Abstract
Latent transforming growth factor-beta-1 binding protein-2 (LTBP-2) is a member of the fibrillin/LTBP super family of extracellular matrix proteins, found to be overexpressed in certain malignant tumors. However, the clinical significance and biological role of LTBP-2 in cervical adenocarcinoma has remains unclear. We found that the expression of LTBP2 was higher in cervical adenocarcinoma than in normal cervical epithelial tissue as assessed by immunohistochemistry. Expression of LTBP2 is related to clinical stage, cervical tumor size, depth of cervical stromal invasion and lymph node metastasis. Knockdown of LTBP2 expression can inhibit the proliferation and migration of HeLa cells. Moreover, LTBP2 knockdown affected multiple tumor-related pathway genes including: the MAPK signaling pathway, the PI3K-AKT signaling pathway, receptor tyrosine kinase signaling and the P53 pathway. Taken together, this work suggests that LTBP2 may promote the development of cervical adenocarcinoma and serve as a prognostic factor in the clinical evaluation of patients with cervical adenocarcinoma. Our findings provide a new strategy for the diagnosis and treatment of cervical adenocarcinoma.
Keywords: LTPB2, prognostic factor, cervical adenocarcinoma
Introduction
Cervical cancer is the fourth most common cause of cancer and the fourth most common cause of cancer death in women worldwide [1]. In the past three decades, cervical cancer screening using the Pap smear or acetic acid has resulted in a significant decline in the incidence of cervical cancer. Nonetheless, it is estimated that there are 528,000 new cases of cervical cancer and 266,000 deaths each year representing [1]. 8% of the total mortality attributed to cancer [2]. Histologic subtypes of invasive cervical carcinoma include squamous cell carcinoma and cervical adenocarcinoma. Although squamous cell is most often observed (80% incidence), the incidence of adenocarcinoma of the cervix has also been increasing in recent decades [3]. Human papillomavirus (HPV) infection appears to be involved in the etiology of cervical intraepithelial neoplasia (CIN), squamous cell carcinoma and cervical adenocarcinoma. However, fewer than 1% of HPV infections result in cervical cancer [4,5]. Therefore, it remains unclear what factors result in HPV clearance versus neoplastic transformation.
In recent years, extracellular matrix proteins have been implicated in various facets of cancer biology. In cervical cancer specifically, examples include the extracellular matrix proteins NOV/CCN3, osteopontin (OPN) and Type IIB Procollagen NH2-propeptide (PIIBNP) [5-7]. Latent transforming growth factor (TGF)-beta binding proteins (LTBP) includes four subtypes (LTBP1-4) that belong to the fibrillin/LTBP super family of extracellular matrix proteins. Latent transforming growth factor-beta-1 binding protein-2 (LTBP-2) is the largest member of the LTBP family with a multi-domain structure and with greatest similarity to the fibrillins. Multiple functions for LTBP-2 have been proposed, including being a member of the TGF-beta latent complex, a structural component of microfibrils, and having a role in cell adhesion. LTBP2 is mainly expressed in the lung [8], however residual expression has been observed in other tissues including the liver, spleen, skeletal muscle, embryonic tissue and heart [9]. LTBP2 also associates with microfibrils and has a structural role in the developing neointimal lesion [10]. It has been reported that the LTBP2 knockout mice have a preimplantation genetic lethal defect, highlighting its critical role for cell integrity and survival [11]. LTBP2 expression has been observed in various tumor cells, such as in lung cancer, liver cancer, esophageal cancer and ovarian cancer, and associated with poor prognosis [12-14].
There are few studies that delineate the function and mechanisms of LTBP2 in cancer progression, particularly in cervical adenocarcinoma. In the present study, we analyzed the relationship between LTBP2 and clinical pathological characteristics of cervical adenocarcinoma by studying LTBP2 expression in cervical adenocarcinoma and cervical cancer cell lines. We also interrogated the functional role of LTBP2 using cell culture experiments and microarray expression analysis. This work aims to provide a new strategy for the clinical diagnosis and treatment of cervical adenocarcinoma.
Materials and methods
Patients
A total of 59 cervical adenocarcinoma tissue samples were obtained from the Department of Gynecology, Changzhou Maternal and Child Care Hospital and Department of Gynecology and Fengxian Hospital, Southern Medical University. The cases of cervical adenocarcinoma were selected in this study only if follow up was obtained and clinical data were available. All patients underwent a modified radical hysterectomy or complete hysterectomy, bilateral salpingo-oophorectomy and pelvic lymphadenectomy with or without para-aortic lymph node sampling. None of the patients had received radiotherapy, chemotherapy, hormone therapy or other related anti-tumor therapies prior to surgical resection. Thirty (30) cases of normal cervical epithelium were selected as a control group. All tissue samples were obtained with informed consent and all procedures were performed in accordance with the Human Investigation Ethical Committee of the two hospitals.
Immunohistochemistry (IHC)
Tissue samples were fixed in phosphate-buffered neutral formalin and routinely embedded in paraffin, and then cut into 5-μm-thick sections. The sections were incubated with 0.3% hydrogen peroxide/phosphate-buffered saline for 30 minutes and blocked with 10% BSA (Sangon, Shanghai, China), then were detected with primary polyclonal antibody for LTBP2 (Proteintech Group, Chicago, USA), overnight at 4°C in a moist chamber. After incubation with the secondary antibody (Thermo Scientific, US) labeled with HRP (rabbit) for 1 hour at room temperature, the sections were treated with diaminobenzidine and counterstained with hematoxylin. All the sections were observed and photographed with a microscope (Carl Zeiss). Scoring was conducted according to the ratio and intensity of positive-staining cells as follows: strongly staining (score 1) designated as high expression and weakly staining (score 0) designated as low expression. All the LTBP2 expression levels were quantified double-blindly by two independent pathologists.
Cell line and cell culture
Human EC cell lines HeLa, Hela229, Siha, Caski, and Ms751 were purchased from Cell Bank of the Chinese Academy of Sciences.AN3CA, Hela, Hela229, Siha, and Ms751 Cells were cultured in Eagle’s Minimum Essential Medium (EMEM); Caski cells were cultured in RPMI-1640; and all of these cells were supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin, and incubated at 37°C in a humidified incubator under 5% CO2 condition.
siRNA transfection
SiRNA duplexes targeted against LTBP2 and scrambled control siRNA duplex were obtained from GenePharma (Shanghai, China). Small interfering RNAs duplexes for LTBP2 were as follows:
siRNA1 sense, 5’-GCACCAACCACUGUAUCAATT-3’, anti-sense, 5’-UUGAUACAGUGGUUGGUGCTT-3’; siRNA2 sense, 5’-GGAGUGUCAAGAUAUCAAUTT-3’, anti-sense, 5’-AUUGAUAUCUUGACACUCCTT-3’; siRNA3 sense, 5’-CGGAUCACCAAGCAGAUAUTT-3’, anti-sense, 5’-AUAUCUGCUUGGUGAUCCGTT-3’.
The scramble control siRNA duplex were as sense, 5’-UUCUCCGAACGUGUCACGUTT-3’, antisense, 5’-ACGUGACACGUUCGGAGAATT-3’.
Transfection was performed by using the Lipofectamine RNAiMAX Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction.
Cell proliferation and invasion assay by x’Celligence Biosensor System
Experiments were carried out using the RTCA DPinstrument (Roche Diagnostics GmbH, Germany), which was placed in humidified incubator maintained at a 5% CO2 humidified incubator at 37°C. Growth curves were constructed using 16-well plates (E-plate 16, Roche Diagnostics GmbH). For the proliferation assay, cells were seeded in E-plates 16 at 5,000 cells/well in fetal calf serum (FCS)-containing medium, and the plate was then monitored once every 15 min. Cell invasion was assessed using specifically designed 16-well plates (CIM-plate 16, Roche DiagnosticsGmbH) with 8 m pores. These plates are similar to conventional transwells with the microelectrodes located on the underside of the membrane of the upper chamber. Over the membrane a thin layer of 20 μl 1:8 diluted extracellular matrigel (BD Bioscience, Franklin Lakes, NJ) is dried. The 10% FCS medium was added in the lower chamber, and cells were seeded into the upper chamber at 30,000 cells/well in serum-free medium. The CIM-plate 16 was monitored every 15 min. Data analysis was carried out using RTCA software 1.2 supplied with the instrument.
Transwell assay
For the Transwell Invasion assay, we placed 100,00 HeLa cells in 100 μl matrigel on the top chamber of each transwell (Millicell) of each insert (BD Bioscience, Franklin Lakes, NJ). After 48 hours, any cells remaining in the top chambers or on the upper membrane of the inserts were carefully removed. After fixation and staining in a dye solution containing 0.1% crystal violet and 20% methanol, cells adhering to the lower membrane of the inserts were counted and imaged through an I×71 inverted microscope (Olympus Corp. Tokyo, Japan).
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from cells and tissues using Trizol reagent (Takara, Dalian, China) and reverse transcribed using the Prime Script RT reagent kit (Takara, Dalian, China) according to the manufacturer’s instruction. The quantitative real-time polymerase chain reaction (qRT-PCR) was subsequently performed with SYBR Premix Ex Taq (Takara, Dalian, China) using an ABI7300 instrument (Applied Biosystems). And the primers for LTBP2 were as follows, forward: 5’-AGCACCAACCACTGTATCAAAC-3’; reverse: 5’-CTCATCGGGAATGACCTCCTC-3’. The relative expression of LTBP2 was analyzed by the comparative cycle threshold method (ΔΔCt method) which was normalized to 18 s RNA (forward: 5’-TGCGAGTACTCAACACCAACA-3’, reverse: 5’-GCATATCTTCGGCCCACA-3’).
Analysis of cDNA microarray
HeLa cell lines were transfected with small interfering RNA (siRNA) to knockdown LTBP2 expression. The following samples were also analyzed by cDNA microarray: LTBP2-siRNA2-HeLa and Control-siRNA-HeLa. (Shanghai Biotechnology Co. Ltd). The cDNA microarray data was analyzed using the KEGG database.
Statistical analysis
Data were presented as the means ± standard error of the mean (SEM). Statistical analyses were done using SPSS 16.0 for windows (IBM). The chi-square was used to analyze the correlations between LTBP2 expression and clinicopathologic features in patients with Cervical cancer. The student’s t-test was used for comparison between groups in cellular functions of LTBP2. Values of P<0.05 were considered statistically significant.
Results
LTBP2 expression in cervical adenocarcinoma by immunohistochemistry
Basic patient characteristics are shown in Table 1. The immunohistochemistry of tissue microarrays demonstrated that expression of LTBP2 was significantly higher in cervical adenocarcinoma than normal cervical epithelial tissue (P=0.002) (Figure 1). Further analysis identified a significant relationship between LTBP2 expression and clinical stage, cervical tumor size, depth of cervical stromal invasion and lymph node metastasis, which is associated with poor prognosis (Table 2). These results suggest that LTBP2 may be a prognostic marker of cervical adenocarcinoma.
Table 1.
Characteristics of cervical adenocarcinoma patients and control group
| Cervical adenocarcinoma (n=59) | Control group (n=30) | |
|---|---|---|
| Age (years ) | ||
| Mean (Std) | 47.1 (9.7) | 44.2 (7.8) |
| Min, Max | (24, 73) | (28, 61) |
| Clinical stage | ||
| I | 38 | N/A |
| II | 14 | N/A |
| III | 7 | N/A |
| Histology | ||
| Grade 1 | 11 | N/A |
| Grade 2 | 26 | N/A |
| Grade 3 | 22 | N/A |
| Lymphatic metastasis | ||
| Absent | 42 | N/A |
| Present | 17 | N/A |
| Interstitial invasion | ||
| ≤1/2 | 22 | N/A |
| >1/2 | 37 | N/A |
| Tumor size | ||
| ≤4 cm | 36 | N/A |
| >4 cm | 23 | N/A |
| Vascular invasion | ||
| Absent | 31 | N/A |
| Present | 28 | N/A |
Figure 1.

LTBP2 is frequently up-regulated in cervical adenocarcinoma tissues. Representative photomicrographs of the LTBP2 immunoreactivity in normal cervix (A), down-expression of cervical adenocarcinoma (B), up-expression of cervical adenocarcinoma (C) and the distribution of down-expression and up-expression of normal cervix and cervical cancer (D) (Scale bar: 50 um).
Table 2.
Correlations between LTBP2 expression and clinicopathologic features in patients with cervical cancer
| Expression of LTBP2 | ||||
|---|---|---|---|---|
|
|
||||
| Clinicopathological feature | Total 59 | Low (n=29, 49.2%) | High (n=30, 51.8%) | P value (÷2 test/fisher’s exact test) |
| Age (years) | ||||
| 45 | 30 | 18 (60.0) | 12 (40.0) | 0.090 |
| >45 | 29 | 11 (37.93) | 18 (62.07) | |
| Clinical stage | ||||
| I | 38 | 23 (60.53) | 15 (39.47) | 0.041 (0.043) |
| II | 14 | 5 (35.71) | 9 (64.29) | |
| III | 7 | 1 (14.29) | 6 (85.71) | |
| Histology | ||||
| Grade 1 | 11 | 7 (63.64) | 4 (36.36) | 0.274 (0.302) |
| Grade 2 | 26 | 14 (53.85) | 12 (46.15) | |
| Grade 3 | 22 | 8 (36.36) | 14 (63.64) | |
| Lymphatic metastasis | ||||
| Absent | 42 | 25 (59.52) | 17 (40.48) | 0.012 (0.020) |
| Present | 17 | 4 (23.53) | 13 (76.47) | |
| Interstitial invasion | ||||
| ≤1/2 | 22 | 15 (68.18) | 7 (31.82) | 0.024 |
| >1/2 | 37 | 14 (37.84) | 23 (62.16) | |
| Tumor size | ||||
| ≤4 cm | 36 | 23 (63.89) | 13 (36.11) | 0.005 |
| >4 cm | 23 | 6 (26.09) | 17 (73.91) | |
| Vascular invasion | ||||
| Absent | 31 | 19 (61.29) | 12 (38.71) | 0.050 |
| Present | 28 | 10 (35.71) | 18 (64.29) | |
Values in parentheses indicate percentage values. The bold number represents the P-values with significant differences.
We an increasing prevalence of high-LTBP2 expressors associated with advanced stages of disease (stage I=39.47% (n=15), stage II=64.29% (n=9) and stage III=85.71% (n=6); P=0.043). Moreover, our data showed that 73.91% cases (n=17) had high expression of LTBP2 in the >4 cm group while 36.11% (n=13) had high expression of LTBP2 in the ≤4 cm group (P=0.005). With respect to interstitial invasion, high LTBP2 expression was seen in only 31.82% in the invasion ≤1/2 group (n=7), while high LTBP2 expression was significantly increased in the interstitial invasion >1/2 group (n=23, 62.16%) (P=0.024). We also found an increased rate of high LTBP2 expression in the lymphatic metastasis positive group as compared with the non-metastatic group (n=13, 76.47% vs. n=17, 40.48%) (P=0.020). These results suggest that LTBP2 expression is associated the clinical progression of cervical cancer, and that LTBP2 may be related to cervical adenocarcinoma invasion and metastasis.
There was, however, no statistically significant relationship between LTBP2 expression and tumor (Grade I=36.36%, Grade II=46.15% and Grade III=63.64%; P=0.302), nor vascular invasion (P=0.050). Specifically, 38.71% of the vascular invasion positive group demonstrated high LTBP2 expression, while high-LTBP2 was observed in 64.29% of vascular invasion negative group.
mRNA expression of LTBP2and siRNA transfection
LTBP2 expression was detected in five cervical cancer cell lines: Hela, Hela229, Siha, Caski and, and Ms751by qRT-PCR (Figure 2A). Using small interfering RNA (siRNA), LTBP2 expression was knocked down in Hela cells. As shown in Figure 2B, mRNA level of LTBP2 was successfully decreased after siRNA transfection. We proceeded with siRNA1 and siRNA2 for additional experiments as these siRNAs exhibited the best interference efficacy.
Figure 2.
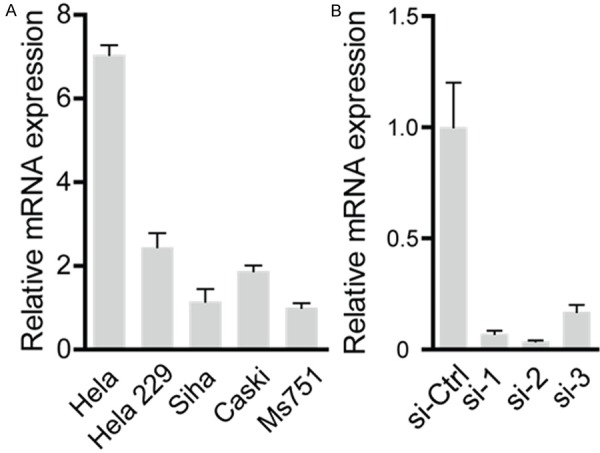
The expression level of LTBP2 in different cervical cancer cells lines and validation of siRNA interference efficiency. A. The mRNA level of LTBP2 in Hela is higher than other cervical cancer cells. B. The siRNA1, siRNA2 and siRNA3 could notably restrain LTBP2 by real-time PCR (sh-NC versus sh-1 or sh-2 or sh-3, *P<0.05, **P<0.01).
Knockdown of LTBP2 inhibits cancer cells proliferation in vitro
We observed a suppressive effect on cell growth in HeLa cells treated with LTBP2-specific siRNAs1 and 2 (Figure 3A, 3B). Cell numbers were measured using the x’Celligence Biosensor System. We infer from these results that cervical adenocacinoma cells with low LTBP2 expression have a growth disadvantage.
Figure 3.
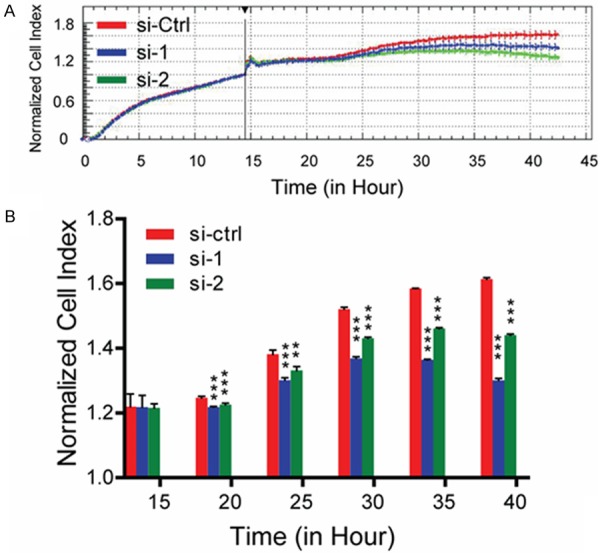
Silencing of LTBP2 inhibits proliferation in cervical adenocarcinoma cells. A. Cell numbers are detected by x’Celligence Biosensor System. B. Effect of LTBP2 knockdown on cell survival of Hela cells is analyzed by x’Celligence Biosensor System (sh-NC versus sh-1 and sh-NC versus sh-2, *P<0.05, **P<0.01, ***P<0.0001).
Knockdown of LTBP2 inhibited cervical cancer cell migration in vitro
Next, we conducted in vitro migration assays to evaluate the impact of LTBP2-depleiont on HeLa cells migration. We observed a significant decrease in HeLa cell migration following LTBP2-siRNA silencing using in a transwell assay (Figure 4A, 4B). These results suggest that LTBP2 expression is associated with the migration potential of cervical adenocacinoma cell lines in vitro.
Figure 4.
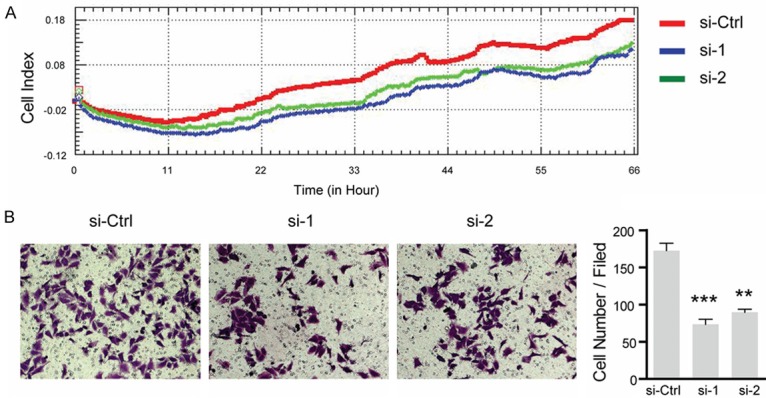
Silencing of LTBP2 inhibits cervical cancer cells migration. A. Cell numbers are detected by x’Celligence Biosensor System. B. Hela cells that migrate through the transwell chambers and are photographed at ×100 magnification. The in vitro migration ability of Hela is measured by determining the number of uncoated cells that penetrated through the transwell chambers (Columns, mean values; bar, SD. *P<0.05, **P<0.01, ***P<0.001).
Gene ontology analysis of cDNA microarray
To further elucidate the role off LTBP2 in cervical cancer cells, we performed a transcriptional analysis of control versus LTBP2-silenced HeLA cells. Following knockdown, the most down-regulated gene sets include: apoptotic cell clearance (75%), positive regulation of Rho GTPase activity (60%), Camp catabolic process (60%) and activation of RacGTPase activity (50%) (Figure 5A). All of these pathways affect cell survival and migration. The heat map images showing reduction of tumor-related genes after knockdown of LTBP2 in HeLa are shown in Figure 5B. These heat maps illustrated the changes in the aforementioned pathways upon LTBP2 knockdown (Figure 5B). Upon interrogation of the KEGG database, we found that the following networks were altered after LTBP2 was silenced: MAPK family (MAPK1, MAPK3, MAPK14), PI3K-AKTsignalling pathway, Tyrosine receptor kinase and cancer tumor suppressor gene P53 (Figure 5C; Table 3).
Figure 5.
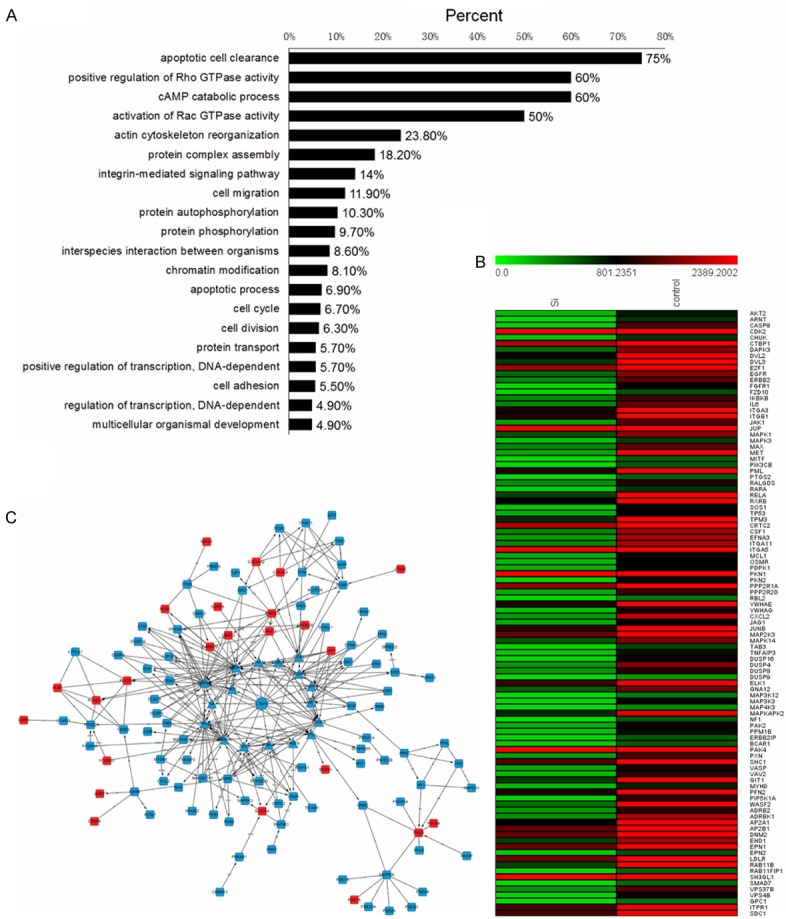
Transcriptional changes induced by LTBP2inactivation. A. Gene ontology analysis of cDNA microarray shows gene set enrichment in cellular functions after LTBP2 knockdown. B. Heat maps of the genes enriched in indicated pathways illustrate the changes in gene expression upon LTBP2 knockdown. Red signal denotes higher expression and green signal denotes lower expression. C. Alternations of gene network after LTBP2 was silenced. Red rectangle represents up-regulated and blue rectangle represents down-regulated.
Table 3.
Genes selected from interaction network sequenced by Betweenness Centrality after LTBP2 was knockdown
| Gene name | NCBI_GENE_ID | Betweenness centrality | Degree | Indegree | Outdegree |
|---|---|---|---|---|---|
| RELA | 5970 | 0.14132 | 21 | 11 | 10 |
| PRKCG | 5582 | 0.090808 | 11 | 4 | 7 |
| PPP2R1A | 5518 | 0.06473 | 7 | 2 | 5 |
| MAPK1 | 5594 | 0.062679 | 29 | 19 | 10 |
| MAPK3 | 5595 | 0.062679 | 29 | 19 | 10 |
| PIK3CB | 5291 | 0.05494 | 20 | 10 | 10 |
| MAPK14 | 1432 | 0.043934 | 22 | 11 | 11 |
| PXN | 5829 | 0.041517 | 7 | 3 | 4 |
| AKT2 | 208 | 0.041256 | 14 | 9 | 5 |
| VEGF | 7422 | 0.040769 | 6 | 2 | 4 |
| TP53 | 7157 | 0.036519 | 8 | 4 | 4 |
| ITGB1 | 3688 | 0.034905 | 6 | 4 | 2 |
| EGFR | 1956 | 0.030959 | 15 | 8 | 7 |
| JAK1 | 3716 | 0.022494 | 7 | 5 | 2 |
| PDPK1 | 5170 | 0.019201 | 11 | 4 | 7 |
| RRM2 | 6241 | 0.018393 | 3 | 2 | 1 |
| BCAR1 | 9564 | 0.017686 | 7 | 6 | 1 |
| IL6 | 3569 | 0.017563 | 7 | 5 | 2 |
| PKLR | 5313 | 0.016393 | 8 | 5 | 3 |
| FGFR1 | 2260 | 0.014549 | 9 | 4 | 5 |
Discussion
The identification of tumor-specific markers may have important implications for the diagnosis and prognosis of human malignancies. Although a few biomarkers of cervical cancer exist (e.g. SCCS, CA125, p16ink4a, and MCM-5), most of these are not suitable for prognostication or screening [15-17]. As such, there remains an urgent need to identify additional markers of cervical adenocarcinoma.
LTPB2 is a recently discovered cell exocrine protein whose expression is associated with poor prognosis in many tumors. For example, Turtoi et al., identified LTBP2 using 2D-nano-HPLC_MS/MS in pancreatic ductal adenocarcinoma tissue and found that it was significantly expressed in a large group of clinical pancreas ductal adenocarcinoma samples by Western blot [18]. Torres et al., found that, in mouse and human samples, expression of LTBP2 was significantly increased in the tumor stroma, without significant expression in the cancer epithelial cells [19]. Yoshiharar et al. described LTBP2 as a novel prognostic factors associated with progression-free survival using gene ontology analysis and Ingenuity Pathway Analysis. However, the finding was not confirmed by real-time RT-PCR experiments [14]. Chan et al. found that LTPB2 was highly expressed in poorly differentiated squamous cell carcinoma and that those with low expression of LTBP-2 had longer survival times [13]. To date, LTBP2 has not been studied in the context of cervical cancer.
Our data demonstrated that, compared with normal cervical epithelial tissue, LTBP2 is significantly overexpressed in cervical adenocarcinoma. HeLa cells have the highest expression of LTBP2 as compared to other cell lines tested in this investigation Our results suggest that initial results suggested that LTBP2 may participate in the pathogenesis of cervical adenocarcinoma.
In cervical cancer, clinical stage, tumor size, cervical invasive interstitial depth, lymph node metastasis, histological level and lymphatic vascular invasion are adverse prognostic markers. FIGO staging of cervical cancer is the most used clinical staging system, but does not take into account uterine transfer and lymph node metastasis for clinical outcomes [20]. It is reported that tumour volume and stromal invasion depth maintained their significance as indicators of an adverse prognosis regarding the disease-free survival interval. When the greatest tumour dimension exceeded 2 cm, prognosis worsened and continued to do so more markedly when a dimension of 4 cm was exceeded. Patients who presented a stromal invasion depth in excess of 7 mm also saw their prognosis adversely affected [21,22]. A large number of research studies have confirmed the relationship between lymph node metastasis and prognosis of cervical cancer. Girardi et al., reported that lymph node metastasis is an independent prognostic feature of cervical cancer. The 5-year survival rate of patients with negative nodes was 89.3%. Survival dropped to 69.8% and 37% in patients with 1 or >/4 positive nodes, respectively [23]. In our study, the rate of LTBP2 high expression in clinical stage II and III, tumour size >4 cm, interstitial invasion >1/2 and lymphatic metastasis was higher than clinical stage I, tumour size <4 cm, interstitial invasion ≤1/2 and no lymphatic metastasis, respectively (P<0.05). Together, this suggests that high LTBP2 expression may be a poor prognostic marker in cervical adenocarcinoma.
The function of LTBP2 in tumors has not yet been clearly characterized. The protein appears to function differently across various tumor types. It is reported that all of melanoma cell lines adhered to LTBP2 very efficiently and in a concentration-dependent manner. For example, Bowes melanoma cells bound most efficiently to LTBP2. The adhesive site is located in an N-terminal region of LTBP2. The attachment of melanoma cells to LTBP2 was prevented with monoclonal antibody against beta 1 integrin in a concentration-dependent manner. LTBP-2 also supported Bowes cell migration in modified Boyden chamber assays in a manner similar to the migration on fibronectin. Current data therefore indicate that LTBP-2 can play a role in melanoma cell adhesion [12]. Consistent with this, LTBP2 has been found to be highly expressed in the distant metastasis of esophageal squamous cell carcinoma [13].
In this study, the proliferation and invasive ability of HeLa cells following LTBP2 knockdown was significantly affected, which is consistent with a role for LTBP2 in stromal invasion and lymph node metastasis. This further supports the notion that LTBP2 may promote the growth and progression of the cervical adenocarcinoma cells. Therefore, LTBP2 may be a potential prognostic marker and a therapeutic targets as well for cervical adenocarcinoma.
We analyzed gene expression in HeLa cells after knockdown of LTBP2 using a cDNA microarray. The microarray analysis following LTBP2 silencing revealed reduced expression of genes sets implicated in: apoptotic cell clearance (75%), positive regulation of Rho GTPase activity (60%), cAMP catabolic process (60%) and activation of RacGTPase activity (50%). We specifically found changes in some important signaling pathways after knockdown of LTBP2 in HeLa cells, including the MAPK1 family pathway (MAPK1, MAPK3, MAPK14), PI3K-AKT pathway, and receptor tyrosine kinase pathway (VEGF, EGFR, FGFR1) and TP53 (Figure 5C). We speculate that LTBP2 influences the invasive and metastatic potential of cervical adenocarcinoma by modulating classical oncogenic signaling pathways. However, more studies are needed to explore the underlying mechanism of cervical cancer invasion and metastasis mediated by LTBP2.
In conclusion, LTBP2 is a potential prognostic marker for cervical adenocarcinoma and may promote local invasion and lymph node metastasis of cervical cancer oncogenic signaling pathways. LTBP2 may provide a therapeutic value for cervical adenocarcinoma, and potentially helps with the diagnosis and treatment of cervical adenocarcinoma.
Acknowledgements
This study was supported by a grant (no. 81272879 to C-J Xu) from the Natural Science Foundation of China (NSFC), a grant (no. 81472445 to R. Zhang) from the National Natural Science Foundation of China, a grant (no. 2010249 to R. Zhang) from the Shanghai Municipal Health Bureau Project, a grant (no. F201311 to Y. Ren) from the Jiangsu provincial commission of health and family planning.
Disclosure of conflict of interest
None.
References
- 1.World Cancer Report 2014. World Health Organization. 2014 Chapter 5.12.
- 2.World Cancer Report 2014. World Health Organization. 2014 Chapter 1.1.
- 3.Smith HO, Tiffany MF, Qualls CR, Key CR. The rising incidence of adenocarcinoma relative to squamous cell carcinoma of the uterine cervix in the United States--a 24-year population-based study. Gynecol Oncol. 2000;78:97–105. doi: 10.1006/gyno.2000.5826. [DOI] [PubMed] [Google Scholar]
- 4.Galic V, Herzog TJ, Lewin SN, Neugut AI, Burke WM, Lu YS, Hershman DL, Wright JD. Prognostic significance of adenocarcinoma histology in women with cervical cancer. Gynecol Oncol. 2012;125:287–291. doi: 10.1016/j.ygyno.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Zhang T, Zhao C, Luo L, Xiang J, Sun Q, Cheng J, Chen D. The clinical and prognostic significance of CCN3 expression in patients with cervical cancer. Adv Clin Exp Med. 2013;22:839–845. [PubMed] [Google Scholar]
- 6.Kumar V, Behera R, Lohite K, Karnik S, Kundu GC. p38 kinase is crucial for osteopontin-induced furin expression that supports cervical cancer progression. Cancer Res. 2010;70:10381–10391. doi: 10.1158/0008-5472.CAN-10-1470. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Bryan J, Franz C, Havlioglu N, Sandell LJ. Type IIB procollagen NH(2)-propeptide induces death of tumor cells via interaction with integrins alpha(V)beta(3) and alpha(V)beta(5) J Biol Chem. 2010;285:20806–20817. doi: 10.1074/jbc.M110.118521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kingsley DM. The TGF-p superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 9.Gibson MA, Hatzinikolas G, Davis EC, Baker E, Sutherland GR, Mecham RP. Bovine Latent Transforming Growth Factor beta 1-Binding Protein 2: Molecular Cloning, Identification of Tissue Isoforms, and Immunolocalization to Elastin-Associated Microfibrils. Mol Cell Biol. 1995;15:6932–6942. doi: 10.1128/mcb.15.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha S, Heagerty AM, Shuttleworth CA, Kielty CM. Expression of latent TGF-beta binding proteins and association with TGF-beta1 and fibrillin-1 following arterial injury. Cardiovasc Res. 2002;53:971–983. doi: 10.1016/s0008-6363(01)00512-0. [DOI] [PubMed] [Google Scholar]
- 11.Shipley JM, Mecham RP, Maus E, Bonadio J, Rosenbloom J, McCarthy RT, Baumann ML, Frankfater C, Segade F, Shapiro SD. Developmental Expression of Latent Transforming Growth Factor beta Binding Protein 2 and Its Requirement Early in Mouse Development. Mol Cell Biol. 2000;20:4879–4887. doi: 10.1128/mcb.20.13.4879-4887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vehvilainen P, Hyytiainen M, Keski-Oja J. Latent transforming growth factor-beta-binding protein 2 is an adhesion protein for melanoma cells. J Biol Chem. 2003;278:24705–24713. doi: 10.1074/jbc.M212953200. [DOI] [PubMed] [Google Scholar]
- 13.Chan SH, Yee Ko JM, Chan KW, Chan YP, Tao Q, Hyytiainen M, Keski-Oja J, Law S, Srivastava G, Tang J, Tsao SW, Chen H, Stanbridge EJ, Lung ML. The ECM protein LTBP-2 is a suppressor of esophageal squamous cell carcinoma tumor formation but higher tumor expression associates with poor patient outcome. Int J Cancer. 2011;129:565–573. doi: 10.1002/ijc.25698. [DOI] [PubMed] [Google Scholar]
- 14.Yoshihara K, Tajima A, Komata D, Yamamoto T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Fujiwara H, Kudo Y, Inoue I, Tanaka K. Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci. 2009;100:1421–1428. doi: 10.1111/j.1349-7006.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadducci A, Tana R, Cosio S, Genazzani AR. The serum assay of tumour markers in the prognostic evaluation, treatment monitoring and follow-up of patients with cervical cancer: A review of the literature. Crit Rev Oncol Hematol. 2008;66:10–20. doi: 10.1016/j.critrevonc.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Malinowski DP. Multiple biomarkers in molecular oncology. I. Molecular diagnostics applications in cervical cancer detection. Expert Rev Mol Diagn. 2007;7:117–131. doi: 10.1586/14737159.7.2.117. [DOI] [PubMed] [Google Scholar]
- 17.Tiltman AJ. The pathology of cervical tumours. Best Pract Res Clin Obstet Gynaecol. 2005;19:485–500. doi: 10.1016/j.bpobgyn.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Turtoi A, Musmeci D, Wang Y, Dumont B, Somja J, Bevilacqua G, De Pauw E, Delvenne P, Castronovo V. Identification of Novel Accessible Proteins Bearing Diagnostic and Therapeutic Potential in Human Pancreatic Ductal Adenocarcinoma. J Proteome Res. 2011;10:4302–4313. doi: 10.1021/pr200527z. [DOI] [PubMed] [Google Scholar]
- 19.Torres S, Bartolome RA, Mendes M, Barderas R, Fernandez-Acenero MJ, Pelaez-Garcia A, Pena C, Lopez-Lucendo M, Villar-Vazquez R, de Herreros AG, Bonilla F, Casal JI. Proteome profiling of cancer-associated fibroblasts identifies novel proinflammatory signatures and prognostic markers for colorectal cancer. Clin Cancer Res. 2013;19:6006–6019. doi: 10.1158/1078-0432.CCR-13-1130. [DOI] [PubMed] [Google Scholar]
- 20.Kodaira T, Fuwa N, Toita T, Nomoto Y, Kuzuya K, Tachibana H, Furutani K, Ogawa K. Comparison of prognostic value of MRI and FIGO stage among patients with cervical carcinoma treated with radiotherapy. Int J Radiat Oncol Biol Phys. 2003;56:769–777. doi: 10.1016/s0360-3016(03)00007-5. [DOI] [PubMed] [Google Scholar]
- 21.Martin-Loeches M, Ortí RM, Cazorla E, Asins E, Llixiona J. Multivariate analysis of the morphometric characteristics of tumours as prognostic factors in the survival of patients with uterine cervix cancer treated with radical surgery. Eur J Obstet Gynecol Reprod Biol. 2002;105:170–176. doi: 10.1016/s0301-2115(02)00156-2. [DOI] [PubMed] [Google Scholar]
- 22.Rotman M, Sedlis A, Piedmonte MR, Bundy B, Lentz SS, Muderspach LI, Zaino RJ. A phase III randomized trial of postoperative pelvic irradiation in Stage IB cervical carcinoma with poor prognostic features: follow-up of a gynecologic oncology group study. Int J Radiat Oncol Biol Phys. 2006;65:169–176. doi: 10.1016/j.ijrobp.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Girardi F, Haas J. The importance of the histologic processing of pelvic lymph nodes in the treatment of cervical cancer. Int J Gynecol Cancer. 1993;3:12–17. doi: 10.1046/j.1525-1438.1993.03010012.x. [DOI] [PubMed] [Google Scholar]


