Abstract
During development, oligodendrocytes (OLGs), the myelinating cells of the CNS, undergo a stepwise progression during which OLG progenitors, specified from neural stem/progenitor cells, differentiate into fully mature myelinating OLGs. This progression along the OLG lineage is characterized by well synchronized changes in morphology and gene expression patterns. The latter have been found to be particularly critical during the early stages of the lineage, and they have been well described to be regulated by epigenetic mechanisms, especially by the activity of the histone deacetylases HDAC1 and HDAC2. The data presented here identify the extracellular factor autotaxin (ATX) as a novel upstream signal modulating HDAC1/2 activity and gene expression in cells of the OLG lineage. Using the zebrafish as an in vivo model system as well as rodent primary OLG cultures, this functional property of ATX was found to be mediated by its lysophospholipase D (lysoPLD) activity, which has been well characterized to generate the lipid signaling molecule lysophosphatidic acid (LPA). More specifically, the lysoPLD activity of ATX was found to modulate HDAC1/2 regulated gene expression during a time window coinciding with the transition from OLG progenitor to early differentiating OLG. In contrast, HDAC1/2 regulated gene expression during the transition from neural stem/progenitor to OLG progenitor appeared unaffected by ATX and its lysoPLD activity. Thus, together, our data suggest that an ATX–LPA–HDAC1/2 axis regulates OLG differentiation specifically during the transition from OLG progenitor to early differentiating OLG and via a molecular mechanism that is evolutionarily conserved from at least zebrafish to rodent.
SIGNIFICANCE STATEMENT The formation of the axon insulating and supporting myelin sheath by differentiating oligodendrocytes (OLGs) in the CNS is considered an essential step during vertebrate development. In addition, loss and/or dysfunction of the myelin sheath has been associated with a variety of neurologic diseases in which repair is limited, despite the presence of progenitor cells with the potential to differentiate into myelinating OLGs. This study characterizes the autotaxin–lysophosphatidic acid signaling axis as a modulator of OLG differentiation in vivo in the developing zebrafish and in vitro in rodent OLGs in culture. These findings provide novel insight into the regulation of developmental myelination, and they are likely to lead to advancing studies related to the stimulation of myelin repair under pathologic conditions.
Introduction
The differentiation of the myelinating cells of the CNS, namely oligodendrocytes (OLGs), follows a stepwise program that is characterized by well coordinated changes in gene expression and cellular morphology (Wegner, 2008; Bauer et al., 2009; Mitew et al., 2014). The developmental timing of this progression along the OLG lineage is finely tuned by extracellular signals and intracellular pathways. In particular, and at the transition from OLG progenitor to differentiating OLG, epigenetic mechanisms associated with a progressive compaction of chromatin have been found to play a central role (Mori and Leblond, 1970; Shen and Casaccia-Bonnefil, 2008; Copray et al., 2009; Liu and Casaccia, 2010; Jacob et al., 2011; Yu et al., 2013; Bischof et al., 2015; Liu et al., 2015). The current understanding of the above mechanisms affecting the chromatin landscape involves histone deacetylation via the action of the class I histone deacetylase (HDAC) members HDAC1 and HDAC2 (Shen et al., 2005, 2008; Ye et al., 2009; Wu et al., 2012). In general, HDAC1/2-containing complexes are considered to remove acetyl groups from histone tails, thereby favoring a condensed chromatin structure and limiting DNA access for transcription factors (Grunstein, 1997). In the case of the OLG lineage, it has been demonstrated that the target genes affected by HDAC1/2 deacetylation include clusters of coregulated genes implicated in transcriptional repression (Swiss et al., 2011). Thus, it is deacetylation-mediated repression of transcriptional inhibitors of OLG differentiation, such as Egr1, Id2, and Sox11, that is thought to promote progression along the OLG lineage. Although the intracellular aspects of this mechanism have been well characterized, little is known about the extracellular signals that may be modulating it (Wu et al., 2012).
In our previous studies, we identified autotaxin (ATX), also known as ENPP2, phosphodiesterase-Iα/ATX, or lysophospholipase D (lysoPLD), as an extracellular factor that promotes OLG differentiation at various steps along the progression of the lineage and via the action of two distinct functionally active domains (Dennis et al., 2005, 2008; Yuelling and Fuss, 2008; Nogaroli et al., 2009; Yuelling et al., 2012). In this regard, the so-called modulator of OLG remodeling and focal adhesion organization (MORFO) domain of ATX was found to regulate primarily the morphological aspects of OLG differentiation (Fox et al., 2003; Dennis et al., 2008, 2012), while its lysoPLD-active site, which generates the lipid signaling molecule lysophosphatidic acid (LPA; Tokumura et al., 2002; Umezu-Goto et al., 2002; van Meeteren and Moolenaar, 2007; Nakanaga et al., 2010), may be predominantly modulating gene expression (Nogaroli et al., 2009). Intriguingly, studies assessing the functions of ATX as a whole and in vivo in the developing zebrafish revealed a lineage promoting, and likely gene expression regulatory, role at the transition from OLG progenitor to differentiating OLG (Yuelling et al., 2012). Thus, we investigated here a possible role of, in particular, the lysoPLD activity of ATX in modulating histone deacetylation and gene expression during the early stages of the OLG lineage.
Materials and Methods
Animals.
Zebrafish embryos were obtained through natural matings, raised at 28.5°C and staged according to morphological criteria and hours postfertilization (hpf; Kimmel et al., 1995). Wild-type fish were of the AB strain and Tg(nkx2.2a:megfp)vu17 (Kirby et al., 2006; Kucenas et al., 2008), abbreviated Tg(nkx2.2a:megfp), as well as Tg(olig2:dsred2)vu19 (Kucenas et al., 2008), abbreviated Tg(olig2:dsred2), fish were kindly provided by Sarah Kucenas (University of Virginia, Charlottesville, VA) and Bruce Appel (University of Colorado Anschutz Medical Campus, Aurora, CO). Sprague Dawley female rats with early postnatal litters were obtained from Harlan Laboratories. All animal studies were approved by the institutional animal care and use committee at Virginia Commonwealth University.
Antibodies.
Supernatants from cultured hybridoma cells (clone A2B5; American Type Culture Collection) were used for immunopanning. Anti-acetyl-histone H3 (Lys 9) antibodies (Cell Signaling Technology) were used for Western blot analysis and immunocytochemistry. Anti-histone H3 antibodies (Cell Signaling Technology) and IRDye 680RD- and 800CW-conjugated secondary antibodies (LI-COR Biosciences) were used for Western blot analysis. Supernatants from cultured hybridoma cells (clone O4; gift from S. E. Pfeiffer, University of Connecticut Health Center, Farmington, CT), anti-caspase-3, active (cleaved) form (EMD Millipore), anti-Ki67 (Leica), and Alexa Fluor 488- and/or Alexa Fluor 568-conjugated secondary antibodies (Life Technologies) were used for immunocytochemistry.
Pharmacological compounds.
Zebrafish embryos and primary cultures of differentiating OLGs were treated with the following compounds: ATX–lysoPLD activity inhibitors S32826 (P-[[4-[(1-oxotetradecyl)amino]phenyl]methyl]-phosphonic acid; Cayman Chemical Company) and HA130 (B-[3-[[4-[[3-[(4-fluorophenyl)methyl]-2,4-dioxo-5-thiazolidinylidene]methyl]phenoxy]methyl]phenyl]-boronic acid; Tocris Bioscience/Bio-Techne), HDAC1/2/3 inhibitor CI994 (4-(acetylamino)-N-(2-aminophenyl)benzamide; Selleckchem) and HDAC6 inhibitor Tubastatin A (N-hydroxy-4-((2-methyl-1,2,3,4-tetrahydropyrido[4,3-b]indol-5-yl)methyl)benzamide hydrochloride; Selleckchem), all of which were dissolved in dimethylsulfoxide (DMSO), resulting in a final experimental concentration of 0.1% DMSO, which is well tolerated in both the zebrafish and the cell culture system (Lyssiotis et al., 2007; Maes et al., 2012). LPA (18:1; Sigma) was dissolved in DMEM containing 0.1% fatty acid-free bovine serum albumin (BSA). Control in all experiments in which pharmacological compounds were used refers to vehicle treatment.
Cell culture.
Primary OLG progenitors were isolated from postnatal day 2 (P2) rat brains by A2B5 immunopanning and cultured as described previously (Barres et al., 1992; Lafrenaye and Fuss, 2010; Martinez-Lozada et al., 2014). Briefly, immunopanned OLG progenitors were plated onto fibronectin (10 μg/ml)-coated tissue culture dishes, glass coverslips, or 96-well plates. Plated OLG progenitors were cultured in serum-free proliferation medium (DMEM containing human PDGF and basic fibroblast growth factor; Gemini Bio-Products) for 24 h, after which cells were allowed to differentiate in serum-free medium [DMEM containing 40 ng/ml tri-iodo-thyronine (T3; Sigma) and N2 supplement (Life Technologies); DMEM/T3/N2] over the time periods indicated. Differentiating OLGs were either directly analyzed or treated as indicated. Typically, at least three independent experiments were performed, whereby an independent experiment refers to an experiment in which cells were isolated from a separate P2 rat litter at an independent time point (day) and treated separately from all other independent experiments.
CRISPR/Cas9 genome editing.
Clustered regularly interspaced short palindromic repeat (CRISPR) target sequences for atx (enpp2), as depicted in Figure 1, were identified using the online tools http://crispr.mit.edu and https://chopchop.rc.fas.harvard.edu (Montague et al., 2014). Short-guide RNAs (sgRNAs) were generated using the cloning-free/short oligonucleotide approach described by Talbot and Amacher (2014). Briefly, short-guide oligonucleotides containing a T7 promoter and genomic target sites were synthesized by Eurofins MWG Operon. Guide-constant oligonucleotides were obtained from Life Technologies. Full-length templates for sgRNA synthesis were generated by PCR using short-guide and guide-constant oligonucleotides, and PCR templates were purified using ISOLATE II PCR cleanup columns (Bioline). The plasmid pT7tyrgRNA (Addgene 46761; kindly provided by Wenbiao Chen; Jao et al., 2013) was used as a template for the generation of the tyrosinase (tyr) control sgRNA. sgRNAs were transcribed from their templates using the MEGAshortscript T7 kit (Life Technologies). Cas9 mRNA was synthesized from the plasmid pT3TS–nCas9n (Addgene 46757; kindly provided by Wenbiao Chen, Vanderbilt University School of Medicine, Nashville, TN; Jao et al., 2013) using the mMESSAGE mMACHINE T3 Transcription kit (Life Technologies). One nanoliter of a mixture of Cas9 mRNA (100 ng/μl) and sgRNA (80 ng/μl for atx–E2; 95 ng/μl for atx–E7; 100 ng/μl for tyr) was injected per one-cell stage zebrafish embryo. Zebrafish were analyzed at 48 hpf. Bright-field images of whole embryos were taken using an Olympus SZX12 zoom stereo microscope (Olympus America) equipped with an Olympus DP70 digital camera system.
Figure 1.
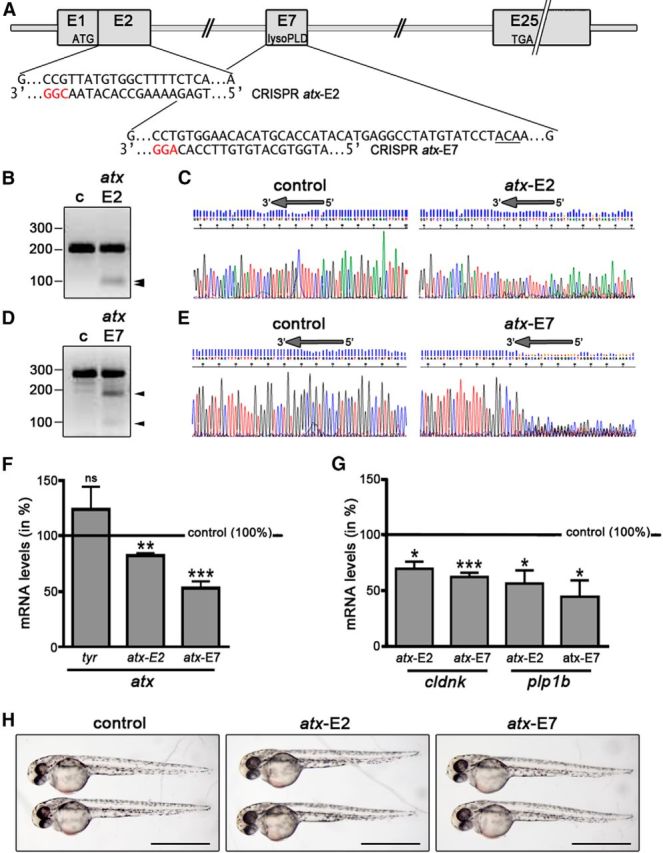
In the developing zebrafish, CRISPR/Cas9-mediated mutagenesis of atx leads to a reduction in the mRNA levels for OLG marker genes. A, atx (enpp2) genomic structure and CRISPR target sequences. ATG and TGA indicate the locations of the translation start and stop sites. Protospacer adjacent motif sequences are highlighted in red. The sequence encoding T210, the threonine residue shown to be obligatory for the enzymatic activity of ATX, is underlined. B, D, Representative images of agarose gels showing DNA fragments subsequent to Surveyor nuclease treatment of control homoduplexes (c) and control/atx-E2 (B) or control/atx-E7 (D) homo/heteroduplexes. Numbers on the left indicate DNA sizes in base pairs. Arrowheads indicate cleaved PCR amplicon fragments at the expected sizes indicative of indel mutations generated by genome editing. C, E, Representative images of sequencing traces obtained from genomic DNA-derived PCR amplicons from control and Cas9 mRNA/sgRNA injected zebrafish embryos. Arrows at the top of each trace indicate the location of the target sequence, over and upstream of which a composite sequence trace indicates the presence of indel mutations generated by genome editing. F, G, Bar graphs illustrating mRNA levels for atx (F) or the OLG marker genes cldnk and plp1b (G) in whole embryos at 48 hpf and as determined by real-time RT-qPCR analysis. Control (uninjected embryos) values were set to 100% (see horizontal line) and experimental values were calculated accordingly. Data shown represent means ± SEM. ns, Not significant; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. H, Representative bright-field images of control (uninjected) and Cas9 mRNA/sgRNA injected embryos at 48 hpf. Scale bars, 1 mm.
Genomic DNA analysis.
Genomic DNA was isolated from control and Cas9 mRNA/sgRNA injected embryos using the DNeasy Blood and Tissue kit (Qiagen). DNA fragments spanning genomic target sites were amplified by PCR using the following primer pairs: atx–E2(ZDB-GENE-040426-1156), forward, 5′-AAATTGTTGAAACCATATGCGA-3′) and reverse, 5′-GCATCCTACTTTTTGAGAGCGA; and atx–E7(ZDB-GENE-040426-1156), forward, 5′-TGTTTAGGTGTGTGCTTAGTGC-3′ and reverse, 5′-AATCTGCAATTAATACATACCGTTG-3′.
To assess for indels generated by CRISPR/Cas9 genome editing, PCR fragments were sequenced (Virginia Commonwealth University Nucleic Acid Research Facilities), and the Surveyor Mutation Detection Kit (Integrated DNA Technologies) was used.
ATX–lysoPLD activity assay.
ATX–lysoPLD activity was determined using the fluorogenic assay as described by Ferguson et al. (2006). Whole zebrafish homogenates diluted in 140 mm NaCl, 5 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 5 mm Tris/HCl, pH 8.0, 1 mg/ml fatty acid-free BSA (Sigma-Aldrich) or concentrated (40×) conditioned medium from primary OLG cultures grown in phenol-red-free DMEM were incubated with 2.5 μm FS-3 substrate (Echelon Biosciences) for 4 h at 37°C. Increase in fluorescence with time was measured at an excitation wavelength of 485 nm and an emission wavelength of 520 nm using a PHERAstar multimode microplate reader (BMG Labtech).
RNA isolation and real-time RT-qPCR analysis.
For the isolation of RNA from whole zebrafish embryos or cultured rodent OLGs, samples were collected in TRIzol (Invitrogen) and homogenized immediately. RNA was extracted as described by Chomczynski and Sacchi (1987). For the isolation of RNA from fluorescence-activated cell sorting (FACS)-isolated cells, cells were collected directly in TRIzol and then immediately processed using the RNeasy Micro Kit from Qiagen. All RNA samples were treated with DNase (DNA-Free Kit; Applied Biosystems/Ambion. RNA concentrations were determined using a Nanodrop ND 1000 Spectrophotometer (Thermo Fisher Scientific/NanoDrop), and purity was assessed by the ratio of absorbance at 260 and 280 nm (OD260/280 ≥ 1.8). Oligo-dT-primed cDNAs were synthesized from 50 ng to 1 μg of RNA using the Omniscript or Sensiscript RT Kit (Qiagen) according to the guidelines of the manufacturer. RNA samples were normalized to the same approximate concentration, and the same amount of RNA was used for all conditions of an individual independent experiment. As control, cDNA reactions without reverse transcriptase were performed for all samples. PCR reactions performed with such no-reverse-transcriptase cDNAs as template yielded quantitation cycle (Cq) numbers that were at least five cycles below the lowest Cq for any of the experimental samples (unknowns), whereby Cq values for unknowns were within the linear quantifiable range as determined by calibration curves. Gene-specific primers were designed and in silico tested for specificity using National Center for Biotechnology Information/Primer-BLAST (basic local alignment search tool; Ye et al., 2012). All primers were designed to amplify all known splice variants. For each primer pair listed below, amplicon length is noted in base pairs. In addition, PCR efficiencies are given in percentage and correlation coefficients (r2 values) as single numbers, whereby the latter were determined from calibration curves using RNA samples derived from species and tissue/cell type equivalent sources.
The following unmodified zebrafish gene-specific primer pairs were used: atx/enpp2(ZDB-GENE-040426-1156), forward, 5′-CAATGTATGCAGCATTCAAACGAGTG-3′ (exon 22), and reverse, 5′-CACCATTTTTCTCACTAGCGTAACG-3′ (exon 23), 79 bp, 107%, 0.996; plp1b(ZDB-GENE-030710-6), forward, 5′-TGCCATGCCAGGGGTTGTTTGTGGA-3 (exon 5), and reverse, 5′-GGCGACCATGTAAACGAACAGGGC-3 (exon 6/7), 143 bp, 96%, 1000; cldnk(ZDB-GENE-040801-201), forward, 5′-TGGCATTTCGGCTCAAGCTCTGGA-3′ (exon 1), and reverse, 5′-GGTACAGACTGGGCAATGGACCTGA-3 (exon 2), 135 bp, 93%, 0.999; inab (gefiltin)(ZDB-GENE-990415-83), forward, 5′-CTCTCCATCCCGGCTGAGCAGCT-3′ (exon 1), and reverse, 5′-AGCCTGGGATGTTGGGGCGGTT-3′ (exon 1), 74 bp, 105%, 0.997; gfap(ZDB-GENE-990914-3), forward, 5′-TGAGACAAGCGAAGCAGGAGGCCA-3′ (exon 5), and reverse, 5′-GGCGCTCCAGGGACTCATTAGACCC-3′ (exon5/6), 102 bp, 101%, 0.997; prph (plasticin)(ZDB-GENE-990415-207), forward, 5′-ACTCCTACCGCACCTCTCACCACCG-3′ (exon 1), and reverse, 5′-TAGCGGCCCCCGATGCGACTG-3′ (exon 1), 70 bp, 93%, 0.997; and sox10(ZDB-GENE-011207-1), forward, 5′-TGGACACCACCCTCACGCTA-3′ (exon 3), and reverse, 5′-CTGCAGTTCCGTCTTGGGGG-3′ (exon 4), 76 bp, 94%, 0.966. β-Actin [actb2(ZDB-GENE-000329-3); Buckley et al., 2010] was used as reference gene (60 bp, 95%, 0.999); eef1a1/1(ZDB-GENE-990415-52) has been assessed as an additional reference gene for the analysis of RNA derived from the developing zebrafish, and no pronounced differences were noted (Yuelling et al., 2012).
The following unmodified rat gene-specific primer pairs were used: Cnp(NM_012809.2), forward, 5′-ATGCCCAACAGGATGTGGTG-3′ (exon 3/4) and reverse, 5′-AGGGCTTGTCCAGGTCACTT-3′ (exon 4), 150 bp, 103%, 0.990; Ugt8(NM_019276.3), forward, 5′-AGGAGCTCTGGGGAGATTGC-3′ (exon 3) and reverse, 5′-TTTGAATGGCCAAGCAGGTCA-3′ (exon 4/5), 126 bp, 108%, 0.960; Egr1(NM_012551.2), forward, 5′-CCTGACCACAGAGTCCTTTTCT-3′ (exon 1/2) and reverse, 5′-AAAGTGTTGCCACTGTTGGG-3′ (exon 2), 150 bp, 93%, 0.996; and Pgk1(NM_053291.3) (as reference gene), forward, 5′-ATGCAAAGACTGGCCAAGCTAC-3′ (exon 8) and reverse, 5′-AGCCACAGCCTCAGCATATTTC-3′ (exon 9), 103 bp, 103%, 0.997.
Pgk1 was used as reference gene due to its previously established expression stability in rat OLGs (Nelissen et al., 2010). In addition, Ppia(NM_017101.1) has been assessed as additional reference gene for the analysis of RNA derived from differentiating OLG cultures, and no pronounced differences were noted.
RT-qPCR reactions with at least two technical replicates per sample were performed on a CFX96 real-time PCR detection system (Bio-Rad) using the iQ SYBR Green Supermix (Bio-Rad). PCR conditions were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 58°C for 30 s, and 95°C for 10 s. For all primer pairs, melting curves were used to ensure specificity. Relative expression levels were determined using the ΔΔCT method (Livak and Schmittgen, 2001).
Whole-mount in situ hybridization.
Zebrafish embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C and stored in methanol at 22°C for at least 1 d. Colorimetric in situ hybridizations using digoxigenin-labeled antisense cRNA probes were performed by standard methods and as described previously (Thisse and Thisse, 2008, 2014; Yuelling et al., 2012). In situ hybridized embryos were mounted in 90% glycerol/PBS, and whole-mount images were acquired using the extended focus module of the Axiovision software package in combination with an Axio Observer Z.1 or SteREO Discovery.V20 microscope equipped with an AxioCam MRc digital camera (Carl Zeiss). Once captured, images were imported into Adobe Photoshop, and adjustments were limited to cropping and brightness/contrast adjustments that were applied equally across the entire image and to controls. In situ-labeled cells were counted using the Cell Counter plugin to the NIH ImageJ software package (Abramoff et al., 2004).
Isolation of OLG progenitors from zebrafish embryos using FACS.
Tg(nkx2.2a:megfp;olig2:dsred2) zebrafish embryos (up to 300 per sample) were anesthetized with tricaine methanesulfonate (MS-222; Sigma-Aldrich) and subjected to enzymatic (trypsin and DNase) and mechanical (20 and 26 gauge needle) dissociation. Cell suspensions were resuspended first in 1 ml and then in 500 μl of ice-cold 5% goat serum in PBS after centrifugation (at 956 × g for 2 min at 4°C) and then subjected once again to mechanical dissociation (26 gauge needle). Cell suspensions were passed through a 40 μm filter and then through a fluorescence-activated cell sorter (FACS Aria II; BD Biosciences) using an 85 μm nozzle. Cell sorting was performed at room temperature using a 488 nm laser (>20 mW power, solid state; Coherent) for excitation and 530/30 and 585/42 nm bandpass filters for the collection of emission. Settings were carefully determined empirically and reproduced exactly in each experiment. Gates were demarcated to sort only mEGFP/DsRed2 double-positive cells. The speed of sorting (usually no more than one to two drops per event) was adjusted to obtain a purity of >95%. Cells were suspended in either PBS (HDAC activity assay) or TRIzol (RNA isolation).
HDAC activity assay.
FACS-isolated cells were plated into 96-well plates (1 × 104 cells per well) and assayed directly. Rodent OLG progenitor cells were cultured and treated in fibronectin-coated 96-well plates (1 × 105 cells per well) and then assayed. HDAC activity was determined using the fluorometric HDAC Activity Assay Kit from BioVision. Briefly, the cell-permeable HDAC substrate Boc-Lys(AC)-AMC was added, and cells were incubated for 3 h at 37°C. Subsequent incubation with the lysine developer (30 min at 37°C) was used to produce a fluorophore from substrate sensitized through deacetylation. Fluorescence, as a measure of HDAC activity, was determined using a Spectra Max M5 fluorescent plate reader (Molecular Devices) with excitation/emission at 360/460 nm. In each experiment, triplicates were prepared for all conditions and treatments.
Immunocytochemistry.
For immunocytochemistry using O4 hybridoma supernatants, cells were fixed in 4% paraformaldehyde/PBS, nonspecific binding sites were blocked in 10% FCS/DMEM, and cells were incubated with the supernatant (1:1 diluted in 10% FCS/DMEM) overnight. Cells were fixed and, in case of dual staining, permeabilized using 0.5% Triton X-100/0.4 m sucrose/PBS and then incubated for 30 min in blocking solution (10% FCS/DMEM). Subsequently, cells were incubated overnight with anti-acetyl-histone H3 (Lys 9), anti-caspase-3 [active (cleaved) form], or anti-Ki67 antibodies overnight. Primary antibodies were detected using Alexa Fluor 488- or Alexa Fluor 568-conjugated secondary antibodies (Life Technologies), and nuclei were counterstained using Hoechst 33342 (EMD Millipore). For the generation of representative images, confocal laser scanning microscopy was used (LSM 700; Carl Zeiss). Images represent 2D maximum projections of stacks of 0.5 μm optical sections.
Nuclear histone acetylation analysis.
Cells double-labeled for O4 and acetyl-histone H3 (Lys 9) were imaged using confocal laser scanning microscopy (LSM 700; Carl Zeiss). Single focal plane images were taken using a 40×/1.3 numerical aperture plan-apochromat objective lens. Images were taken from O4-positive cells and focused on the region of the nucleus as marked by staining with Hoechst. Laser intensity, detector gain, and amplifier offset were optimized and kept constant throughout the imaging of all samples from one experiment. Fluorescence intensity, i.e., average pixel intensity, for the anti-acetyl histone H3 (Lys 9) signal, as a measure for the level of histone acetylation, was determined over the area of the nucleus using IP Lab imaging software (BD Biosciences). Unprocessed confocal images were used for analysis, and threshold settings were kept constant once determined.
Western blot analysis.
Cells were homogenized in lysis buffer (150 mm NaCl, 10 mm KCl, 20 mm HEPES, pH 7.0, 1 mm MgCl2, 20% glycerol, and 1% Triton X-100) including the complete protease and phosphatase inhibitor mixture (Thermo Fisher Scientific). Five micrograms per sample were used for Western blot analysis to detect acetyl histone H3 (Lys 9) and total H3. Bound primary antibodies were detected using IRDye 680RD- and 800CW-conjugated secondary antibodies (LI-COR Biosciences). Signal intensities were determined using an Odyssey infrared imaging system (LI-COR Biosciences).
Cell survival and proliferation analysis.
Cells were subjected to immunostaining using O4 hybridoma cell supernatants and antibodies specifically recognizing the active (cleaved) form of caspase-3 or the Ki67 antigen. To determine the number of caspase-3 or Ki67 immunopositive cells, images of four fields per coverslip were taken with a 20×/0.8 numerical aperture plan-apochromat objective lens using a confocal laser scanning microscope (LSM 700; Carl Zeiss). Three coverslips per condition for each of three independent experiments were analyzed, and Hoechst 33342-positive nuclei as well as caspase-3 or Ki67 immunopositive OLGs were counted using the Cell Counter plugin to the NIH ImageJ software package (Abramoff et al., 2004).
Statistical analysis.
To determine significance, GraphPad Prism (GraphPad Software) or SigmaPlot (Systat Software) software was used. Data composed of two groups were analyzed using the two-tailed Student's t test or the Mann–Whitney U test and presented in bar graphs depicting means ± SEMs or box and whisker plots depicting medians and quartiles. Data compared with a set control value lacking variability were analyzed using the one-sample t test (Skokal and Rohlf, 1995; Dalgaard, 2008) and presented in bar graphs.
Results
In the developing zebrafish hindbrain, ATX promotes the timely appearance of differentiating OLGs via its lysoPLD activity
Our previous studies, using antisense morpholino oligonucleotide-mediated knock-down of atx expression, revealed that, in the developing zebrafish hindbrain, ATX promotes the timely appearance of cells committed to the OLG lineage without affecting the number of olig2-positive progenitor cells, the overall morphology of the axonal network, or the differentiation of somatic abducens motor neurons (Yuelling et al., 2012). Because recent studies have questioned the correlation between morpholino oligonucleotide-induced and mutant phenotypes in the zebrafish (Kok et al., 2015), we used CRISPR/Cas9 genome editing to substantiate the above proposed in vivo role of ATX. For these studies, we used the codon-optimized Cas9 with nuclear localization signals system described by Jao et al. (2013). This optimized system has been described to lead to an increased frequency of biallelic disruption so that injected (F0) embryos phenocopy known mutant phenotypes (Jao et al., 2013). As shown in Figure 1A–E, injection of guide RNAs against exon 2 or 7 of the atx (enpp2) gene and in vitro transcribed, capped, polyadenylated nls–zCas9–nls RNA into one-cell stage zebrafish embryos resulted in the generation of indel mutations over and upstream of the CRISPR targeting sequence. These mutations resulted in reduced levels of atx mRNA (Fig. 1F; atx–E2, n = 4, t test, p = 0.0038; atx–E7, n = 6, t test, p = 0.0006), an effect that was not observed when using a guide RNA against tyr (tyr, n = 3, t test, p = 0.3468). Most importantly, the CRISPR/Cas9-mediated reduction in atx expression was found associated with a decrease in the transcript levels for the later-stage OLG differentiation marker genes claudin K (cldnk) and proteolipid protein (plp1b; Fig. 1G; cldnk atx–E2, n = 4, t test, p = 0.0188; cldnk atx–E7, n = 5, t test, p = 0.0006; plp1b atx–E2, n = 6, t test, p = 0.0155; plp1b atx–E7, n = 5, t test, p = 0.0203). As shown in Figure 1H, injection of Cas9 mRNA with either of the guide RNAs was not found to be associated with changes in the gross morphology of the embryos. Therefore, the above described data corroborate our previous morpholino oligonucleotide-mediated knock-down results and strongly support a critical in vivo role of ATX in the regulation of OLG differentiation.
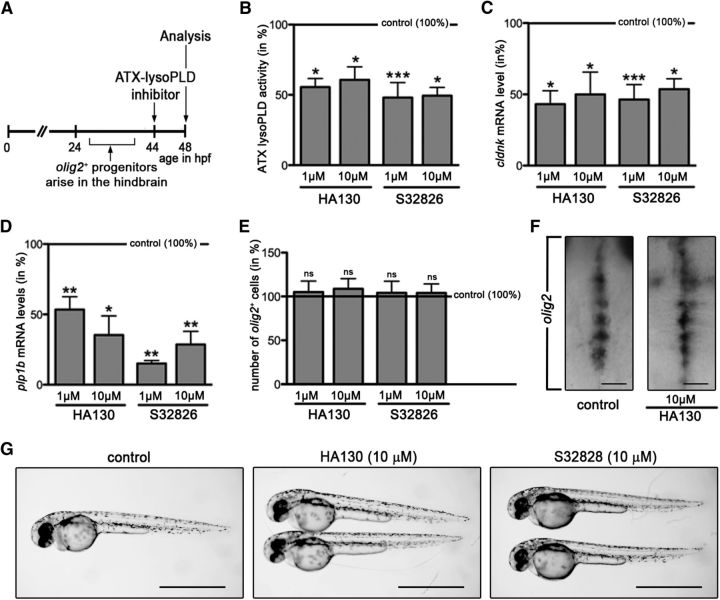
To assess the extent to which this functional role of ATX may be dependent on its lysoPLD activity, the inhibitors HA130 and S32826, which have been well characterized to block the lysoPLD activity of ATX (Ferry et al., 2008; Albers et al., 2010), were used. Of these inhibitors, HA130 has been demonstrated previously to effectively block the lysoPLD activity of ATX in the developing zebrafish at concentrations similar to the ones found to block human ATX (Lai et al., 2012). Two experimental designs were used (Figs. 2A, 3A). First, to determine the effect of an inhibition of the lysoPLD activity of ATX on OLG differentiation, inhibitors were added to zebrafish embryos at 44 h hpf, i.e., at a time point when OLG progenitors arise in the ventral hindbrain and start to differentiate (Zannino and Appel, 2009; Fig. 2A). Zebrafish embryos were analyzed at 48 hpf, a time point at which transcripts for cldnk and plp1b start to become detectable (Brösamle and Halpern, 2002; Takada and Appel, 2010; Münzel et al., 2012). As shown in Figure 2B, treatment with either of the inhibitors at both 1 and 10 μm concentrations significantly reduced the lysoPLD activity of ATX (HA130 at 1 μm, n = 4, t test, p = 0.0166; HA130 at 10 μm, n = 4, t test, p = 0.0132; S32826 at 1 μm, n = 6, t test, p = 0.0008; S32826 at 10 μm, n = 5, t test, p = 0.0131). Importantly, both inhibitors significantly reduced the mRNA levels for cldnk and plp1b at both concentrations used (Fig. 2C,D; cldnk: HA130 at 1 μm, n = 3, t test, p = 0.0144; HA130 at 10 μm, n = 3, t test, p = 0.0413; S32826 at 1 μm, n = 3, t test, p = 0.0006; S32826 at 10 μm, n = 3, t test, p = 0.0166; plp1b: HA130 at 1 μm, n = 5, t test, p = 0.0038; HA130 at 10 μm, n = 5, t test, p = 0.0325; S32826 at 1 μm, n = 5, t test, p = 0.0071; S32826 at 10 μm, n = 5, t test, p = 0.0031). In contrast, there was no change in the number of olig2-positive progenitors (Fig. 2E,F; HA130 at 1 μm, n = 5, t test, p = 0.7184; HA130 at 10 μm, n = 5, t test, p = 0.5050; S32826 at 1 μm, n = 5, t test, p = 0.7679; S32826 at 10 μm, n = 5, t test, p = 0.7277), which at 48 hpf represent early stages of both motor neuron and OLG lineages (Park et al., 2002; Zannino and Appel, 2009). It is notable that neither S32826 nor HA130 were found to exert cytotoxic effects when treating zebrafish embryos or cells in culture at the concentrations used here (Ferry et al., 2008; Albers et al., 2010; Iyer et al., 2012, Lai et al., 2012; see also the description to Fig. 7 below). Consistently, no gross morphological defects were noted during the treatment of zebrafish embryos with HA130 or S32826 (Fig. 2G), and no noticeable increase in 7-aminoactinomycin D staining, as a measure for increased cell death (Philpott et al., 1996), was detected when analyzing single-cell suspensions from control and inhibitor-treated whole fish embryos by flow cytometry (data not shown). Hence, the data shown in Figure 2 suggest that the lysoPLD activity of ATX, as ATX as a whole (Yuelling et al., 2012), regulates the progression of olig2-positive progenitor cells into lineage committed and differentiating OLGs but not the appearance of olig2-positive progenitors itself.
Figure 2.
In the developing zebrafish, inhibition of the lysoPLD activity of ATX during early stages of OLG differentiation leads to a reduction in the levels of OLG-enriched transcripts without apparent effects on the number of olig2-positive cells. A, Experimental design. Embryos were treated with the ATX–lysoPLD inhibitors HA130 and S32826 at a time point when newly specified OLG progenitors start to differentiate. B, Bar graph depicting ATX–lysoPLD activity in whole embryos as assessed by using the fluorogenic substrate FS-3. C, D, Bar graphs illustrating mRNA levels for the OLG marker genes cldnk (C) and plp1b (D) in whole embryos as determined by real-time RT-qPCR analysis. E, Bar graph showing the number of olig2-positive progenitors in whole embryos as determined through whole-mount in situ hybridization. F, Representative extended focus images of whole-mount embryos after in situ hybridization with a probe specific for olig2. Dorsal views over the hindbrain are shown with anterior to the top. Scale bars, 50 μm. G, Representative bright-field images of control (vehicle-treated) and ATX–lysoPLD inhibitor-treated embryos at 48 hpf. Scale bars, 1 mm. For all bar graphs, control (vehicle-treated) values were set to 100% (see horizontal line), and experimental values were calculated accordingly. Data shown represent means ± SEMs. ns, Not significant; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
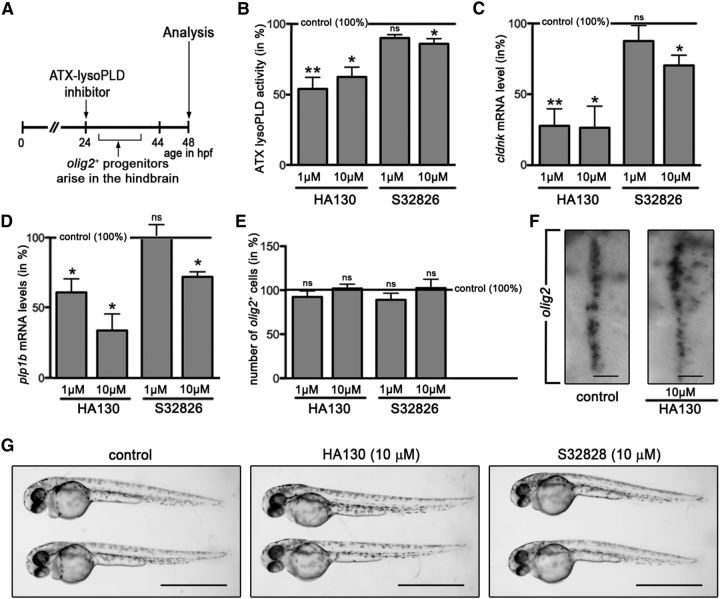
Figure 3.
In the developing zebrafish, inhibition of the lysoPLD activity of ATX during the time window when olig2-positive progenitors arise in the hindbrain leads to a reduction in the levels of OLG-enriched transcripts without apparent effects on the number of olig2-positive cells. A, Experimental design. Embryos were treated with the ATX–lysoPLD inhibitors HA130 and S32826 during the time window when olig2-positive progenitors arise in the ventral hindbrain. B, Bar graph depicting ATX–lysoPLD activity in whole embryos as assessed by using the fluorogenic substrate FS-3. C, D, Bar graphs illustrating mRNA levels for the OLG marker genes cldnk (C) and plp1b (D) in whole embryos as determined by real-time RT-qPCR analysis. E, Bar graph showing the number of olig2-positive progenitors in whole embryos as determined through whole-mount in situ hybridization. F, Representative extended focus images of whole-mount embryos after in situ hybridization with a probe specific for olig2. Dorsal views over the hindbrain are shown with anterior to the top. Scale bars, 50 μm. G, Representative bright-field images of control (vehicle-treated) and ATX–lysoPLD inhibitor-treated embryos at 48 hpf. Scale bars, 1 mm. For all bar graphs, control (vehicle-treated) values were set to 100% (see horizontal line), and experimental values were calculated accordingly. Data shown represent means ± SEMs. ns, Not significant; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
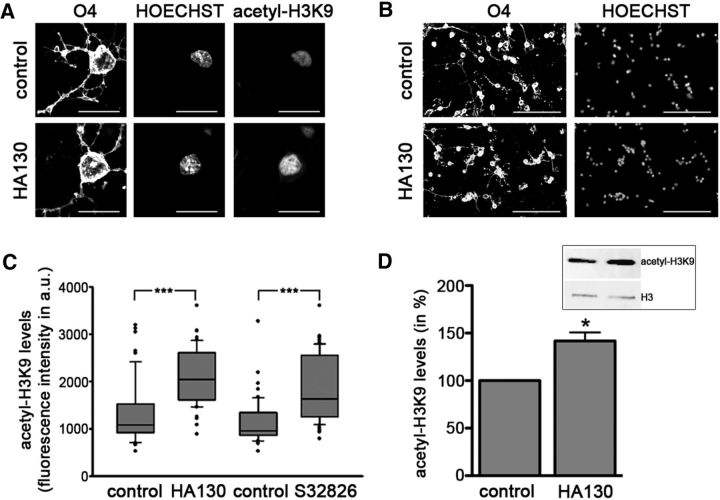
Figure 7.
In rodent OLG cultures, inhibition of the lysoPLD activity of ATX leads to an increase in nuclear histone acetylation at H3K9. A, B, Representative confocal images of differentiating OLGs treated with vehicle (control, top) or HA130 (bottom) and immunolabeled using O4 hybridoma supernatants (A, B) and anti-acetyl-histone H3K9 antibodies (A). Nuclei are visualized via staining with Hoechst (A, B). Scale bars: A, 20 μm; B, 100 μm. C, Box and whisker plot depicting nuclear acetyl-H3K9 levels as assessed by determining fluorescence intensities shown in arbitrary units (a.u.). The plot depicts medians and quartiles of three independent experiments. Whiskers represent the 10th and 90th percentiles. ***p ≤ 0.001. D, Bar graph illustrating acetyl-H3K9 levels as assessed by Western blot analysis. A representative Western blot image is shown in the inset (top right). For the bar graph, control (vehicle-treated) values were set to 100% (see control bar), and experimental values were calculated accordingly. Data represent means ± SEMs. *p ≤ 0.05.
To further confirm the above conclusion, ATX–lysoPLD inhibitors were added at a time point at which olig2-positive progenitors begin to arise in the hindbrain, i.e., at 24 hpf (Zannino and Appel, 2009; Fig. 3A). As shown in Figure 3B–E, treatment with HA130 had a comparable effect in both experimental designs (ATX activity: HA130 at 1 μm, n = 5, t test, p = 0.0051; HA130 at 10 μm, n = 4, t test, p = 0.0130; cldnk: HA130 at 1 μm, n = 4, t test, p = 0.0037; HA130 at 10 μm, n = 5, t test, p = 0.0167; plp1b: HA130 at 1 μm, n = 4, t test, p = 0.0272; HA130 at 10 μm, n = 4, t test, p = 0.0111; olig2: HA130 at 1 μm, n = 5, t test, p = 0.1867; HA130 at 10 μm, n = 5, t test, p = 0.8770). Interestingly, treatment with S32826, when applied at 24 hpf, did not cause a significant effect on the lysoPLD activity of ATX or cldnk/plp1b mRNA levels when used at 1 μm and a much-reduced effect when used at 10 μm (Fig. 3B–E; ATX activity: S32826 at 1 μm, n = 3, t test, p = 0.0566; S32826 at 10 μm, n = 4, t test, p = 0.0322; cldnk: S32826 at 1 μm, n = 5, t test, p = 0.3999; S32826 at 10 μm, n = 5, t test, p = 0.0145; plp1b: S32826 at 1 μm, n = 3, t test, p = 0.9437; S32826 at 10 μm, n = 3, t test, p = 0.0162; olig2: S32826 at 1 μm, n = 5, t test, p = 0.2823; S32826 at 10 μm, n = 5, t test, p = 0.8073). This lack of effective inhibition of the lysoPLD activity of ATX is likely attributable to the known poor in vivo stability and/or bioactivity of S32826 (Ferry et al., 2008, Gupte et al., 2011). As in the studies to the experimental design shown in Figure 2, no gross morphological defects (Fig. 3G) or noticeable increase in cell death (data not shown) were noted.
Together, the above data demonstrate that inhibition of the lysoPLD activity of ATX mimics the effects on the OLG lineage as seen during morpholino oligonucleotide-mediated knockdown of atx expression (Yuelling et al., 2012) or CRISPR/Cas9-mediated genome editing at the atx locus (Fig. 1). Thus, they establish that it is the lysoPLD activity of ATX that is necessary for the timely appearance of differentiating OLGs in the developing zebrafish hindbrain.
In the developing zebrafish, ATX–lysoPLD promoted progression along the early stages of the OLG lineage is associated with an increase in HDAC activity
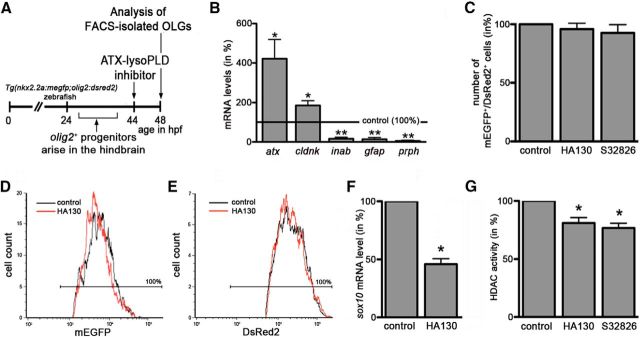
The progression along the early stages of the OLG lineage, as seen regulated by the lysoPLD activity of ATX, has been well established to be associated with epigenetic modifications and in particular with an increase in histone deacetylation (Marin-Husstege et al., 2002; Shen et al., 2005, 2008; He et al., 2007; Liu and Casaccia, 2010; Takada and Appel, 2010; Swiss et al., 2011). In addition, in studies unrelated to ATX, it has been shown that LPA, for which enzymatic lysoPLD activity represents one of the known biosynthetic pathways (Aoki et al., 2008), can increase histone deacetylation in a number of human cancer cell lines (Ishdorj et al., 2008). Thus, and in an effort to identify downstream targets of the lysoPLD activity of ATX involved in the regulation of OLG differentiation, we focused on HDACs and their activity. For these studies, the experimental design as depicted in Figure 4A was used. In addition, and to enable an analysis of HDAC activity explicitly in cells of the OLG lineage, the double-transgenic zebrafish line Tg(nkx2.2a:megfp;olig2:dsred2) was used in which a subset of OLG lineage cells can be identified by the concurrent expression of mEGFP and DsRed2 (Kucenas et al., 2008) and thus be isolated by FACS. In agreement with an OLG lineage commitment, such sorted and double-positive cells were characterized under control conditions by an enriched expression of the OLG marker gene cldnk (Fig. 4B; n = 5, t test, p = 0.0262, compared with whole fish embryo expression). In contrast, there were, if at all, only slight levels of expression detectable for the radial/ependymal/enteric glia marker gfap (Fig. 4B; n = 4, t test, p = 0.0023, compared with whole fish embryo expression; Bernardos and Raymond, 2006; Lam et al., 2009; Doodnath et al., 2010; Grupp et al., 2010; Hagström and Olsson, 2010) and the neuronal intermediate filament encoding genes gefiltin (inab; Fig. 4B; n = 4, t test, p = 0.0014, compared with whole fish embryo expression) and plasticin (prph; Fig. 4B; n = 4, t test, p = 0.0019, compared with whole fish embryo expression; Leake et al., 1999). In addition, and similar to the rodent system (Savaskan et al., 2007), such early stages of the OLG lineage were found to express atx (Fig. 4B; n = 4, t test, p = 0.0464). It is notable that such early, and relatively low, expression during the OLG lineage remained undetected in our previous studies (Yuelling et al., 2012) because of the use of less sensitive detection methods. Upon inhibition of the lysoPLD activity of ATX, and consistent with the previously observed lack of an effect on the number of olig2-expressing progenitor cells (Fig. 2E,F), no change in the number of double-positive cells was observed (Fig. 4C; HA130, n = 4, t test, p = 0.4722; S32826, n = 4, t test, p = 0.3765). To further assess potential effects of ATX–lysoPLD inhibition on progenitor marker expression levels, the fluorescence intensities for mEGFP and DsRed2 were monitored, because they are indicators for nkx2.2a and olig2 promoter activities. As shown in Figure 4, D and E, no changes were observed (n = 5, t test, p = 0.5855, mEGFP; n = 5, t test, p = 0.6436, DsRed2). In contrast, and in agreement with our previous findings (Fig. 2C,D), the expression of sox10, which at the developmental stage analyzed marks olig2/nkx2.2a double-positive cells that are OLG lineage committed and differentiating (Kucenas et al., 2008), was found to be decreased (Fig. 4F; HA130, n = 4, t test, p = 0.0358). Importantly, such a reduction in the expression of a gene characteristic for recently specified and differentiating OLGs was associated with a reduction in overall HDAC activity (Fig. 4G; HA130, n = 4, t test, p = 0.0275; S32826, n = 4, t test, p = 0.0106). This effect was found to not be associated with a noticeable decrease in cell viability as assessed by propidium iodide staining and flow cytometry (Sasaki et al., 1987; data not shown).
Figure 4.
In the developing zebrafish, inhibition of the lysoPLD activity of ATX during early stages of OLG differentiation leads to a reduction in HDAC activity within cells of the OLG lineage. A, Experimental design using Tg(nkx2.2a:megfp;olig2:dsred2) zebrafish embryos, which were treated with the ATX–lysoPLD inhibitors HA130 (10 μm) or S32826 (10 μm) at a time point when newly specified OLG progenitors start to differentiate. B, Bar graph illustrating the mRNA expression profile under control conditions for FACS-isolated mEGFP/DsRed2 double-positive cells as determined by real-time RT-qPCR analysis. mRNA levels in whole fish were set to 100% for each gene individually (marked by the horizontal line), and the values for sorted cells were calculated accordingly. C, Bar graph showing the number of mEGFP/DsRed2 double-positive cells as determined by FACS analysis. Values for vehicle-treated embryos were set to 100% (see control bar), and experimental values were calculated accordingly. D, E, Representative single-parameter fluorescence histograms (x-axis: fluorescence intensity, logarithmic scale; y-axis: cell number) depicting mEGFP (D) or DsRed2 (E) intensities of mEGFP/DsRed2 double-positive cells under control conditions (black line) and after treatment with HA130 (red line). Cells within the area marked by the horizontal line were considered mEGFP (D) or DsRed2 (E) positive. Note the high purity (100%) of the sorted cell population. F, Bar graph depicting sox10 mRNA levels for FACS-isolated mEGFP/DsRed2 double-positive cells as determined by real-time RT-qPCR analysis. Values for vehicle-treated embryos were set to 100% (see control bar), and experimental values were calculated accordingly. G, Bar graph depicting HDAC activity as determined by using the fluorogenic substrate Boc-Lys(Ac)-AMC. Values for vehicle-treated embryos were set to 100% (see control bar), and experimental values were calculated accordingly. Data shown in all bar graphs represent means ± SEMs. *p ≤ 0.05, **p ≤ 0.01.
Together, the above data demonstrate that, in the developing zebrafish, ATX, via its lysoPLD activity, promotes the lineage progression from an olig2/nkx2.2a double-positive progenitor cell to a sox10-expressing fully committed and early differentiating OLG by a mechanism that is associated with an increase in overall HDAC activity.
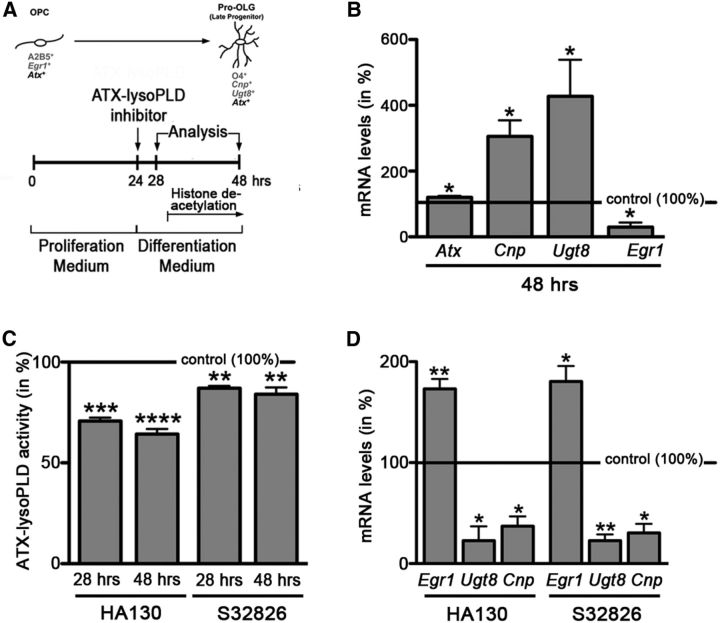
In enriched primary cultures of rodent OLG lineage cells, the lysoPLD activity of ATX promotes OLG differentiation
To more precisely define the functional correlation between the lysoPLD activity of ATX and changes in HDAC activity/histone acetylation and to assess evolutionary conservation of this mechanism, we turned to the well established culture system of enriched primary rodent OLG lineage cells. In this culture system, OLG differentiation follows the same sequence of events as observed in vivo but provides the advantage of studying intrinsic cellular mechanisms (Temple and Raff, 1985). In addition, the role of HDACs and histone acetylation has been well characterized in this system (Marin-Husstege et al., 2002; Swiss et al., 2011). More specifically, an increase in histone deacetylation as early as 6 h after mitogen withdrawal has been found crucial for the transition from a proliferating OLG progenitor to a differentiating OLG (Marin-Husstege et al., 2002). To establish the role of the lysoPLD activity of ATX on OLG differentiation during this developmental time window, we used the experimental design as depicted in Figure 5A. First, we established the expression profile of transcriptional targets shown previously to be regulated by histone deacetylation, namely 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (Cnp), UDP glycosyltransferase 8 (Ugt8), and the transcriptional regulator Egr1 (Swiss et al., 2011). In agreement with the findings described by Swiss et al. (2011), OLG differentiation was found in our system to be characterized by an increase in the expression of the OLG marker genes Cnp and Ugt8 and a decrease in the expression of Egr1 (Fig. 5B; Cnp, n = 5, t test, p = 0.0134; Ugt8, n = 5, t test, p = 0.0429; Egr1, n = 3, t test, p = 0.0362). In addition, there was a slight increase in the expression of Atx (Fig. 5B; n = 5, t test, p = 0.0118). As shown in Figure 5C, treatment with either of the ATX–lysoPLD activity inhibitors HA130 or S32826 significantly reduced ATX–lysoPLD activity after 4 and 24 h (Fig. 5C; HA130 at 10 μm 28 h, n = 4, t test, p = 0.0004; HA130 at 10 μm 48 h, n = 5, t test, p = 0.0001; S32826 at 10 μm 28 h, n = 4, t test, p = 0.0078; S32826 at 10 μm 48 h, n = 5, t test, p = 0.0095). This inhibition of ATX–lysoPLD activity resulted in an attenuation of both the upregulation of Cnp and Ugt8 expression and the downregulation of Egr1 expression (Fig. 5D; Cnp: HA130 at 10 μm, n = 3, t test, p = 0.0224; S32826 at 10 μm, n = 3, t test, p = 0.0159; Ugt8: HA130 at 10 μm, n = 3, t test, p = 0.0321; S32826 at 10 μm, n = 3, t test, p = 0.0063; Egr1: HA130 at 10 μm, n = 4, t test, p = 0.0051; S32826 at 10 μm, n = 3, t test, p = 0.0345). The above findings demonstrate that ATX, via its lysoPLD activity and similar to what we have seen in the developing zebrafish (Figs. 1–4), promotes gene expression changes associated with OLG differentiation in primary cultures of rodent OLGs.
Figure 5.
In rodent OLG cultures, inhibition of the lysoPLD activity of ATX leads to a reduction in the levels of mRNAs encoding OLG differentiation genes and to an increase in the level of mRNA encoding the transcriptional OLG differentiation inhibitor Egr1. A, Experimental design. Timing of histone deacetylation is marked as described by Marin-Husstege et al. (2002). OPC, OLG progenitor. B, Bar graph depicting mRNA levels for the OLG differentiation genes Cnp and Ugt8 and the transcriptional inhibitor Egr1, as well as Atx as determined by real-time RT-qPCR analysis. mRNA levels at 24 h were set to 100% for each gene, and 48 h values were calculated accordingly. C, Bar graph illustrating ATX–lysoPLD activity as assessed by using the fluorogenic substrate FS-3. Control (vehicle-treated) values were set to 100% (see horizontal line), and experimental values were calculated accordingly. D, Bar graph depicting mRNA levels for the OLG differentiation genes Cnp and Ugt8 and the transcriptional inhibitor Egr1 as determined by real-time RT-qPCR analysis. Control (vehicle-treated) values were set to 100% (see horizontal line), and experimental values were calculated accordingly. Data shown in all bar graphs represent means ± SEMs. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
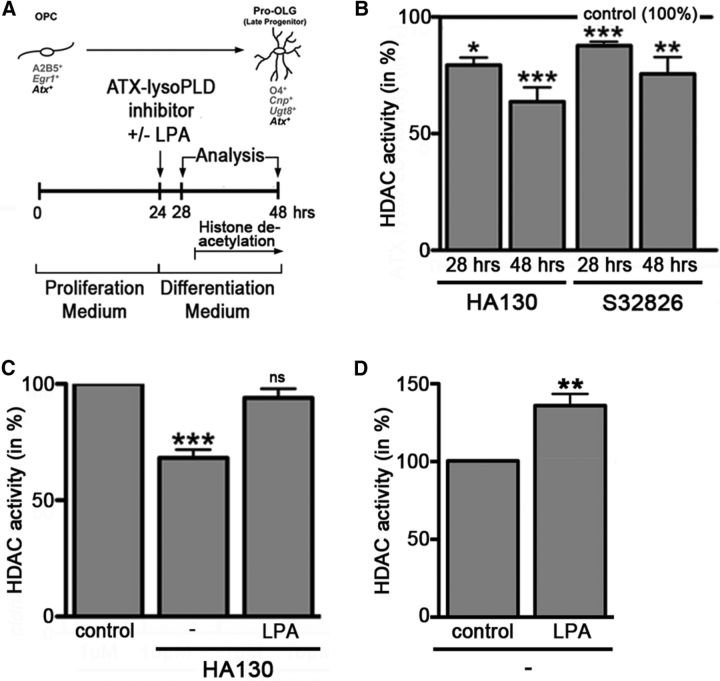
In primary cultures of rodent OLGs, ATX–lysoPLD promoted progression along the early stages of the OLG lineage is associated with an increase in HDAC activity
The gene expression changes observed during inhibition of the lysoPLD activity of ATX (Fig. 5D) mimicked those changes described previously to occur during inhibition of HDAC activity (Swiss et al., 2011), thus suggesting that, as in the zebrafish, these changes may be associated with a decrease in overall HDAC activity. To assess this possibility, the experimental design as shown in Figure 6A was used. Indeed, HDAC activity was found to be decreased as early as 4 h and up to 24 h after application of ATX–lysoPLD inhibitors (Fig. 6B) (HA130 at 10 μm 28 h, n = 5, t test, p = 0.0280; HA130 at 10 μm 48 h, n = 7, t test, p = 0.0001; S32826 at 10 μm 28 h, n = 3, t test, p = 0.0004; S32826 at 10 μm 48 h, n = 5, t test, p = 0.0034). To provide additional evidence for a direct link between the lysoPLD activity of ATX and the regulation of HDAC activity, we performed rescue experiments in which LPA, the enzymatic product of the lysoPLD activity of ATX, was added concurrent with ATX–lysoPLD inhibitors. As shown in Figure 6C, addition of LPA (10 μm) attenuated the effect of HA130 on HDAC activity (n = 7, t test, p = 0.0003). Consistent with the suggested HDAC activity-promoting role of LPA in differentiating OLGs, LPA treatment alone was found to lead to an increase in HDAC activity (Fig. 6D; n = 4, t test, p = 0.0052). Together, the above data confirm that, as in the developing zebrafish, progression along the early stages of the OLG lineage in rodents is promoted by the lysoPLD activity of ATX via the generation of LPA and a downstream mechanism that involves an increase in HDAC activity.
Figure 6.
In rodent OLG cultures, inhibition of the lysoPLD activity of ATX leads to a reduction in HDAC activity. A, Experimental design. Timing of histone deacetylation is marked as described by Marin-Husstege et al. (2002). OPC, OLG progenitor. B–D, Bar graphs showing HDAC activity as determined by using the fluorogenic substrate Boc-Lys(Ac)-AMC. Control (vehicle-treated) values were set to 100% (see horizontal line in B and control bars in C, D), and experimental values were calculated accordingly. Data represent means ± SEMs. ns, Not significant; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
ATX–lysoPLD-promoted increase in HDAC activity is associated with a decrease in nuclear histone acetylation
It has been shown previously that HDAC activity in differentiating OLGs is directed primarily to lysine residues of histones H4 and H3, including the lysine residue on the tail of histone H3 at position 9 (H3K9) (Swiss et al., 2011). To assess changes in acetylation at this site, cells treated as depicted in Figure 5A were analyzed at 48 h by immunocytochemistry combined with confocal microscopy. As shown in Figure 7A, nuclear histone acetylation can be readily detected in O4-positive differentiating OLGs by this method. Most importantly, inhibition of the lysoPLD activity of ATX led to a significant increase in the levels of H3K9 acetylation (Fig. 7C; HA130, n = 3, Mann–Whitney U test, p = 0.00005; S32826, n = 3, Mann–Whitney U test, p = 0.0003). This effect was not found to be associated with changes in cell survival or proliferation as determined by immunostaining using antibodies specifically recognizing the active (cleaved) form of caspase-3 (n = 3, t test, p = 0.1355) or the Ki67 antigen (n = 3, t test, p = 0.1189). In addition, no prominent changes in cellular morphology or nuclear appearance were noted (Fig. 7B). The above described increase in H3K9 acetylation during ATX–lysoPLD inhibition could be further confirmed by Western blot analysis of cell homogenates (Fig. 7D; n = 3, t test, p = 0.0442). Together, our data so far demonstrate that attenuation of OLG differentiation via inhibition of the lysoPLD activity of ATX (Figs. 2, 3, 5) is associated with a decrease in HDAC activity (Figs. 4, 6) and an increase in histone acetylation (Fig. 7).
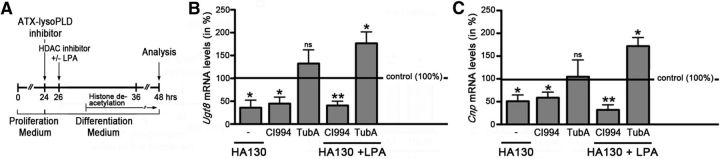
ATX–lysoPLD-promoted progression along the early stages of the OLG lineage is dependent on the activity of HDAC1/2 but not HDAC6
The general requirement of HDAC activity for OLG differentiation has been shown previously to involve particularly the class I HDAC members HDAC1 and HDAC2 (Shen et al., 2008; Ye et al., 2009; Wu et al., 2012). To investigate the extent to which the lysoPLD activity of ATX may represent an upstream signal regulating especially the activity of the class I HDAC members HDAC1 and HDAC2, we used a pharmacological HDAC1/2/3 inhibitor, namely CI994 (Kraker et al., 2003; Beckers et al., 2007), in the experimental design as depicted in Figure 8A. It is notable that HDAC3 was found to not be involved in the regulation of OLG differentiation at the stages analyzed here (Shen et al., 2008; Wu et al., 2012). In addition, at the concentration used (2.5 μm), CI994 is considered to effectively inhibit HDAC1 (Beckers et al., 2007). The effectiveness of HDAC1/2 inhibition in our system is corroborated by the observed reduction in Ugt8 mRNA levels (Fig. 8B), because its expression has been shown previously to be regulated by direct targets of HDAC1/2-mediated histone deacetylation (Swiss et al., 2011). Furthermore, and as a control, inhibition of HDAC6, which represents a class II HDAC member, was performed by using the inhibitor Tubastatin A (Butler et al., 2010). At the concentration used (5 μm), Tubastatin A is considered to effectively inhibit HDAC6 with additional but only moderate activity at HDAC8 (Beckers et al., 2007). Neither HDAC6 nor HDAC8 have been found involved in the regulation of gene expression changes associated with OLG differentiation (Shen et al., 2005, 2008; Wu et al., 2012; Noack et al., 2014), and HDAC6 has in OLGs functionally been implicated primarily in contributing to the clearing process of misfolded protein aggregates (Noack et al., 2014; Leyk et al., 2015). As indicated above and shown in Figure 8, application of CI994, but not Tubastatin A, attenuated the expression of the OLG differentiation genes Ugt8 and Cnp to a similar extent as the application of the ATX–lysoPLD activity inhibitor HA130 (Ugt8: HA130, n = 3, t test, p = 0.0299; CI994, n = 3, t test, p = 0.0176; Tubastatin A, n = 3, t test, NS; Cnp: HA130, n = 3, t test, p = 0.0382; CI994, n = 3, t test, p = 0.0270; Tubastatin A, n = 3, t test, NS). To assess the extent to which HDAC1/2 activity may be required for the observed ATX–lysoPLD-mediated changes in gene expression, the effect of HDAC1/2/3 or HDAC6 inhibition on LPA-stimulated rescue was determined. In agreement with a critical role of HDAC1/2, but not HDAC6, the addition of LPA was unable to rescue the effect of ATX–lysoPLD inhibition on Ugt8 and Cnp mRNA levels in the presence of CI994 but not Tubastatin A (Fig. 8B,C; Ugt8: HA130 + CI994 + LPA, n = 3, t test, p = 0.0033; HA130 + Tubastatin A + LPA, n = 3, t test, p = 0.0373; Cnp, HA130 + CI994 + LPA, n = 3, t test, p = 0.0036; HA130 + Tubastatin A + LPA, n = 3, t test, p = 0.0325). It is notable that ATX–lysoPLD inhibition could be observed as early as 2 h after application of HA130 (data not shown) and was thus fully effective at the time point of HDAC inhibition. Furthermore, no additive or synergistic effects were observed during additional inhibition of HDAC1/2/3 or HDAC6 (data not shown). Moreover, CI994 (Kraker et al., 2003), Tubastatin A (Parab et al., 2015), and LPA (our own data; data not shown) have been shown to exert their effects within the first 2 h of treatment, thus mostly excluding a timely separation of their actions. Thus, the above data provide strong support for a crucial role of the class I HDAC members HDAC1 and HDAC2 as downstream targets mediating the OLG differentiation promoting effect of the lysoPLD activity of ATX.
Figure 8.
In rodent OLG cultures, LPA rescue of ATX–lysoPLD activity inhibition requires the activity of class I HDAC members HDAC1 and HDAC2 but not the class II HDAC member HDAC6. A, Experimental design. Timing of histone deacetylation is marked as described by Marin-Husstege et al. (2002). OPC, OLG progenitor. B, C, Bar graphs depicting mRNA levels for the OLG differentiation genes Ugt8 (B) and Cnp (C) as determined by real-time RT-qPCR analysis. Control (vehicle-treated) values were set to 100% (see horizontal line), and experimental values were calculated accordingly. Data represent means ± SEMs. ns, Not significant; *p ≤ 0.05, **p ≤ 0.01. TubA, Tubastatin A.
Discussion
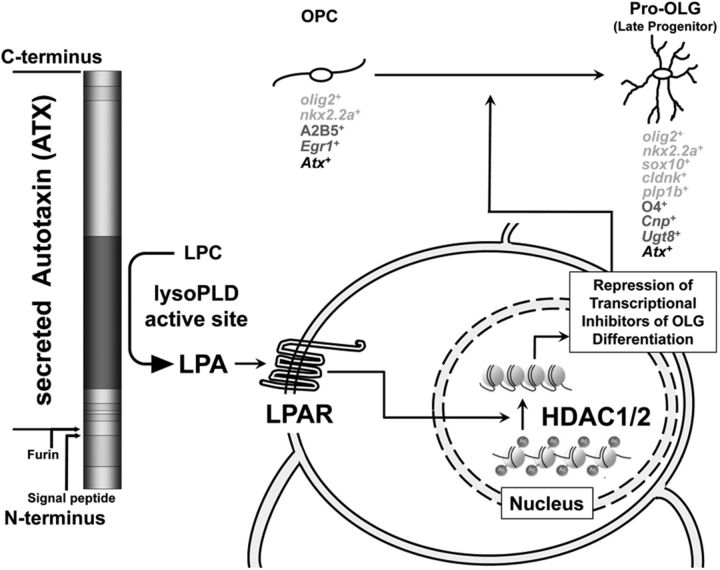
Our data presented here suggest a modulatory role of ATX during the transition from an OLG progenitor to an early-stage differentiating OLG as depicted in our proposed model shown in Figure 9. In this model, LPA, generated via the lysoPLD activity of ATX, activates one (or more) of its cognate G-protein-coupled receptors, which are referred to as LPA receptors (Chun et al., 2010; Choi and Chun, 2013; Kihara et al., 2014) and have been found expressed by cells of the OLG lineage (Weiner et al., 1998; Stankoff et al., 2002; Dawson et al., 2003; Nogaroli et al., 2009; Zhang et al., 2014). Activation of the above ATX–LPA axis initiates a downstream signaling cascade leading to the activation of HDAC1/2, which in turn mediates histone deacetylation and repression of transcriptional inhibitors of OLG differentiation, thereby promoting gene expression changes that are associated with the transition from an OLG progenitor to an early-stage differentiating OLG. ATX has been found to be expressed and secreted by OLG progenitors and differentiating OLGs (Figs. 4, 5; Fuss et al., 1997; Fox et al., 2003; Savaskan et al., 2007), thus suggesting an autocrine signaling mechanism. However, and during development, paracrine regulation may also occur via the secretion of ATX by cells of the floor plate (Yuelling et al., 2012). Importantly, the regulatory mechanism described here appears evolutionarily conserved, at least from zebrafish to rodents.
Figure 9.
Proposed model for the role of the ATX–LPA axis in OLG differentiation. ATX, which has been found secreted by OLGs throughout the early stages of the lineage, generates the lipid signaling molecule LPA via its enzymatically active lysoPLD site. LPA, in turn, activates one (or more) of its cognate receptors (LPARs) on the surface of OLG progenitors, which leads to the downstream activation of HDAC1/2 in the nucleus. Histone deacetylation mediates the repression of transcriptional inhibitors of OLG differentiation and thereby promotes gene expression changes associated with the differentiation from an OLG progenitor (OPC) to an early differentiating OLG (Pro-OLG). Expression markers identifying individual stages of the OLG lineage are listed below each OLG stage. Markers in black were used in both the zebrafish and rodent studies, whereas markers in light and dark gray are unique for the zebrafish and rodent studies, respectively.
In the context of histone deacetylation during the early stages of OLG differentiation, it is notable that HDAC1/2 activity has not only been implicated in modulating the transition from an OLG progenitor to an early-stage differentiating OLG but also the specification of an OLG progenitor from a multipotent neural progenitor cell (Copray et al., 2009; Jacob et al., 2011). However, and based on the data shown here, HDAC1/2-mediated regulation of OLG progenitor specification is unlikely to be modulated by ATX and its lysoPLD activity. First, inhibition of ATX–lysoPLD activity was not found to affect the expression of nkx2.2.a (Fig. 4), which has been described to be upregulated upon early developmental loss of HDAC1 activity (Cunliffe and Casaccia-Bonnefil, 2006). Second, ATX and its lysoPLD activity were not found to affect the expression of olig2, which during OLG progenitor specification has been proposed to be modulated via HDAC1/2 activity and downstream of sonic hedgehog signaling (Cunliffe and Casaccia-Bonnefil, 2006; Ye et al., 2009). Hence, it appears that ATX, via its lysoPLD activity, modulates HDAC1/2-regulated gene expression explicitly during a developmental time window that coincides with the transition from OLG progenitor to early-stage differentiating OLG and that likely occurs after sonic hedgehog-modulated HDAC1/2-regulated gene expression implicated in OLG progenitor specification. However, future work will be necessary to more precisely define the relationships between the ATX–LPA axis, sonic hedgehog signaling, and their downstream effects on HDAC1/2-regulated gene expression.
In general, it is becoming increasingly clear that histone acetylation and deacetylation events represent dynamic modifications (Peserico and Simone, 2011). Consistent with this notion, histone deacetylation in cells of the OLG lineage has been found to be transient and reversible (Shen et al., 2005). In a search for more stable repressive histone modifications, marks of trimethylation of H3K9 have been found recently to coincide with the transition from OLG progenitor to differentiating OLG (Liu et al., 2015). Thus, it will be interesting to explore in subsequent studies a potential role of the ATX–LPA axis in modulating H3K9 methyltransferases.
ATX has been demonstrated to possess two functionally distinct domains, the lysoPLD active site and the MORFO domain. Our data presented here demonstrate that the lysoPLD active site of ATX affects epigenetic and gene expression aspects of OLG differentiation. In contrast, and based on our previous data, the function of the MORFO domain of ATX, as it relates to OLG differentiation, appears restricted to promoting cytoskeletal and thus morphological changes (Fox et al., 2003; Dennis et al., 2008, 2012). This concept that OLG morphogenesis is regulated, at least in part, by mechanisms distinct from the ones affecting gene expression is supported by previous findings (Osterhout et al., 1999; Buttery and ffrench-Constant, 1999; Sloane and Vartanian, 2007; Younes-Rapozo et al., 2009; Ishii et al., 2012). However, at the same time, it is also well known that both of the above stated aspects of OLG differentiation occur well synchronized during development (Wegner, 2008; Bauer et al., 2009; Mitew et al., 2014). Together, these observations raise the exciting possibility that ATX, via the concerted action of its two distinct functional activities, may be one of the critical players coordinating gene expression and morphological changes during OLG differentiation.
The functional effects of the lysoPLD activity of ATX have been well described to be mediated by signaling through a family of G-protein-coupled receptors, the so-called LPA receptors. To date, in mammals there are six receptors that are recognized as bona fide LPA receptors, all of which appear to be expressed, at least to some degree, in cells of the OLG lineage (Zhang et al., 2014). Notably, LPA1 has long been known to be present on OLGs (Weiner et al., 1998; Stankoff et al., 2002; Dawson et al., 2003; Nogaroli et al., 2009), and a number of functional roles have been proposed based on tissue culture studies. Interestingly, Lpar1 knock-out mice have initially not been found to display CNS myelin-related pathologies (Contos et al., 2000, 2002). However, myelin defects were reported in more recent studies analyzing the so-called Málaga variant (maLpar1 null mice), a stable variant of the original Lpar1 knock-out mouse strain in which defects are more pronounced likely as a result of interactions of LPA1 with currently unknown genetic modifiers (Estivill-Torrús et al., 2008; Garcia-Diaz et al., 2014). In maLpar1 null mice, impaired transport of one of the major myelin proteins, namely PLP, was found to be impaired, leading to loss of OLGs most likely because of stress-induced apoptosis. No deficits in OLG differentiation were noted in these mice. Thus, and despite its prominent expression in OLGs, LPA1 appears unlikely to play a unique and major role in mediating the ATX–LPA–HDAC1/2 axis described here.
To our knowledge, null mice have been characterized for all currently recognized LPA receptors with the exception of LPA6 (Contos et al., 2000, 2002; Ye et al., 2005; Liu et al., 2010; Lin et al., 2012). In addition, knockdown studies have been performed in the zebrafish and in Xenopus laevis that cover all known bona fide LPA receptors (Yukiura et al., 2011; Geach et al., 2014). In neither of the studies have myelin defects been reported. This does not exclude a prominent role of LPA receptor signaling in OLGs but rather highlights the complication of functional redundancy and compensation when eliminating a single LPA receptor. For example, it has been shown that combined knockdown of lpar1 and lpar4 in the zebrafish leads to vascular defects, whereas single lpar knockdown has no effect (Yukiura et al., 2011). Similarly, male reproductive defects have been observed in triple Lpar1/2/3 knock-out mice but not any of the single Lpar knock-out mice (Ye et al., 2008). In the context of our studies, it is interesting that, for the triple knock-out mice, no myelin defects have been reported, thus suggesting an involvement of at least one of the remaining LPA receptors, namely LPA4, LPA5, and/or LPA6. Of these, LPA4 has been shown to activate PKA via coupling to the heterotrimeric G-protein Gαs and increase in cAMP levels (Gardell et al., 2006). PKA, in turn, has been implicated in regulating HDAC activity via phosphorylation (Sengupta and Seto, 2004). Although LPA5 was found to not couple to Gαs, its stimulation was nevertheless found to be associated with an increase in cAMP levels possibly through involvement of G-protein βγ subunits (Lee et al., 2006). LPA6 may couple to Gαs but with likely much lower potencythan LPA4 (Yanagida et al., 2009). Clearly, more comprehensive analyses will be necessary to dissect the exact roles for each of the LPA receptors in the regulation of OLG differentiation and via the ATX–LPA axis.
Our data presented here reveal a novel functional role for the lysoPLD activity of ATX, namely the promotion of OLG differentiation via an increase in HDAC1/2 activity and associated changes in gene expression. In the context of human diseases and in particular the major demyelinating disease in human, multiple sclerosis (MS), it is worth mentioning that ATX mRNA levels have been found reduced within the MS CNS (Raddatz et al., 2014) and that a shift toward a decrease in histone deacetylation has been implicated in contributing to the limitations in myelin repair seen in MS (Pedre et al., 2011). These findings further support a critical role of ATX in the regulation of CNS myelination, and they suggest that misregulation of the ATX–LPA–HDAC1/2 axis may contribute to the pathology seen in MS.
Footnotes
This work was supported by a grant from the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS; B.F.). Microscopy was performed at the Virginia Commonwealth University (VCU) Department of Anatomy and Neurobiology Microscopy Facility, supported, in part, with funding from NIH/NINDS Center Core Grant P30 NS047463. Fluorescent-activated cell sorting (FACS) was performed at the VCU Massey Cancer Center Flow Cytometry Core supported, in part, by funding from the NIH/National Cancer Institute Cancer Center Support Grant P30 CA016059. We thank Sarah Kucenas and Bruce Appel for providing transgenic fish and invaluable discussions and Wenbio Chen for providing plasmid constructs for CRISPR/Cas9 genome editing. We also thank Patrizia Casaccia and Gregory Walsh for helpful advice, Scott Henderson for assistance with the nuclear histone acetylation analysis, and Julie Farnsworth and Unsong Oh for their help with FACS analysis.
The authors declare no competing financial interests.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Albers HM, Dong A, van Meeteren LA, Egan DA, Sunkara M, van Tilburg EW, Schuurman K, van Tellingen O, Morris AJ, Smyth SS, Moolenaar WH, Ovaa H. Boronic acid-based inhibitor of autotaxin reveals rapid turnover of LPA in the circulation. Proc Natl Acad Sci U S A. 2010;107:7257–7262. doi: 10.1073/pnas.1001529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki J, Inoue A, Okudaira S. Two pathways for lysophosphatidic acid production. Biochim Biophys Acta. 2008;1781:513–518. doi: 10.1016/j.bbalip.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD, Raff MC. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:31–46. doi: 10.1016/0092-8674(92)90531-G. [DOI] [PubMed] [Google Scholar]
- Bauer NG, Richter-Landsberg C, ffrench-Constant C. Role of the oligodendroglial cytoskeleton in differentiation and myelination. Glia. 2009;57:1691–1705. doi: 10.1002/glia.20885. [DOI] [PubMed] [Google Scholar]
- Beckers T, Burkhardt C, Wieland H, Gimmnich P, Ciossek T, Maier T, Sanders K. Distinct pharmacological properties of second generation HDAC inhibitors with the benzamide or hydroxamate head group. Int J Cancer. 2007;121:1138–1148. doi: 10.1002/ijc.22751. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Raymond PA. GFAP transgenic zebrafish. Gene Expr Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Bischof M, Weider M, Küspert M, Nave KA, Wegner M. Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation. J Neurosci. 2015;35:21–35. doi: 10.1523/JNEUROSCI.1468-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brösamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Buckley CE, Marguerie A, Roach AG, Goldsmith P, Fleming A, Alderton WK, Franklin RJ. Drug reprofiling using zebrafish identifies novel compounds with potential pro-myelination effects. Neuropharmacology. 2010;59:149–159. doi: 10.1016/j.neuropharm.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc. 2010;132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery PC, ffrench-Constant C. Laminin-2/integrin interactions enhance myelin membrane formation by oligodendrocytes. Mol Cell Neurosci. 1999;14:199–212. doi: 10.1006/mcne.1999.0781. [DOI] [PubMed] [Google Scholar]
- Choi JW, Chun J. Lysophospholipids and their receptors in the central nervous system. Biochim Biophys Acta. 2013;1831:20–32. doi: 10.1016/j.bbalip.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Chun J, Hla T, Lynch KR, Spiegel S, Moolenaar WH. International Union of Basic and Clinical Pharmacology. LXXVIII. Lysophospholipid receptor nomenclature. Pharmacol Rev. 2010;62:579–587. doi: 10.1124/pr.110.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contos JJ, Fukushima N, Weiner JA, Kaushal D, Chun J. Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc Natl Acad Sci U S A. 2000;97:13384–13389. doi: 10.1073/pnas.97.24.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contos JJ, Ishii I, Fukushima N, Kingsbury MA, Ye X, Kawamura S, Brown JH, Chun J. Characterization of lpa(2) (Edg4) and lpa(1)/lpa(2) (Edg2/Edg4) lysophosphatidic acid receptor knockout mice: signaling deficits without obvious phenotypic abnormality attributable to lpa(2) Mol Cell Biol. 2002;22:6921–6929. doi: 10.1128/MCB.22.19.6921-6929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copray S, Huynh JL, Sher F, Casaccia-Bonnefil P, Boddeke E. Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia. 2009;57:1579–1587. doi: 10.1002/glia.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Dalgaard P. Introductory statistics with R. New York: Springer; 2008. [Google Scholar]
- Dawson J, Hotchin N, Lax S, Rumsby M. Lysophosphatidic acid induces process retraction in CG-4 line oligodendrocytes and oligodendrocyte precursor cells but not in differentiated oligodendrocytes. J Neurochem. 2003;87:947–957. doi: 10.1046/j.1471-4159.2003.02056.x. [DOI] [PubMed] [Google Scholar]
- Dennis J, Nogaroli L, Fuss B. Phosphodiesterase-Ialpha/autotaxin (PD-Ialpha/ATX): a multifunctional protein involved in central nervous system development and disease. J Neurosci Res. 2005;82:737–742. doi: 10.1002/jnr.20686. [DOI] [PubMed] [Google Scholar]
- Dennis J, White MA, Forrest AD, Yuelling LM, Nogaroli L, Afshari FS, Fox MA, Fuss B. Phosphodiesterase-Ialpha/autotaxin's MORFO domain regulates oligodendroglial process network formation and focal adhesion organization. Mol Cell Neurosci. 2008;37:412–424. doi: 10.1016/j.mcn.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis J, Morgan MK, Graf MR, Fuss B. P2Y(12) receptor expression is a critical determinant of functional responsiveness to ATX's MORFO domain. Purinergic Signal. 2012;8:181–190. doi: 10.1007/s11302-011-9283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doodnath R, Dervan A, Wride MA, Puri P. Zebrafish: an exciting model for investigating the spatio-temporal pattern of enteric nervous system development. Pediatr Surg Int. 2010;26:1217–1221. doi: 10.1007/s00383-010-2746-7. [DOI] [PubMed] [Google Scholar]
- Estivill-Torrús G, Llebrez-Zayas P, Matas-Rico E, Santín L, Pedraza C, De Diego I, Del Arco I, Fernández-Llebrez P, Chun J, De Fonseca FR. Absence of LPA1 signaling results in defective cortical development. Cereb Cortex. 2008;18:938–950. doi: 10.1093/cercor/bhm132. [DOI] [PubMed] [Google Scholar]
- Ferguson CG, Bigman CS, Richardson RD, van Meeteren LA, Moolenaar WH, Prestwich GD. Fluorogenic phospholipid substrate to detect lysophospholipase D/autotaxin activity. Org Lett. 2006;8:2023–2026. doi: 10.1021/ol060414i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry G, Moulharat N, Pradère JP, Desos P, Try A, Genton A, Giganti A, Beucher-Gaudin M, Lonchampt M, Bertrand M, Saulnier-Blache JS, Tucker GC, Cordi A, Boutin JA. S32826, a nanomolar inhibitor of autotaxin: discovery, synthesis and applications as a pharmacological tool. J Pharmacol Exp Ther. 2008;327:809–819. doi: 10.1124/jpet.108.141911. [DOI] [PubMed] [Google Scholar]
- Fox MA, Colello RJ, Macklin WB, Fuss B. Phosphodiesterase-Ialpha/autotaxin: a counteradhesive protein expressed by oligodendrocytes during onset of myelination. Mol Cell Neurosci. 2003;23:507–519. doi: 10.1016/S1044-7431(03)00073-3. [DOI] [PubMed] [Google Scholar]
- Fuss B, Baba H, Phan T, Tuohy VK, Macklin WB. Phosphodiesterase I, a novel adhesion molecule and/or cytokine involved in oligodendrocyte function. J Neurosci. 1997;17:9095–9103. doi: 10.1523/JNEUROSCI.17-23-09095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz B, Riquelme R, Varela-Nieto I, Jimenez AJ, de Diego I, Gomez-Conde AL, Matas-Rico E, Aguirre JA, Chun J, Pedraza C, Santin LJ, Fernandez O, Rodriguez de Fonseca F, Estivill-Torrus G. Loss of lysophosphatidic acid receptor LPA alters oligodendrocyte differentiation and myelination in the mouse cerebral cortex. Brain Struct Funct. 2014 doi: 10.1007/s00429-014-0885-7. doi: 10.1007/s00429-014-0885-7. Advance online publication. Retrieved July 9, 2015. [DOI] [PubMed] [Google Scholar]
- Gardell SE, Dubin AE, Chun J. Emerging medicinal roles for lysophospholipid signaling. Trends Mol Med. 2006;12:65–75. doi: 10.1016/j.molmed.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Geach TJ, Faas L, Devader C, Gonzalez-Cordero A, Tabler JM, Brunsdon H, Isaacs HV, Dale L. An essential role for LPA signalling in telencephalon development. Development. 2014;141:940–949. doi: 10.1242/dev.104901. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Grupp L, Wolburg H, Mack AF. Astroglial structures in the zebrafish brain. J Comp Neurol. 2010;518:4277–4287. doi: 10.1002/cne.22481. [DOI] [PubMed] [Google Scholar]
- Gupte R, Patil R, Liu J, Wang Y, Lee SC, Fujiwara Y, Fells J, Bolen AL, Emmons-Thompson K, Yates CR, Siddam A, Panupinthu N, Pham TC, Baker DL, Parrill AL, Mills GB, Tigyi G, Miller DD. Benzyl and naphthalene methylphosphonic acid inhibitors of autotaxin with anti-invasive and anti-metastatic activity. Chem Med Chem. 2011;6:922–935. doi: 10.1002/cmdc.201000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagström C, Olsson C. Glial cells revealed by GFAP immunoreactivity in fish gut. Cell Tissue Res. 2010;341:73–81. doi: 10.1007/s00441-010-0979-3. [DOI] [PubMed] [Google Scholar]
- He Y, Sandoval J, Casaccia-Bonnefil P. Events at the transition between cell cycle exit and oligodendrocyte progenitor differentiation: the role of HDAC and YY1. Neuron Glia Biol. 2007;3:221–231. doi: 10.1017/S1740925X08000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishdorj G, Graham BA, Hu X, Chen J, Johnston JB, Fang X, Gibson SB. Lysophosphatidic acid protects cancer cells from histone deacetylase (HDAC) inhibitor-induced apoptosis through activation of HDAC. J Biol Chem. 2008;283:16818–16829. doi: 10.1074/jbc.M710177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Fyffe-Maricich SL, Furusho M, Miller RH, Bansal R. ERK1/ERK2 MAPK signaling is required to increase myelin thickness independent of oligodendrocyte differentiation and initiation of myelination. J Neurosci. 2012;32:8855–8864. doi: 10.1523/JNEUROSCI.0137-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer P, Lalane R, 3rd, Morris C, Challa P, Vann R, Rao PV. Autotaxin-lysophosphatidic acid axis is a novel molecular target for lowering intraocular pressure. PLoS One. 2012;7:e42627. doi: 10.1371/journal.pone.0042627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Lebrun-Julien F, Suter U. How histone deacetylases control myelination. Mol Neurobiol. 2011;44:303–312. doi: 10.1007/s12035-011-8198-9. [DOI] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara Y, Maceyka M, Spiegel S, Chun J. Lysophospholipid receptor nomenclature review: IUPHAR Review 8. Br J Pharmacol. 2014;171:3575–3594. doi: 10.1111/bph.12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B. In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci. 2006;9:1506–1511. doi: 10.1038/nn1803. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraker AJ, Mizzen CA, Hartl BG, Miin J, Allis CD, Merriman RL. Modulation of histone acetylation by [4-(acetylamino)-N-(2-amino-phenyl) benzamide] in HCT-8 colon carcinoma. Mol Cancer Ther. 2003;2:401–408. [PubMed] [Google Scholar]
- Kucenas S, Snell H, Appel B. nkx2.2a promotes specification and differentiation of a myelinating subset of oligodendrocyte lineage cells in zebrafish. Neuron Glia Biol. 2008;4:71–81. doi: 10.1017/S1740925X09990123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafrenaye AD, Fuss B. Focal adhesion kinase can play unique and opposing roles in regulating the morphology of differentiating oligodendrocytes. J Neurochem. 2010;115:269–282. doi: 10.1111/j.1471-4159.2010.06926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Yao WL, Tsao KC, Houben AJ, Albers HM, Ovaa H, Moolenaar WH, Lee SJ. Autotaxin/Lpar3 signaling regulates Kupffer's vesicle formation and left-right asymmetry in zebrafish. Development. 2012;139:4439–4448. doi: 10.1242/dev.081745. [DOI] [PubMed] [Google Scholar]
- Lam CS, März M, Strähle U. gfap and nestin reporter lines reveal characteristics of neural progenitors in the adult zebrafish brain. Dev Dyn. 2009;238:475–486. doi: 10.1002/dvdy.21853. [DOI] [PubMed] [Google Scholar]
- Leake D, Asch WS, Canger AK, Schechter N. Gefiltin in zebrafish embryos: sequential gene expression of two neurofilament proteins in retinal ganglion cells. Differentiation. 1999;65:181–189. doi: 10.1046/j.1432-0436.1999.6540181.x. [DOI] [PubMed] [Google Scholar]
- Lee CW, Rivera R, Gardell S, Dubin AE, Chun J. GPR92 as a new G12/13- and Gq-coupled lysophosphatidic acid receptor that increases cAMP, LPA5. J Biol Chem. 2006;281:23589–23597. doi: 10.1074/jbc.M603670200. [DOI] [PubMed] [Google Scholar]
- Leyk J, Goldbaum O, Noack M, Richter-Landsberg C. Inhibition of HDAC6 modifies tau inclusion body formation and impairs autophagic clearance. J Mol Neurosci. 2015;55:1031–1046. doi: 10.1007/s12031-014-0460-y. [DOI] [PubMed] [Google Scholar]
- Lin ME, Rivera RR, Chun J. Targeted deletion of LPA5 identifies novel roles for lysophosphatidic acid signaling in development of neuropathic pain. J Biol Chem. 2012;287:17608–17617. doi: 10.1074/jbc.M111.330183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Magri L, Zhang F, Marsh NO, Albrecht S, Huynh JL, Kaur J, Kuhlmann T, Zhang W, Slesinger PA, Casaccia P. Chromatin landscape defined by repressive histone methylation during oligodendrocyte differentiation. J Neurosci. 2015;35:352–365. doi: 10.1523/JNEUROSCI.2606-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YB, Kharode Y, Bodine PV, Yaworsky PJ, Robinson JA, Billiard J. LPA induces osteoblast differentiation through interplay of two receptors: LPA1 and LPA4. J Cell Biochem. 2010;109:794–800. doi: 10.1002/jcb.22471. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lyssiotis CA, Walker J, Wu C, Kondo T, Schultz PG, Wu X. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc Natl Acad Sci U S A. 2007;104:14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes J, Verlooy L, Buenafe OE, de Witte PA, Esguerra CV, Crawford AD. Evaluation of 14 organic solvents and carriers for screening applications in zebrafish embryos and larvae. PLoS One. 2012;7:e43850. doi: 10.1371/journal.pone.0043850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lozada Z, Waggener CT, Kim K, Zou S, Knapp PE, Hayashi Y, Ortega A, Fuss B. Activation of sodium-dependent glutamate transporters regulates the morphological aspects of oligodendrocyte maturation via signaling through calcium/calmodulin-dependent kinase IIbeta's actin-binding/-stabilizing domain. Glia. 2014;62:1543–1558. doi: 10.1002/glia.22699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitew S, Hay CM, Peckham H, Xiao J, Koenning M, Emery B. Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience. 2014;276:29–47. doi: 10.1016/j.neuroscience.2013.11.029. [DOI] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Leblond CP. Electron microscopic identification of three classes of oligodendrocytes and a preliminary study of their proliferative activity in the corpus callosum of young rats. J Comp Neurol. 1970;139:1–28. doi: 10.1002/cne.901390102. [DOI] [PubMed] [Google Scholar]
- Münzel EJ, Schaefer K, Obirei B, Kremmer E, Burton EA, Kuscha V, Becker CG, Brösamle C, Williams A, Becker T. Claudin k is specifically expressed in cells that form myelin during development of the nervous system and regeneration of the optic nerve in adult zebrafish. Glia. 2012;60:253–270. doi: 10.1002/glia.21260. [DOI] [PubMed] [Google Scholar]
- Nakanaga K, Hama K, Aoki J. Autotaxin–an LPA producing enzyme with diverse functions. J Biochem. 2010;148:13–24. doi: 10.1093/jb/mvq052. [DOI] [PubMed] [Google Scholar]
- Nelissen K, Smeets K, Mulder M, Hendriks JJ, Ameloot M. Selection of reference genes for gene expression studies in rat oligodendrocytes using quantitative real time PCR. J Neurosci Methods. 2010;187:78–83. doi: 10.1016/j.jneumeth.2009.12.018. [DOI] [PubMed] [Google Scholar]
- Noack M, Leyk J, Richter-Landsberg C. HDAC6 inhibition results in tau acetylation and modulates tau phosphorylation and degradation in oligodendrocytes. Glia. 2014;62:535–547. doi: 10.1002/glia.22624. [DOI] [PubMed] [Google Scholar]
- Nogaroli L, Yuelling LM, Dennis J, Gorse K, Payne SG, Fuss B. Lysophosphatidic acid can support the formation of membranous structures and an increase in MBP mRNA levels in differentiating oligodendrocytes. Neurochem Res. 2009;34:182–193. doi: 10.1007/s11064-008-9772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout DJ, Wolven A, Wolf RM, Resh MD, Chao MV. Morphological differentiation of oligodendrocytes requires activation of Fyn tyrosine kinase. J Cell Biol. 1999;145:1209–1218. doi: 10.1083/jcb.145.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parab S, Shetty O, Gaonkar R, Balasinor N, Khole V, Parte P. HDAC6 deacetylates alpha tubulin in sperm and modulates sperm motility in Holtzman rat. Cell Tissue Res. 2015;359:665–678. doi: 10.1007/s00441-014-2039-x. [DOI] [PubMed] [Google Scholar]
- Park HC, Mehta A, Richardson JS, Appel B. olig2 is required for zebrafish primary motor neuron and oligodendrocyte development. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- Pedre X, Mastronardi F, Bruck W, López-Rodas G, Kuhlmann T, Casaccia P. Changed histone acetylation patterns in normal-appearing white matter and early multiple sclerosis lesions. J Neurosci. 2011;31:3435–3445. doi: 10.1523/JNEUROSCI.4507-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott NJ, Turner AJ, Scopes J, Westby M, Marsh JC, Gordon-Smith EC, Dalgleish AG, Gibson FM. The use of 7-amino actinomycin D in identifying apoptosis: simplicity of use and broad spectrum of application compared with other techniques. Blood. 1996;87:2244–2251. [PubMed] [Google Scholar]
- Raddatz BB, Hansmann F, Spitzbarth I, Kalkuhl A, Deschl U, Baumgärtner W, Ulrich R. Transcriptomic meta-analysis of multiple sclerosis and its experimental models. PLoS One. 2014;9:e86643. doi: 10.1371/journal.pone.0086643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki DT, Dumas SE, Engleman EG. Discrimination of viable and non-viable cells usingpropidium iodide in two color immunofluorescence. Cytometry. 1987;8:413–420. doi: 10.1002/cyto.990080411. [DOI] [PubMed] [Google Scholar]
- Savaskan NE, Rocha L, Kotter MR, Baer A, Lubec G, van Meeteren LA, Kishi Y, Aoki J, Moolenaar WH, Nitsch R, Bräuer AU. Autotaxin (NPP-2) in the brain: cell type-specific expression and regulation during development and after neurotrauma. Cell Mol Life Sci. 2007;64:230–243. doi: 10.1007/s00018-006-6412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- Shen S, Casaccia-Bonnefil P. Post-translational modifications of nucleosomal histones in oligodendrocyte lineage cells in development and disease. J Mol Neurosci. 2008;35:13–22. doi: 10.1007/s12031-007-9014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJ, Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokal RR, Rohlf FJ. Biometry: the principle and practice in biological research. New York: Freeman; 1995. [Google Scholar]
- Sloane JA, Vartanian TK. Myosin Va controls oligodendrocyte morphogenesis and myelination. J Neurosci. 2007;27:11366–11375. doi: 10.1523/JNEUROSCI.2326-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankoff B, Barron S, Allard J, Barbin G, Noël F, Aigrot MS, Premont J, Sokoloff P, Zalc B, Lubetzki C. Oligodendroglial expression of Edg-2 receptor: developmental analysis and pharmacological responses to lysophosphatidic acid. Mol Cell Neurosci. 2002;20:415–428. doi: 10.1006/mcne.2002.1129. [DOI] [PubMed] [Google Scholar]
- Swiss VA, Nguyen T, Dugas J, Ibrahim A, Barres B, Androulakis IP, Casaccia P. Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS One. 2011;6:e18088. doi: 10.1371/journal.pone.0018088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N, Appel B. Identification of genes expressed by zebrafish oligodendrocytes using a differential microarray screen. Dev Dyn. 2010;239:2041–2047. doi: 10.1002/dvdy.22338. [DOI] [PubMed] [Google Scholar]
- Talbot JC, Amacher SL. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish. 2014;11:583–585. doi: 10.1089/zeb.2014.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple S, Raff MC. Differentiation of a bipotential glial progenitor cell in a single cell microculture. Nature. 1985;313:223–225. doi: 10.1038/313223a0. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. In situ hybridization on whole-mount zebrafish embryos and young larvae. Methods Mol Biol. 2014;1211:53–67. doi: 10.1007/978-1-4939-1459-3_5. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Tokumura A, Majima E, Kariya Y, Tominaga K, Kogure K, Yasuda K, Fukuzawa K. Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. J Biol Chem. 2002;277:39436–39442. doi: 10.1074/jbc.M205623200. [DOI] [PubMed] [Google Scholar]
- Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, Yamori T, Mills GB, Inoue K, Aoki J, Arai H. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol. 2002;158:227–233. doi: 10.1083/jcb.200204026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meeteren LA, Moolenaar WH. Regulation and biological activities of the autotaxin-LPA axis. Prog Lipid Res. 2007;46:145–160. doi: 10.1016/j.plipres.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Weiner JA, Hecht JH, Chun J. Lysophosphatidic acid receptor gene vzg-1/lpA1/edg-2 is expressed by mature oligodendrocytes during myelination in the postnatal murine brain. J Comp Neurol. 1998;398:587–598. doi: 10.1002/(SICI)1096-9861(19980907)398:4%3C587::AID-CNE10%3E3.0.CO%3B2-5. [DOI] [PubMed] [Google Scholar]
- Wu M, Hernandez M, Shen S, Sabo JK, Kelkar D, Wang J, O'Leary R, Phillips GR, Cate HS, Casaccia P. Differential modulation of the oligodendrocyte transcriptome by sonic hedgehog and bone morphogenetic protein 4 via opposing effects on histone acetylation. J Neurosci. 2012;32:6651–6664. doi: 10.1523/JNEUROSCI.4876-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida K, Masago K, Nakanishi H, Kihara Y, Hamano F, Tajima Y, Taguchi R, Shimizu T, Ishii S. Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. J Biol Chem. 2009;284:17731–17741. doi: 10.1074/jbc.M808506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Hama K, Contos JJ, Anliker B, Inoue A, Skinner MK, Suzuki H, Amano T, Kennedy G, Arai H, Aoki J, Chun J. LPA3-mediated lysophosphatidic acid signalling in embryo implantation and spacing. Nature. 2005;435:104–108. doi: 10.1038/nature03505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Skinner MK, Kennedy G, Chun J. Age-dependent loss of sperm production in mice via impaired lysophosphatidic acid signaling. Biol Reprod. 2008;79:328–336. doi: 10.1095/biolreprod.108.068783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes-Rapozo V, Felgueiras LO, Viana NL, Fierro IM, Barja-Fidalgo C, Manhães AC, Barradas PC. A role for the MAPK/ERK pathway in oligodendroglial differentiation in vitro: stage specific effects on cell branching. Int J Dev Neurosci. 2009;27:757–768. doi: 10.1016/j.ijdevneu.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, Chan JR, Wu J, Lu QR. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuelling LM, Fuss B. Autotaxin (ATX): a multi-functional and multi-modular protein possessing enzymatic lysoPLD activity and matricellular properties. Biochim Biophys Acta. 2008;1781:525–530. doi: 10.1016/j.bbalip.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuelling LW, Waggener CT, Afshari FS, Lister JA, Fuss B. Autotaxin/ENPP2 regulates oligodendrocyte differentiation in vivo in the developing zebrafish hindbrain. Glia. 2012;60:1605–1618. doi: 10.1002/glia.22381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukiura H, Hama K, Nakanaga K, Tanaka M, Asaoka Y, Okudaira S, Arima N, Inoue A, Hashimoto T, Arai H, Kawahara A, Nishina H, Aoki J. Autotaxin regulates vascular development via multiple lysophosphatidic acid (LPA) receptors in zebrafish. J Biol Chem. 2011;286:43972–43983. doi: 10.1074/jbc.M111.301093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannino DA, Appel B. Olig2+ precursors produce abducens motor neurons and oligodendrocytes in the zebrafish hindbrain. J Neurosci. 2009;29:2322–2333. doi: 10.1523/JNEUROSCI.3755-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]