transcription factor Npas4. Journal of Neurochemistry. 2010;114:1840–1851. doi: 10.1111/j.1471-4159.2010.06893.x. [DOI] [PubMed] [Google Scholar]
- 50.Maya-Vetencourt JF. Activity-dependent NPAS4 expression and the regulation of gene programs underlying plasticity in the central nervous system. Neural Plasticity. 2013;2013:683909. doi: 10.1155/2013/683909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ploski JE, Monsey MS, Nguyen T, DiLeone RJ, Schafe GE. The neuronal PAS domain protein 4 (Npas4) is required for new and reactivated fear memories. PloS One. 2011;6:e23760. doi: 10.1371/journal.pone.0023760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schreurs BG, Smith-Bell CA, Burhans LB. Incubation of conditioning-specific reflex modification: implications for Post Traumatic Stress Disorder. Journal of Psychiatric Research. 2011;45:1535–1541. doi: 10.1016/j.jpsychires.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siegmund A, Wotjak CT. A mouse model of posttraumatic stress disorder that distinguishes between conditioned and sensitised fear. Journal of Psychiatric Research. 2007;41:848–860. doi: 10.1016/j.jpsychires.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 54.Kamprath K, Wotjak CT. Nonassociative learning processes determine expression and extinction of conditioned fear in mice. Learning & Memory. 2004;11:770–786. doi: 10.1101/lm.86104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lafaye C, Barbier E, Miscioscia A, Saint-Pierre C, Kraut A, Coute Y, et al. DNA binding of the p21 repressor ZBTB2 is inhibited by cytosine hydroxymethylation. Biochemical and Biophysical Research Communications. 2014;446:341–346. doi: 10.1016/j.bbrc.2014.02.122. [DOI] [PubMed] [Google Scholar]
- 56.Koch SM, Ullian EM. Neuronal pentraxins mediate silent synapse conversion in the developing visual system. The Journal of Neuroscience: the official journal of the Society for Neuroscience. 2010;30:5404–5414. doi: 10.1523/JNEUROSCI.4893-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nature Biotechnology. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milad MR, Pitman RK, Ellis CB, Gold AL, Shin LM, Lasko NB, et al. Neurobiological basis of failure to recall extinction memory in posttraumatic stress disorder. Biological Psychiatry. 2009;66:1075–1082. doi: 10.1016/j.biopsych.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai WH, Blundell J, Han J, Greene RW, Powell CM. Postreactivation glucocorticoids impair recall of established fear memory. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26:9560–9566. doi: 10.1523/JNEUROSCI.2397-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miracle AD, Brace MF, Huyck KD, Singler SA, Wellman CL. Chronic stress impairs recall of extinction of conditioned fear. Neurobiology of Learning and Memory. 2006;85:213–218. doi: 10.1016/j.nlm.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 61.Wilber AA, Walker AG, Southwood CJ, Farrell MR, Lin GL, Rebec GV, et al. Chronic stress alters neural activity in medial prefrontal cortex during retrieval of extinction. Neuroscience. 2011;174:115–131. doi: 10.1016/j.neuroscience.2010.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey KR, Crawley JN. Anxiety-Related Behaviors in Mice. In: Buccafusco JJ, editor. Methods of Behavior Analysis in Neuroscience. 2nd Boca Raton (FL): 2009. [Google Scholar]
- 63.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genetics. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Reports. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stetler RA, Gan Y, Zhang W, Liou AK, Gao Y, Cao G, et al. Heat shock proteins: cellular and molecular mechanisms in the central nervous system. Progress in Neurobiology. 2010;92:184–211. doi: 10.1016/j.pneurobio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kalmar B, Greensmith L. Induction of heat shock proteins for protection against oxidative stress. Advanced Drug Delivery Reviews. 2009;61:310–318. doi: 10.1016/j.addr.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 68.Wistow G. The human crystallin gene families. Human genomics. 2012;6:26. doi: 10.1186/1479-7364-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez-Gonzalez I, Carmona M, Arregui L, Kovacs GG, Ferrer I. alphaB-crystallin and HSP27 in glial cells in tauopathies. Neuropathology: official journal of the Japanese Society of Neuropathology. 2014 doi: 10.1111/neup.12134. [DOI] [PubMed] [Google Scholar]
- 70.Chang LY, Lowe J, Ardiles A, Lim J, Grey AC, Robertson K, et al. Alzheimer's disease in the human eye. Clinical tests that identify ocular and visual information processing deficit as biomarkers. Alzheimer's & Dementia: the journal of the Alzheimer's Association. 2014;10:251–261. doi: 10.1016/j.jalz.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 71.Leng XY, Wang S, Cao NQ, Qi LB, Yan YB. The N-terminal extension of betaB1-crystallin chaperones beta-crystallin folding and cooperates with alphaA-crystallin. Biochemistry. 2014;53:2464–2473. doi: 10.1021/bi500146d. [DOI] [PubMed] [Google Scholar]
- 72.Garbett KA, Hsiao EY, Kalman S, Patterson PH, Mirnics K. Effects of maternal immune activation on gene expression patterns in the fetal brain. Translational Psychiatry. 2012;2:e98. doi: 10.1038/tp.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gill M, Vallada H, Collier D, Sham P, Holmans P, Murray R, et al. A combined analysis of D22S278 marker alleles in affected sib-pairs: support for a susceptibility locus for schizophrenia at chromosome 22q12. Schizophrenia Collaborative Linkage Group (Chromosome 22) American journal of medical genetics. 1996;67:40–45. doi: 10.1002/(SICI)1096-8628(19960216)67:1<40::AID-AJMG6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 74.Takahashi S, Ohtsuki T, Yu SY, Tanabe E, Yara K, Kamioka M, et al. Significant linkage to chromosome 22q for exploratory eye movement dysfunction in schizophrenia. American Journal of Medical Genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2003;123B:27–32. doi: 10.1002/ajmg.b.10046. [DOI] [PubMed] [Google Scholar]
- 75.Pulver AE, Karayiorgou M, Wolyniec PS, Lasseter VK, Kasch L, Nestadt G, et al. Sequential strategy to identify a susceptibility gene for schizophrenia: report of potential linkage on chromosome 22q12-q13.1: Part 1. American Journal of Medical Genetics. 1994;54:36–43. doi: 10.1002/ajmg.1320540108. [DOI] [PubMed] [Google Scholar]
- 76.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biology. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meng S, Luo M, Sun H, Yu X, Shen M, Zhang Q, et al. Identification and characterization of Bmi-1-responding element within the human p16 promoter. The Journal of Biological Chemestry. 2010;285:33219–33229. doi: 10.1074/jbc.M110.133686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Orsini CA, Maren S. Neural and cellular mechanisms of fear and extinction memory formation. Neuroscience & Biobehavioral Reviews. 2012;36:1773–1802. doi: 10.1016/j.neubiorev.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Berkowitz RL, Coplan JD, Reddy DP, Gorman JM. The human dimension: how the prefrontal cortex modulates the subcortical fear response. Reviews in the Neurosciences. 2007;18:191–207. doi: 10.1515/revneuro.2007.18.3-4.191. [DOI] [PubMed] [Google Scholar]
- 85.Indovina I, Robbins TW, Nunez-Elizalde AO, Dunn BD, Bishop SJ. Fear-conditioning mechanisms associated with trait vulnerability to anxiety in humans. Neuron. 2011;69:563–571. doi: 10.1016/j.neuron.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yuen EY, Wei J, Liu W, Zhong P, Li X, Yan Z. Repeated stress causes cognitive impairment by suppressing glutamate receptor expression and function in prefrontal cortex. Neuron. 2012;73:962–977. doi: 10.1016/j.neuron.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ponder CA, Kliethermes CL, Drew MR, Muller J, Das K, Risbrough VB, et al. Selection for contextual fear conditioning affects anxiety-like behaviors and gene expression. Genes, Brain, and Behavior. 2007;6:736–749. doi: 10.1111/j.1601-183X.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 88.Arnsten AF. Prefrontal cortical network connections: key site of vulnerability in stress and schizophrenia. International journal of developmental neuroscience: the official journal of the International Society for Developmental Neuroscience. 2011;29:215–223. doi: 10.1016/j.ijdevneu.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hains AB, Arnsten AF. Molecular mechanisms of stress-induced prefrontal cortical impairment: implications for mental illness. Learning & Memory. 2008;15:551–564. doi: 10.1101/lm.921708. [DOI] [PubMed] [Google Scholar]
- 90.Muller JE, Koen L, Soraya S, Emsley RA, Stein DJ. Anxiety disorders and schizophrenia. Current Psychiatry Reports. 2004;6:255–261. doi: 10.1007/s11920-004-0074-0. [DOI] [PubMed] [Google Scholar]
- 91.Nebioglu M, Altindag A. The prevalence of comorbid anxiety disorders in outpatients with schizophrenia. International Journal of Psychiatry in Clinical Practice. 2009;13:312–317. doi: 10.3109/13651500903094559. [DOI] [PubMed] [Google Scholar]
- 92.Lysaker PH, Salyers MP. Anxiety symptoms in schizophrenia spectrum disorders: associations with social function, positive and negative symptoms, hope and trauma history. Acta Psychiatrica Scandinavica. 2007;116:290–298. doi: 10.1111/j.1600-0447.2007.01067.x. [DOI] [PubMed] [Google Scholar]
- 93.Achim AM, Maziade M, Raymond E, Olivier D, Merette C, Roy MA. How prevalent are anxiety disorders in schizophrenia? A meta-analysis and critical review on a significant association. Schizophrenia Bulletin. 2011;37:811–821. doi: 10.1093/schbul/sbp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Young S, Pfaff D, Lewandowski KE, Ravichandran C, Cohen BM, Ongur D. Anxiety disorder comorbidity in bipolar disorder, schizophrenia and schizoaffective disorder. Psychopathology. 2013;46:176–185. doi: 10.1159/000339556. [DOI] [PubMed] [Google Scholar]
- 95.Pallanti S, Cantisani A, Grassi G. Anxiety as a core aspect of schizophrenia. Current Psychiatry Reports. 2013;15:354. doi: 10.1007/s11920-013-0354-7. [DOI] [PubMed] [Google Scholar]
- 96.Braga RJ, Reynolds GP, Siris SG. Anxiety comorbidity in schizophrenia. Psychiatry Research. 2013;210:1–7. doi: 10.1016/j.psychres.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 97.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. Journal of Cell Science. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 98.Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aberg K, Saetre P, Lindholm E, Ekholm B, Pettersson U, Adolfsson R, et al. Human QKI, a new candidate gene for schizophrenia involved in myelination. American Journal of Medical Genetics Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2006;141B:84–90. doi: 10.1002/ajmg.b.30243. [DOI] [PubMed] [Google Scholar]
- 100.Haroutunian V, Katsel P, Dracheva S, Davis KL. The human homolog of the QKI gene affected in the severe dysmyelination "quaking" mouse phenotype: downregulated in multiple brain regions in schizophrenia. The American Journal of Psychiatry. 2006;163:1834–1837. doi: 10.1176/ajp.2006.163.10.1834. [DOI] [PubMed] [Google Scholar]
- 101.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou YP, Lu YL, Tian WD. Epigenetic features are significantly associated with alternative splicing. BMC Genomics. 2012:13. doi: 10.1186/1471-2164-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mercer TR, Edwards SL, Clark MB, Neph SJ, Wang H, Stergachis AB, et al. DNase I-hypersensitive exons colocalize with promoters and distal regulatory elements. Nature Genetics. 2013;45:852–859. doi: 10.1038/ng.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mercer TR, Mattick JS. Understanding the regulatory and transcriptional complexity of the genome through structure. Genome Research. 2013;23:1081–1088. doi: 10.1101/gr.156612.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Khan DH, Jahan S, Davie JR. Pre-mRNA splicing: role of epigenetics and implications in disease. Advances in Biological Regulation. 2012;52:377–388. doi: 10.1016/j.jbior.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 106.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiological Reviews. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 107.DeLisi LE, Shaw SH, Crow TJ, Shields G, Smith AB, Larach VW, et al. A genome-wide scan for linkage to chromosomal regions in 382 sibling pairs with schizophrenia or schizoaffective disorder. The American Journal of Psychiatry. 2002;159:803–812. doi: 10.1176/appi.ajp.159.5.803. [DOI] [PubMed] [Google Scholar]
- 108.Meyer E, Rahman F, Owens J, Pasha S, Morgan NV, Trembath RC, et al. Initiation codon mutation in betaB1-crystallin (CRYBB1) associated with autosomal recessive nuclear pulverulent cataract. Molecular vision. 2009;15:1014–1019. [PMC free article] [PubMed] [Google Scholar]
- 109.Deng H, Yuan L. Molecular genetics of congenital nuclear cataract. European Journal of Medical Genetics. 2014;57:113–122. doi: 10.1016/j.ejmg.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 110.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & Development. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gargiulo G, Cesaroni M, Serresi M, de Vries N, Hulsman D, Bruggeman SW, et al. In vivo RNAi screen for BMI1 targets identifies TGF-beta/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell. 2013;23:660–676. doi: 10.1016/j.ccr.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 112.Nakamura S, Oshima M, Yuan J, Saraya A, Miyagi S, Konuma T, et al. Bmi1 confers resistance to oxidative stress on hematopoietic stem cells. PloS One. 2012;7:e36209. doi: 10.1371/journal.pone.0036209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Calao M, Sekyere EO, Cui HJ, Cheung BB, Thomas WD, Keating J, et al. Direct effects of Bmi1 on p53 protein stability inactivates oncoprotein stress responses in embryonal cancer precursor cells at tumor initiation. Oncogene. 2013;32:3616–3626. doi: 10.1038/onc.2012.368. [DOI] [PubMed] [Google Scholar]
- 114.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes & Development. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 117.Fasano CA, Phoenix TN, Kokovay E, Lowry N, Elkabetz Y, Dimos JT, et al. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes & Development. 2009;23:561–574. doi: 10.1101/gad.1743709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.He S, Iwashita T, Buchstaller J, Molofsky AV, Thomas D, Morrison SJ. Bmi-1 over-expression in neural stem/progenitor cells increases proliferation and neurogenesis in culture but has little effect on these functions in vivo. Developmental Biology. 2009;328:257–272. doi: 10.1016/j.ydbio.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chatoo W, Abdouh M, David J, Champagne MP, Ferreira J, Rodier F, et al. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. The Journal of Neuroscience: the official journal of the Society for Neuroscience. 2009;29:529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abdouh M, Chatoo W, El Hajjar J, David J, Ferreira J, Bernier G. Bmi1 is down-regulated in the aging brain and displays antioxidant and protective activities in neurons. PloS One. 2012;7:e31870. doi: 10.1371/journal.pone.0031870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li L, Feng T, Lian Y, Zhang G, Garen A, Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.