Abstract
Huntington's disease (HD) is an inherited, neurodegenerative disorder caused by a single-gene mutation: a CAG expansion in the huntingtin (HTT) gene that results in production of a mutated protein, mutant HTT, with a polyglutamine tail (polyQ-HTT). Although the molecular pathways of polyQ-HTT toxicity are not fully understood, because protein misfolding and aggregation are central features of HD, it has long been suspected that cellular housekeeping processes such as autophagy might be important to disease pathology. Indeed, multiple lines of research have identified abnormal autophagy in HD, characterized generally by increased autophagic induction and inefficient clearance of substrates. To date, the origin of autophagic dysfunction in HD remains unclear and the search for actors involved continues. To that end, recent studies have suggested a bidirectional relationship between autophagy and primary cilia, signaling organelles of most mammalian cells. Interestingly, primary cilia structure is defective in HD, suggesting a potential link between autophagic dysfunction, primary cilia and HD pathogenesis. In addition, because polyQ-HTT also accumulates in primary cilia, the possibility exists that primary cilia might play additional roles in HD: perhaps by disrupting signaling pathways or acting as a reservoir for secretion and propagation of toxic, misfolded polyQ-HTT fragments. Here, we review recent research suggesting potential links between autophagy, primary cilia and HD and speculate on possible pathogenic mechanisms and future directions for the field.
Facts
Autophagy is increased, but inefficient, in HD
Primary cilia dysfunction impairs autophagy
Autophagic disruption impairs regulation of primary cilia biogenesis and growth
PolyQ-HTT exhibits increased interaction with huntingtin-associated protein 1 (HAP1), resulting in PCM1 accumulation at the centrosome, aberrant ciliogenesis and altered primary cilia structure
Multiple types of neuronal aggregates containing misfolded disease-associated proteins, such as α-synuclein, tau, ALS-associated proteins and polyglutamine proteins, have been shown to spread in a prion-like manner––mutant huntingtin can undergo cell-to-cell transmission in vitro and in vivo
Open Questions
How does altered ciliary structure affect primary cilia function in HD?
What is the link between elongated primary cilia morphology and aberrant corticostriatal signaling in HD?
Can normalizing primary ciliogenesis reverse neurodegeneration in HD?
Are primary cilia secretory organelles in neurons or astrocytes and do they contribute to propagation of misfolded polyQ-HTT in the brain?
Huntington's disease (HD) is an inherited, autosomal dominant neurodegenerative disease characterized by progressive motor dysfunction, cognitive decline and psychiatric disturbances. The prevalence in the Western hemisphere is 4–10 in 100 000 and the average age of onset is 40 years.1, 2 Brain atrophy in HD manifests first in the striatum, and later, in other brain regions including the substantia nigra, hippocampus, cerebellum and cerebral cortex.3 Striatal medium spiny neurons are the most vulnerable, while interneurons are spared.3 There is currently no cure or effective treatment to halt the progression of neurodegeneration in HD.
HD is caused by a CAG repeat expansion in a single gene, huntingtin (HTT), located on chromosome 4, which codes for a large protein with an expanded poly glutamine (polyQ) tail. The presence of 40 or more CAG repeats causes HD, whereas 36–39 repeats cause HD in some cases (often with later onset).1, 4, 5 Evidence suggests that both loss-of-function and gain-of-function mechanisms play a role in polyQ-HTT-mediated pathology. Embryonic lethality of complete Htt knockout in mice6, 7 and the pathological significance of loss of normal functions of HTT, such as transport of brain-derived neurotrophic factor (BDNF),8, 9 are suggestive of a loss-of-function mechanism. However, the dominant inheritance pattern of HD and the ability of full-length and truncated forms of polyQ-HTT to induce HD-like phenotypes in animal models suggest that a toxic gain-of-function mechanism also contributes to HD pathology.10
Although the normal physiological functions of wild-type (WT) HTT are not completely understood, it is essential for neurodevelopment and is believed to function as a scaffolding protein in multiple pathways including microtubule-dependent vesicular transport, neuronal gene transcription and synaptic transmission.1, 3, 8 Important recent findings also describe a potential function as a scaffold for multiple autophagy-associated proteins in selective autophagy.11 Consistent with these putative functions, HTT contains multiple leucine-rich HEAT repeats that are involved in protein–protein interactions.12, 13
The exact nature of the polyQ-HTT toxic gain of function remains elusive, but several possibilities have been identified. It has been suggested that proteolytic cleavage of polyQ-HTT might play an early role in HD pathogenesis, giving rise to toxic N-terminal fragments.2, 14 Cleavage products generated by caspase-3, caspase-6, calpains and matrix metalloproteinase 10 have been shown to accumulate in the HD brain before neurodegeneration.14, 15, 16, 17, 18, 19 However, more recent evidence suggests full-length polyQ-HTT monomers are more abundant and more toxic than fragments.20
Aberrant interactions of polyQ-HTT and other proteins have been shown to disrupt microtubule-based transport and gene transcription. HTT has an important role in vesicular transport via interactions with motor protein cargo adaptors such as HTT-associated protein 1 (HAP1) and dynactin.2, 8, 21 This complex is disrupted by polyQ-HTT, resulting in reduced transport, including vesicular trafficking of BDNF.2, 8 PolyQ-HTT also interferes with transcription factors and abnormally interacts with genomic DNA, disrupting transcription factor binding and DNA conformation.22 One result of the transcriptional disruption is reduced transcription of BDNF.23 In addition, repression of the transcription co-activator, peroxisome proliferator-activated receptor gamma co-activator-1-alpha (PGC-1α), results in reduced mitochondrial biogenesis,24 and polyQ-HTT interaction with mitochondrial proteins results in impaired energy metabolism stemming from mitochondrial dysfunction and altered dynamics.25, 26 The PGC-1α target and regulator of the autophagy-lysosome pathway, transcription factor E-B, is also impaired in HD mice.27
Similar to other neurodegenerative diseases, protein misfolding and aggregation are a hallmark of HD neuropathology. However, the process of polyQ-HTT aggregation is complex and untangling the pathways and determining the pathological significance remains a challenge. Although it was initially hypothesized that large aggregates containing polyQ-HTT were the cause of neuronal death in HD, more recent evidence suggests that soluble, misfolded monomers and oligomers are in fact toxic and aggregate formation is a protective mechanism.28 PolyQ-HTT aggregates correlate poorly with HD severity and progression.29, 30 In vitro studies suggested that polyQ-HTT fragments undergo a conformational change, forming soluble toxic β-sheet monomers and/or oligomers.31, 32 Analysis of human HD brain and cultured neurons revealed that native polyQ-HTT occurs primarily as a soluble, full-length monomer,20 and in human embryonic stem cells (hESCs), there was a quantitative relationship between neurodegeneration and soluble monomeric (but not oligomeric or aggregated) polyQ-HTT.33
Finally, polyQ-HTT impairs proteasomal activity and autophagy by interacting abnormally with proteins involved in these pathways.34, 35 Dysfunctional autophagy in HD is the focus of this review and will be discussed in more detail in the following sections. This pathological pathway of HD has been extensively investigated, but the functional mechanisms of action and cellular regulators of dysfunction have yet to be sufficiently described. Here we review recent data suggesting a potential link between autophagic dysfunction and primary cilia––often overlooked cellular signaling organelles that are novel regulators of autophagy––in HD and neurodegeneration. We conclude by speculating on other potential avenues of primary cilia involvement in the pathogenesis of HD.
Autophagic Dysfunction in HD
There is substantial evidence that autophagy, the cellular process of protein and organelle degradation by lysosomes, is dysfunctional in HD. Autophagy has a vital role throughout the nervous system and loss of this cellular function causes premature aging and neurodegeneration.36, 37 The autophagy-lysosome system compliments the ubiquitin-proteasome system (UPS) in the degradation of misfolded proteins and is upregulated when proteasome function is impaired. In HD, polyQ-HTT overwhelms UPS chaperones such as Hsp70 and DNAJB, decreasing their availability to other misfolded proteins and increasing the flux of proteins to the proteasome.35 An increased cellular load of ubiquitinated proteins, the relatively low basal functionality of striatal proteasomes38 and an overall age-dependent loss of UPS activity39 all might contribute to the increased burden placed on the autophagy pathway in HD. Endoplasmic reticulum (ER) stress also induces autophagy in HD and it has also been shown that polyQ-HTT disrupts calcium homeostasis,40 impairs ER-Golgi-lysosome trafficking41, 42 and inhibits the ER-associated degradation pathway.43 Furthermore, because post-mitotic striatal neurons are unable to dilute accumulated misfolded proteins through cell division, ineffective autophagy might disrupt cellular proteostasis and exacerbate polyQ-HTT toxicity.44
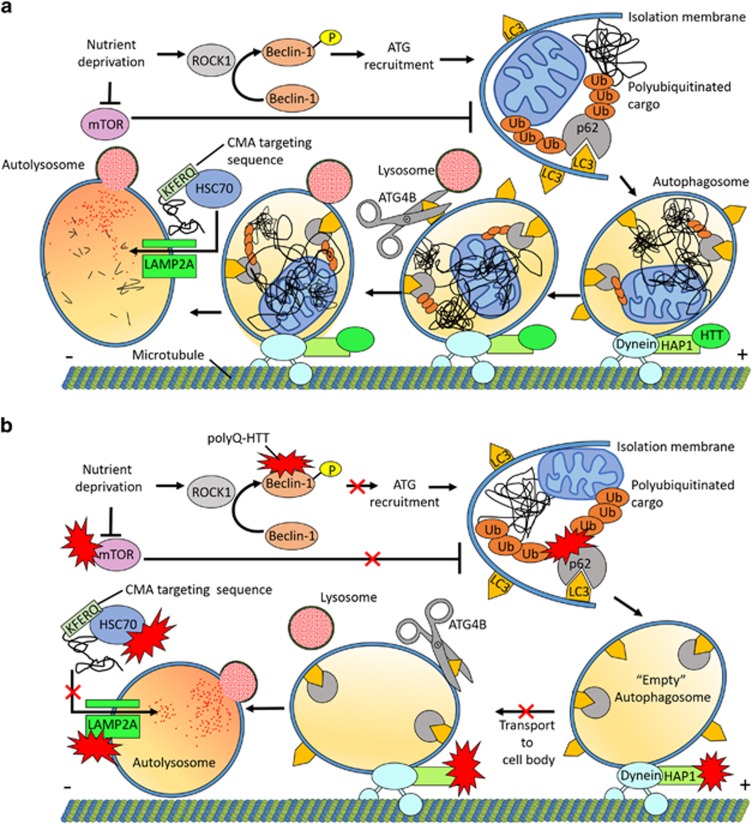
Autophagy can be divided into three subtypes based on cargo recognition and mechanisms of lysosomal delivery––microautophagy, chaperone-mediated autophagy (CMA) and macroautophagy. CMA and macroautophagy are the two primary pathways in mammalian cells (Figure 1a). CMA is a selective form of autophagy that acts upon protein substrates that contain pentapeptide amino-acid targeting sequences biochemically related to KFERQ.45 Chaperones, including heat-shock cognate protein 70 (HSC70), deliver misfolded proteins to lysosomes, where a membrane receptor complex containing lysosome-associated membrane protein 2A (LAMP2A) recognizes and translocates cargo into the lysosomal lumen for degradation.46 Studies have shown that HTT is processed by CMA machinery. HTT contains putative CMA-targeting motifs, interacts with HSC70 and is delivered into the lysosome in a LAMP2A-dependent manner. In HD, CMA function is diminished. PolyQ-HTT excessively interacts with HSC70 and LAMP2A, clogging this pathway47 (Figure 1b).
Figure 1.
PolyQ-HTT impairs autophagy. (a) Nutrient deprivation induces macroautophagy, whereas activated mTOR acts as a major suppressor of this pathway. Upon nutrient deprivation, ROCK1 promotes autophagy by phosphorylating Beclin-1, a key regulator of autophagy. Beclin-1 orchestrates recruitment of additional ATG proteins required for autophagosome formation. The first stage of macroautophagy is formation of the double-membraned autophagosome. Small membrane structures called isolation membranes elongate and surround proteins or organelles destined for degradation. LC3 complexes associate with the isolation membrane early in autophagy. LC3 aids cargo sequestration through interaction with p62, a receptor essential for targeting polyubiquitinated cargo to the autophagosome. Autophagosomes colocalize with HTT and HAP1, enabling dynein-mediated retrograde transport toward the lysosome-enriched cell body. ATG4B cleaves LC3 from the outer autophagosome membrane, allowing fusion with lysosomes and cargo degradation by hydrolytic enzymes. Chaperone-mediated autophagy (CMA) acts upon protein substrates that contain pentapeptide amino-acid targeting sequences biochemically related to KFERQ. Chaperones, including HSC70, deliver misfolded proteins to lysosomes, where a membrane receptor complex containing lysosome-associated membrane protein 2 A (LAMP2A) recognizes and translocates cargo into the lysosomal lumen for degradation. (b) PolyQ-HTT impairs autophagy pathways through aberrant protein interactions. PolyQ-HTT sequesters mTOR, increasing autophagy induction. PolyQ-HTT sequesters Beclin-1, impairing autophagosome formation. PolyQ-HTT binds p62, disrupting LC3 interaction and causing cargo recognition defects with assembly of empty autophagosomes. PolyQ-HTT disrupts retrograde microtubule-based transport through interaction with HAP1 and dynein/dynactin. Autophagosome transport to the cell body is hindered, resulting in formation of fewer autolysosomes and decreasing overall autophagic degradation. CMA is also impaired in HD; polyQ-HTT sequesters the HSC70 chaperone that is important for transporting cargo to lysosomes. PolyQ-HTT also directly interacts with LAMP2A of the lysosomal membrane translocation complex, preventing the transport of cargo into lysosomes
Macroautophagy is a bulk clearance mechanism of proteins or organelles.48, 49 The three main stages of macroautophagy are autophagosome formation, maturation and fusion with lysosomes.39 During the first stage, autophagosome formation, an isolation membrane forms to surround proteins or whole organelles.49 Fifteen autophagy-related (ATG) proteins have key roles in formation of the autophagosome.50, 51 After the autophagosomes are fully formed, they are transported along microtubules and fuse with lysosomes, which results in lysosomal degradation of the contents49 (Figure 1a). Macroautophagy is abnormally induced in HD as evidenced by increased levels of autophagic vacuoles, the cargo adaptor p62 and processed autophagosome membrane marker microtubule-associated protein 1A/1B-light chain 3 (LC3)-II in HD patients and mouse models.34, 52 In addition, polyQ-HTT sequesters and inhibits mammalian target of rapamycin (mTOR), a negative regulator of macroautophagy53 (Figure 1b).
Post-translational modifications of proteins can regulate their autophagic clearance and several such modifications have been shown to contribute to clearance or stabilization of HTT. For example, polyQ-HTT exhibits reduced serine (S13 and S16) phosphorylation, a modification that regulates the balance between ubiquitination and SUMOylation of adjacent lysine residues (K6, K9 and K15).54, 55 Inhibitor of κB kinase-mediated phosphorylation results in increased polyubiquitination and enhanced clearance of WT-HTT. By contrast, decreased phosphorylation of polyQ-HTT leads to SUMOylation, which in turn stabilizes polyQ-HTT and increases neurotoxicity.55, 56 Acetylation is another modification regulating polyQ-HTT clearance. Lysine-444 acetylation, mediated by the acetyltransferase activity of CREB-binding protein and occurring primarily in polyQ-HTT, is an important regulatory mechanism that targets polyQ-HTT to autophagosomes for degradation.57 Myristoylation, another acylation modification, promotes protein–membrane interaction and is relevant to autophagy regulation. After caspase-mediated cleavage of HTT, an N-terminal glycine (G553) undergoes myristoylation.58 Myristoylated HTT553-586 fragments may influence autophagosome formation by sensing and inducing membrane curvature at the ER.49 The ER membrane is the source of the isolation membrane,59 an early structure required for engulfment of cytoplasmic materials.60 Myristoylation has been shown to be significantly less prevalent in polyQ-HTT compared with WT,58 and thus, further examination of this post-translational modification in the context of HD is required.
Despite induction of autophagy, there is inefficient polyQ-HTT clearance in HD. This stems from abnormal autophagic function arising from defects at several key points of the pathway. Normally, upon nutrient deprivation rho-associated, coil-containing protein kinase 1 (ROCK1) promotes autophagy by phosphorylating Beclin-1, a key regulator of autophagy.61 PolyQ-HTT sequesters and inhibits Beclin-1, impairing autophagosome formation and polyQ-HTT clearance.62 In addition, polyQ-HTT disrupts retrograde transport of autophagosomes to the lysosome-rich cell body, resulting in fewer fusion events and defective degradation.63 There are also pronounced cargo recognition defects possibly resulting from aberrant polyQ-HTT–p62 interaction.34 The p62 autophagic adaptor protein has a key role in selective macroautophagy, binding to polyubiquitin chains of targeted cargo and LC3 membrane proteins of the forming autophagosome.64 Interaction with polyQ-HTT decreases p62 availability to other substrates. This results in the production of relatively empty autophagosomes, thereby compromising macroautophagy and further contributing to neurotoxicity in HD34 (Figure 1b).
Results of recent studies in animal models of HD have suggested that HTT plays an important role in autophagy. For example, expression of full-length HTT lacking its polyQ (ΔQ-HTT) in a knock-in mouse model of HD increases autophagy markers and longevity.65 In vitro expression of ΔQ-HTT also increases autophagosome synthesis and ATG5-dependent clearance of HTT aggregates.65 In addition, a new study reported that WT-HTT serves as an important scaffold protein in multiple types of selective macroautophagy (not including starvation-induced autophagy) in Drosophila and mammalian cells (including mouse embryonic fibroblasts and striatal cells).66 WT-HTT is able to bind simultaneously two important ATG proteins, p62 and unc-51-like autophagy activating kinase (ULK1).66
In sum, studies to date indicate abnormal autophagy function in HD and suggest that HTT has a functional role in autophagy. Upregulation of autophagy has been shown to enhance polyQ-HTT clearance53 and drugs targeting autophagy––such as rapamycin/CCI-779, lithium, trehalose and rilmenidine––continue to be of interest as potential therapeutic agents for HD.49 Given the somewhat contradictory findings of polyQ-HTT-mediated autophagy induction being detrimental and autophagy enhancer therapy being beneficial, the role of autophagy in HD appears to be even more complex than previously thought. A key piece of this puzzle might be the primary cilium, a novel regulatory organelle of autophagy.
Primary Cilia and Autophagy
Structure, function and biogenesis of primary cilia
Primary cilia are single, non-motile signaling organelles found on the surface of most mammalian cells (Figure 2). They are required for Sonic hedgehog (Shh) signal transduction and have essential roles in Wnt, platelet-derived growth factor and transforming growth factor—β signaling pathways.67, 68, 69, 70 Present in neurons, astrocytes and progenitors, these structures have an important role in neurodevelopment––acting in neuronal homeostasis, differentiation and survival.71 Neurological defects associated with so-called ciliopathies, multi-system genetic disorders stemming from primary ciliary dysfunction, underscore the critical role of primary cilia in the nervous system. Primary cilia also have important functions in the adult nervous system, including neural stem cell regulation, neuronal signaling and regeneration.72 Interestingly, primary cilia dysfunction has recently been implicated in late-onset neurodegenerative disorders such as Alzheimer's disease and HD.73, 74
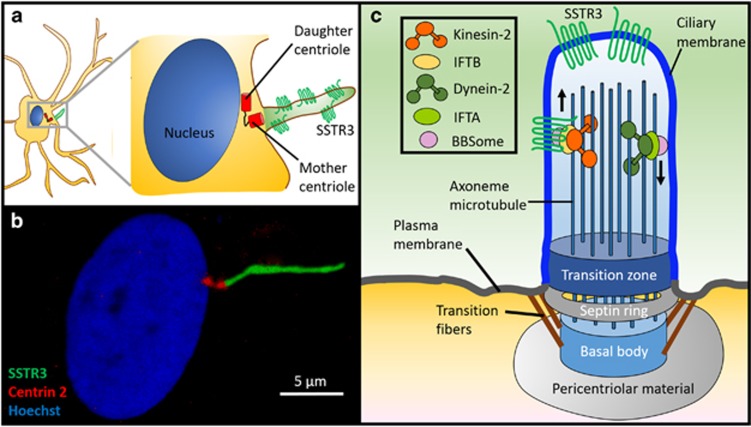
Figure 2.
The primary cilium is a specialized signaling organelle. (a and b) Rat spinal astrocytes expressing dsRed-Centrin 2 (centriole marker) and Somatostatin receptor type III-GFP (primary cilia marker) were fixed and stained with Hoechst dye (nucleus marker). (c) The primary cilium is a specialized non-motile signaling organelle that is compartmentalized from the rest of the cell by unique structures, including an ordered membrane-bound septin ring structure and a transition zone at the base of the cilium. The microtubule-based axoneme nucleates out from the basal body after being anchored to the plasma membrane by transition fibers. Signal transduction is mediated by the receptor dense ciliary membrane and a specialized intraflagellar transport system (IFT) along the axoneme. The primary ciliary membrane is enriched in membrane receptors that include SSTR3. Cargo is transported into the primary cilium via the Bardet-Biedl Syndrome (BBSome) complex (which also coordinates cargo attachment to microtubule motors), IFT complex A (for dynein-2-mediated retrograde transport) and IFT complex B (for kinesin-2-mediated transport to the ciliary tip)
Primary cilia are covered in a specialized receptor-dense membrane and protrude into the extracellular environment like cellular antennae.75 To facilitate their specialized signaling functions, primary cilia are compartmentalized from the rest of the cell. A septin GTPase structured ring embedded within the plasma membrane acts as a diffusion barrier76 and a gate-like transition zone at the ciliary base regulates protein transport from the cytoplasm into the primary cilium.77 A unique bidirectional intraflagellar transport (IFT) system is utilized for trafficking of signaling and structural components along a microtubule-based axoneme.78 IFT employs kinesin-2 motors for anterograde transport (from ciliary base to tip) and specialized cytoplasmic dynein-2 motors for retrograde transport (tip to base). IFT proteins link motor proteins and cargo, creating multi-protein IFT complexes that travel along the axoneme79 (Figure 2).
Primary cilia formation involves axonemal extension from the basal body, a structure originating from the mother centriole in post-mitotic cells.80 Following basal body anchoring to the plasma membrane, vesicles carrying material needed for ciliary growth dock at the ciliary base. Materials, such as ciliary membrane receptors, are incorporated into IFT particles for transport into the cilium,81 and elongation is mediated by bidirectional IFT.82 Bardet-Biedl Syndrome proteins play a key role in vesicular transport of signaling receptors to primary cilia, in addition to their role in regulation of IFT assembly and turnaround.79 Primary cilia are formed in quiescent cells and their disassembly is required for cell cycle entry and progression.83 Aurora A kinase, localized at the basal body, activates the tubulin deacetylase histone deacetylase 6 and promotes deacetylation of α-tubulin, ciliary resorption and axoneme disassembly.84 Subsequent liberation of the centriole, required for reutilization in mitotic spindle bodies, regulates cell cycle re-entry.85
Primary cilia are important cellular environmental sensors, signaling back from the extracellular space information such as nutrient availability.86, 87 Primary cilia communicate external signals from growth factors and nutrients by regulating the activity of mTORC1,88 a protein complex directly linked to the regulation of autophagy,89 which is inhibited by nutrient stress. In cultured cells, primary cilia biogenesis and macroautophagy are both triggered upon growth arrest induced by nutrient withdrawal.90, 91 In addition to this temporal relationship, these processes also share similar sites of origin. Primary cilia have been observed as membrane-derived extensions for well over 100 years and new data now describe the plasma membrane as a source for early autophagosome membranes.92 These findings hint at a functional relationship between these pathways, and recent results from the Cuervo and Zhong groups elegantly describe an intricate crosstalk between primary cilia and autophagy.50
Primary cilia dysfunction impairs autophagy
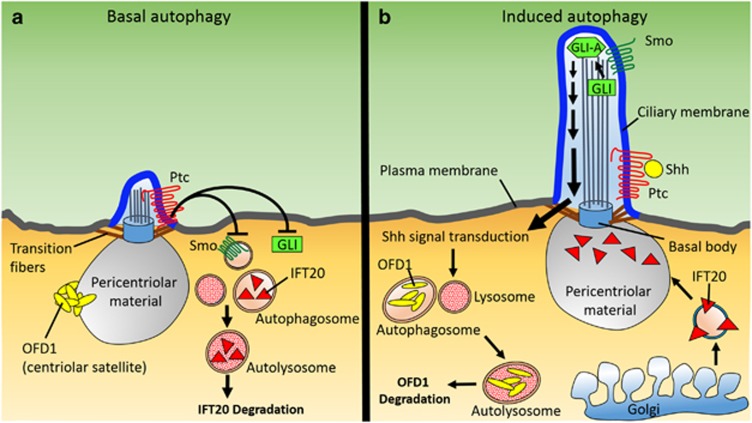
The Cuervo lab recently reported that primary cilia are required for induction of autophagy, as evidenced by compromised autophagosome biogenesis in serum-starved cells deficient in IFT20 or IFT88.93 These proteins modulate protein trafficking between the Golgi complex and primary cilia and are required for primary cilia assembly and growth. In these experiments, autophagy was induced through serum withdrawal, a stimulus that promotes both autophagy and ciliogenesis. The investigators demonstrated that reduced ciliogenesis limited upregulation of autophagy following nutrient deficiency. Further experiments showed that autophagy induction is dependent on Shh signaling,93 a pathway mediated through the primary cilium, and that autophagic dysfunction can be attributed to impaired ciliary signaling (Figure 3). Through activation of Shh signaling, the researchers induced autophagy in serum-supplemented cells to levels observed upon serum removal. Various treatments and conditions increased autophagy, including treatment with purmorpamine, an agonist of the Shh signal transducer membrane protein Smoothened (Smo); knockout of Patched-1 (Ptc), a membrane receptor protein that represses Smo in the absence of Shh; and overexpression of glioma-associated oncogene homolog transcription factor (GLI), the most downstream element within the Shh pathway. Signaling proteins exit the cilium near the basal body, where regulatory ATG proteins required for autophagosome formation are located. One such example is ATG16L, a protein that mediates the activation of membrane-bound LC394 and has an essential role in autophagy through modulation of nascent autophagosomal membrane elongation.95 Upon serum withdrawal, Shh signaling promotes ATG16L recruitment to the ciliary base via IFT20-enriched vesicles,93 demonstrating that Shh signaling and IFT collaboratively promote autophagosome formation and provide an important point of regulation for macroautophagy. Thus, aberrant primary cilia function in the form of altered Shh signaling can hinder cellular homeostasis through misregulation of autophagy. In the context of HD, this may also augment ciliary dysfunction by altering the structural integrity of primary cilia.
Figure 3.
The functional relationship between primary cilia and autophagy. (a) In the absence of Sonic Hedgehog (Shh), the Patched (Ptc) receptor restricts Smoothened (Smo) entry into the primary cilium, which results in processing of the transcriptional effector GLI into its repressor form or targeting for degradation. Autophagy is not induced, OFD1 at centriolar satellites is not degraded and ciliogenesis is impaired. Basal autophagy degrades IFT20 when nutrient levels are normal. This reduces trafficking of IFT components to the pericentriolar material, thus impairing the ciliary transport required for growth of primary cilia. (b) Primary ciliogenesis and autophagy are induced during starvation or periods of low-nutrient availability. Autophagy induction is dependent on Shh signaling through the primary cilium. Shh signal transduction is mediated by Smo, which undergoes localization to the primary cilium following Shh binding to Ptc. Smo accumulation within primary cilia promotes GLI protein processing into its transcriptional activator form (GLI-A), enabling downstream signal transduction. Shh-mediated signal transduction promotes the autophagic degradation of OFD1 at centriolar satellites, which is required for ciliogenesis
Compromised autophagy alters primary cilia growth and structure
Interestingly, autophagy itself has a key role in regulating primary cilia biogenesis and growth––suggesting that the relationship between primary cilia and autophagy is bidirectional. Disruption of either basal or induced autophagy, as seen in HD, alters ciliary regulatory factor availability and causes imbalanced growth of primary cilia. The Zhong lab showed that induced autophagy promotes primary cilia biogenesis through the degradation of oral-facial-digital syndrome type 1 (OFD1) protein at centriolar satellites96––the nonmembranous, electron-dense granules that cluster around centrosomes (Figure 3b). OFD1 also localizes to the centrioles, where it has been shown to be essential for primary ciliogenesis,97 yet the centriolar satellite pool of protein exhibited significantly faster turnover due to induced autophagy. OFD1, a negative regulator of ciliogenesis, is spared during basal autophagy.96 The Cuervo lab demonstrated that basal autophagy regulates primary cilia growth: under normal nutrient conditions, IFT20 is degraded, limiting the ciliary trafficking of components required for lengthening of primary cilia93 (Figure 3a). Thus, disruption of either basal or induced autophagy has the potential to affect primary cilia growth by altering availability of IFT20 and OFD1. Increased autophagy induction results in degradation of OFD1 at the centriolar satellites, removing the block on ciliary growth and allowing continued lengthening of primary cilia. On the other hand, blockage of basal autophagy increases the amount of IFT20, also resulting in enhanced primary cilia growth. Interestingly, elongated ciliary morphology is evident in HD mice and human HD brains, in which autophagic dysfunction has been reported.74
Primary Cilia and HD Pathogenesis
Primary cilia morphology is altered in HD
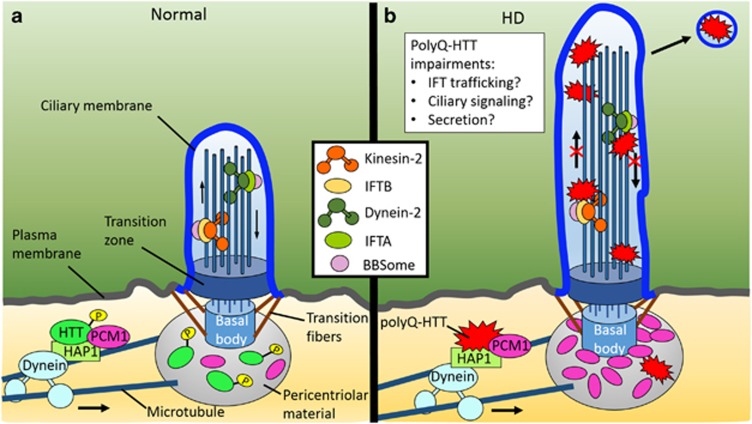
The association between HTT, microtubules and motor proteins is critical for trafficking within the cell. HTT binds directly to dynein98 and interacts indirectly with dynactin through HAP1,99 a neuronal-binding partner of HTT that participates in intracellular trafficking.100, 101 Microtubule-based trafficking is essential for many cellular processes, including primary ciliogenesis. The formation of primary cilia depends on the proper localization of regulatory factors and building components to the pericentriolar material, a matrix of proteins surrounding the centrosome that serves as a binding platform for the microtubule nucleation required for axoneme assembly.102 Proper primary cilia biogenesis is dependent on the centrosomal trafficking of pericentriolar material 1 protein (PCM1), a centriolar satellite protein essential for microtubule organization103 and recruitment of proteins to the centrosome.104 The Saudou group demonstrated that HTT, in conjunction with HAP1, mediates the transport of PCM1 between the cytoplasm and pericentriolar material. Their data elucidate a role for HTT in ciliogenesis through the HAP1-PCM1 pathway74 (Figure 4a). Significantly, they found that polyQ-HTT exhibits increased interaction with HAP1, thereby causing PCM1 accumulation at the centrosome, increased ciliogenesis and primary cilia elongation74 (Figure 4b). The resulting hypermorphic primary cilia may be of particular significance in HD, as altered function caused by changes in ciliary structure may affect autophagic regulation. Primary cilia downregulate mTOR signaling88 and polyQ-HTT sequesters and inhibits mTOR.53 Because mTOR is a negative regulator of autophagy, the combinatorial effects of elongated primary cilia and polyQ-HTT may explain the increased autophagy in HD.
Figure 4.
Abnormal ciliogenesis and hypertrophic cilia are observed in Huntington's disease. (a) HTT and HAP1 regulate primary cilia formation through the transport of PCM1 to the centrosome. (b) PolyQ-HTT exhibits increased binding to HAP1, causing accumulation of PCM1 at the centrosome, increased ciliogenesis and lengthening of the primary cilium. Increased ciliogenesis may perturb neuronal homeostasis and affect neuron survival. Altered polyQ-HTT–HAP1 interaction may also result in aberrant transport of membrane receptors and potential ciliary signaling dysfunction. PolyQ-HTT is also hypophosphorylated within its N17 domain, an area important for intracellular targeting, and exhibits increased localization within the primary cilium. Accumulated polyQ-HTT may interfere with IFT machinery, altering microtubule-based transport along the axoneme and impairing primary ciliary signaling. In light of recent evidence of vesicular secretion from neuronal primary cilia, these organelles may act as a conduit for the intercellular transfer of polyQ-HTT
Primary cilia-mediated signaling is dysfunctional in HD
Changes in primary cilia-mediated signaling may contribute to HD by affecting overall ciliary function, as many signaling pathways relevant to HD operate through primary cilia (Figure 4b). Various pathway impairments are linked to HD pathogenesis, including decreased BDNF signaling at corticostriatal terminals. The clinical manifestations of striatal atrophy have long been attributed to reduced levels of BDNF,105 a secreted neurotrophin vital for neuronal development and synaptic plasticity. Deficits in BDNF-mediated downstream tyrosine-related kinase B (TrkB) signaling have also been reported.106, 107, 108, 109 TrkB receptor localization and activation are primary cilia-dependent processes and primary cilia loss impedes activation of TrkB.110 Conversely, upregulation of p75NTR signaling has been shown to disrupt TrkB signaling.106 Because p75NTR also localizes to primary cilia111 and is the only other known receptor for BDNF, the effect of changes in ciliary-mediated signal transduction in HD merits further examination.
PolyQ-HTT accumulates within primary cilia
PolyQ-HTT may also have a more direct impact on primary cilia structure and function, as recent data have demonstrated that polyQ-HTT accumulates within primary cilia. Transport of HTT between the basal body and primary cilium is mediated by phosphorylation (S13 and S16) within the amino-terminal alpha-helical N17 domain (N17). PolyQ-HTT exhibits lower N17 phosphorylation than WT-HTT, and thus, polyQ-HTT accumulates within the primary cilia stalk112, 113 (Figure 4b). With HTT acting as a scaffolding protein connecting cargo to kinesin or dynactin for microtubule-based transport, polyQ-HTT may convey toxicity by directly interfering with IFT components localized to the primary cilium. In addition to promoting localization to primary cilia, N17 phosphorylation induces clearance of polyQ-HTT through the autophagic pathway,55 again demonstrating the intricate relationship between primary cilia and autophagy in HD pathogenesis.
Primary Cilia: Secretory Organelles?
Another avenue by which primary cilia could participate in HD pathogenesis is by secretion of polyQ-HTT from neurons (Figure 4b). As outlined in the next section, increasing evidence suggests that progression of late-onset neurodegenerative diseases such as HD is driven by transmission and propagation of toxic forms of disease-associated proteins via a mechanism akin to that of prion diseases. Such protein secretion could further overload and overwhelm the protective cellular maintenance systems of vulnerable neurons. Although the secretory capabilities of primary cilia in humans remain uncertain, several lines of evidence suggest that cilia might act as secretory organelles.
First, IFT proteins, key components of the ciliary structure, have been shown to play a role in exocytosis and extracellular signaling in non-ciliated cells.114, 115 T lymphocytes express IFT proteins, including IFT20, and kinesin-2, the IFT-dependent motor. These proteins associate with the centrosome and Golgi.114 Given that transport protein machinery characteristic of and critical to ciliary structure and function is important for protein secretion in cells that do not develop cilia suggests the possibility that this machinery might play a similar protein secretory role in primary cilia.
Second, studies of simple organism model systems have identified release of extracellular vesicles from ciliary structures. EM analysis showed that cilia of Chlamydomonas, a unicellular green alga, secrete bioactive vesicles containing a proteolytic enzyme.116 This finding is of potential relevance to human primary cilia because green algae have traditionally served as a reliable model of mammalian cilia––other ciliary components first identified in algae and later found to be conserved in mammals include the IFT proteins.116, 117 In addition, ciliated sensory neurons in Caenorhabditis elegans were recently shown to release extracellular vesicles.118
Finally, early studies in mammalian cells, although far from conclusive, have yielded indicators of protein secretion from primary cilia. For example, a study of neuroepithelial cells found that the distribution of prominin-1, a stem cell marker, on primary cilia was suggestive of budding of small membrane vesicles from the tips of differentiating cells.119 In addition, the protein contents of purified human urinary exosome-like vesicles suggested that the structures possibly originated from primary cilia.120 To date, evidence of secretion of proteins from primary cilia in humans is at best preliminary, but because of observations in simpler organisms and the potential importance of such a mechanism, further exploration is warranted.
Primary Cilia: Propagation of Unfolded Proteins?
Prion theory
Increasing evidence supports a model of neurodegenerative disease in which pathological progression is driven by prion-like protein seeding and propagation. This prion theory of neurodegeneration postulates that fibrillary protein seeds are taken up from adjacent or connected cells in the brain and induce aggregation of normally structured proteins, leading to spreading of characteristic misfolded proteins.121 This process is analogous to classical prionopathies in which PrPSC, the misfolded form of the normal prion protein PrPC, propagates through neuronal networks by inducing pathological conformational changes of native prion protein. Most neurodegenerative diseases are characterized by dysfunction in discrete brain regions in the early stages and more widespread dysfunction in later stages, suggestive of a spreading pathology.121, 122 For example, in HD, pathophysiological progression is quite predictable, beginning in the striatum and later progressing to the cortex.123 Furthermore, spreading of protein aggregates has been shown to mirror the overall clinical pathophysiology of neurodegeneration.121
In recent years, multiple studies have investigated the prion-like capabilities of pathological proteins and protein aggregates associated with various neurodegenerative diseases.124 Exosomal secretion of small fractions of amyloid-β peptides has been observed.125 Internalization of aggregates containing α-synuclein, tau, ALS-associated proteins and polyglutamine proteins has also been shown in cultured cells.121 Specifically, uptake of α-synuclein has been observed in cultured human dopaminergic neuronal cells by a mechanism that involved endocytosis.126 In addition, cultured HEK and C17.2 cells have been shown to take up tau aggregates and transfer aggregates between cells that are able to interact with native tau directly and convert the native protein to fibrillar forms.127, 128 Furthermore, seeding of cell cultures with aggregated forms of ALS-associated proteins SOD1 and TDP-43 resulted in misfolding of WT proteins129, 130, 131 and cell-to-cell transmission of misfolded WT SOD1 has been observed.132 Finally, polyQ aggregates have been shown to be internalized by COS7, HEK293 and neuro2A cell lines in vitro and internalized aggregates colocalized with cytosolic quality control components such as ubiquitin and proteasome subunits; however, cell-to-cell transmission of aggregates between intact cells was limited and relatively inefficient.133 By contrast, expression of polyQ-HTT fragments in CAD neuronal cells and primary cerebellar granule neurons in vitro did result in spontaneous and efficient transfer to neighboring cells that did not require release from dying cells.134 In this instance, transfer required cell-to-cell contact and aggregates were found in tunneling nanotubes, which were increased by polyQ-HTT expression.134 Perhaps providing the most compelling evidence to date of propagation of polyQ-HTT in vitro, a new study has reported that when hESC-derived neurons were introduced into brain slices from HD disease model R6/2 mice, the hESC-derived neurons developed HTT aggregates and exhibited signs of toxicity.135 Similarly, when R6/2 cortical brain slices were co-cultured with striatal brain slices from WT mice, polyQ-HTT aggregates developed in the WT striatal neurons.135
Evidence of the propagation ability of neuronal aggregates has also been found in vivo in animal models and human patients. For example, multiple studies of animal models of neurodegeneration have reported strong evidence of cell-to-cell transmission of α-synuclein,126, 136 tau137, 138, 139, 140 and amyloid-β.141 Moreover, it was recently shown that lentiviral transduction of polyQ-HTT exon 1 into cortical neurons of WT mice led to the development of polyQ-HTT aggregates in striatal medium spiny neurons located in regions innervated by the transduced cortical neurons135, 142 and polyQ-HTT aggregates in Drosophila were found to effect prion-like conversion of soluble WT-HTT in the cytoplasm of phagocytic glia.143 Finally, transplant studies in PD and HD patients have confirmed propagation in humans. Engrafted fetal mesencephalic dopaminergic neurons developed ubiquitin- and α-synuclein-positive Lewy bodies when transplanted in PD patient brains.121, 123, 144, 145, 146, 147 Similarly, polyQ-HTT aggregates were found in grafted tissue in HD patient brains, although the aggregates were extracellularly located and not within engrafted neurons.148
Mechanism of polyQ-HTT cell-to-cell transmission: primary cilia involvement?
Although the ability of toxic proteins such as polyQ-HTT to propagate and spread in the brain is becoming well-established, the mechanism of such cell-to-cell transmission remains a mystery. To date, the study by Costanzo and colleagues is the only one, to our knowledge, to suggest a mechanism of cell-to-cell transmission of polyQ-HTT––namely, tunneling nanotubes.134 In light of the clues suggesting protein secretion from primary cilia, we speculate that primary cilia might be important, as yet unexplored mediators of prion-like spreading of polyQ-HTT (and potentially other toxic proteins) in the brain.
Remarks and Future Outlook
As discussed in this review, there are multiple avenues through which primary cilia could contribute to HD pathogenesis. It is well documented that autophagy is both increased and inefficient in HD, resulting in wasted energy expenditure without removal of toxic species or recycling of macromolecules. This autophagic increase causes degradation of OFD1 at centriolar satellites, promoting ciliogenesis and extending primary cilia length. Studies also show that autophagy induction depends on primary cilia signaling. With these points in mind, we speculate that elongated primary cilia, enriched in ciliary membrane receptors and signaling components, contribute to HD by exacerbating autophagy induction. Abnormal structure may also alter primary cilia function and affect pathways impaired in HD, such as striatal BDNF signaling. In sum, we propose that primary cilia and autophagic dysfunction have multifaceted and interdependent roles in HD pathogenesis, many of which require further investigation and characterization.
Autophagy and primary cilia
It should be noted that in the experiments investigating interrelationship between primary cilia and autophagy, serum starvation was used to induce autophagy. Although this method is commonly used to induce autophagy in non-neuronal cells, previous in vivo studies show that starvation does not induce autophagy in the brains of mice.149 Therefore, further investigation of primary cilia-autophagy interaction in a system more relevant to primary neuronal cells merits consideration.
Primary cilia in HD
Data from the Saudou group demonstrate that polyQ-HTT affects primary ciliogenesis, altering primary cilia structure and function.74 Based on this finding, we postulate that decreased BDNF-TRkB signaling in HD stems from these structural changes. However, it is possible that signaling through primary cilia is not adversely impacted by increased overall length. There are other potential points along this signaling pathway that may be susceptible to polyQ-HTT. One such area may involve trafficking of TRkB receptors. A recent study shows that HAP1 stabilizes and prevents the degradation of TRkB receptors,150 and in the context of HD, increased polyQ-HTT–HAP1 interaction may interfere with this important function. Establishing these connections will be an important avenue of investigation.
Acknowledgments
We thank Kimberly Kleinschmidt and Austin Kennedy for technical assistance. This work is supported by NIH grant R01NS055193.
Glossary
- ATG
autophagy-related
- BDNF
brain-derived neurotrophic factor
- CMA
chaperone-mediated autophagy
- ER
endoplasmic reticulum
- GLI
glioma-associated oncogene homolog transcription factor
- HAP1
huntingtin-associated protein 1
- HD
Huntington's disease
- HDAC6
histone deacetylase 6
- hESC
human embryonic stem cell
- HSC70
heat-shock cognate protein 70
- HTT/Htt
huntingtin
- IFT
intraflagellar transport
- LAMP2A
lysosome-associated membrane protein 2A
- LC3
microtubule-associated protein 1A/1B-light chain 3
- mTOR
mammalian target of rapamycin
- OFD1
oral-facial-digital syndrome type 1
- PCM1
pericentriolar material 1 protein
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator-1-alpha
- polyQ
poly-glutamine
- polyQ-HTT
mutant huntingtin with a poly-glutamine tail
- Ptc
Patched-1
- ROCK1rho-associated
coiled-coil-containing protein kinase 1
- Shh
Sonic hedgehog
- Smo
Smoothened
- SSTR3
somatostatin receptor type 3
- TrkB
tyrosine-related kinase B
- UPS
ubiquitin-proteasome system
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Edited by D Rubinsztein
References
- Novak MJU, Tabrizi SJ. Huntington's disease. BMJ. 2010;340:c3109–c3109. doi: 10.1136/bmj.c3109. [DOI] [PubMed] [Google Scholar]
- Ross CA, Tabrizi SJ. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Andrew SE, Paul Goldberg Y, Kremer B, Telenius H, Theilmann J, Adam S, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Liu J-P, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Marullo M, Vitali B, Tarditi A, Mariotti C, Valenza M, et al. Brain-derived neurotrophic factor in patients with Huntington's disease. PLoS ONE. 2011;6:e22966. doi: 10.1371/journal.pone.0022966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes CJ, La Spada AR. The many faces of autophagy dysfunction in Huntington's disease: from mechanism to therapy. Drug Discov Today. 2014;19:963–971. doi: 10.1016/j.drudis.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochaba J, Lukacsovich T, Csikos G, Zheng S, Margulis J, Salazar L, et al. Potential function for the huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci USA. 2014;111:16889–16894. doi: 10.1073/pnas.1420103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- Landles C, Sathasivam K, Weiss A, Woodman B, Moffitt H, Finkbeiner S, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J Biol Chem. 2010;285:8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Yi Y, Sapp E, Wang Y, Cuiffo B, Kegel KB, et al. Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc Natl Acad Sci USA. 2001;98:12784–12789. doi: 10.1073/pnas.221451398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Gutekunst C-A, Rogers D, Warby S, Graham RK, et al. Caspase cleavage of mutant huntingtin precedes neurodegeneration in Huntington's disease. J Neurosci. 2002;22:7862–7872. doi: 10.1523/JNEUROSCI.22-18-07862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, Ellerby LM. Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J Biol Chem. 2004;279:20211–20220. doi: 10.1074/jbc.M401267200. [DOI] [PubMed] [Google Scholar]
- Miller JP, Holcomb J, Al-Ramahi I, de Haro M, Gafni J, Zhang N, et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron. 2010;67:199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp E, Valencia A, Li X, Aronin N, Kegel KB, Vonsattel J-P, et al. Native mutant huntingtin in human brain: evidence for prevalence of full-length monomer. J Biol Chem. 2012;287:13487–13499. doi: 10.1074/jbc.M111.286609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Holzbaur ELF. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn CL, Sun T, Sadri-Vakili G, McFarland KN, DiRocco DP, Yohrling GJ, et al. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J Neurosci. 2008;28:10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Brain-derived neurotrophic factor in neurodegenerative diseases. Nat Rev Neurol. 2009;5:311–322. doi: 10.1038/nrneurol.2009.54. [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Panov AV, Gutekunst C-A, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, et al. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- Song W, Chen J, Petrilli A, Liot G, Klinglmayr E, Zhou Y, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RAV, et al. PGC-1α rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra97. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R, Atwal RS, Desmond C, Munsie L, Tran T. Huntington's disease: revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases. FEBS J. 2008;275:4252–4262. doi: 10.1111/j.1742-4658.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Saudou F, Finkbeiner S, Devys D, Greenberg ME. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Inui T, Popiel HA, Fujikake N, Hasegawa K, Urade Y, et al. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14:332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- Zhang QC, Yeh T, Leyva A, Frank LG, Miller J, Kim YE, et al. A compact β MODEL of huntingtin toxicity. J Biol Chem. 2011;286:8188–8196. doi: 10.1074/jbc.M110.192013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Palacino J. A novel human embryonic stem cell-derived Huntington's disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J. 2013;27:1820–1829. doi: 10.1096/fj.12-219220. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, et al. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J Cell Biol. 2012;196:573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Tsvetkov AS, Arrasate M, Barmada S, Ando DM, Sharma P, Shaby BA, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2013;9:586–592. doi: 10.1038/nchembio.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Cao F, Wang Z, Yu Z-X, Nguyen H-P, Evans J, et al. Huntingtin forms toxic NH2-terminal fragment complexes that are promoted by the age-dependent decrease in proteasome activity. J Cell Biol. 2003;163:109–118. doi: 10.1083/jcb.200306038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, et al. The first 17 amino acids of huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- Toro D, del, Alberch J, Lázaro-Diéguez F, Martín-Ibáñez R, Xifró X, Egea G, et al. Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell. 2009;20:1478–1492. doi: 10.1091/mbc.E08-07-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstaetter H, Kruppa AJ, Buss F. Huntingtin is required for ER-to-Golgi transport and for secretory vesicle fusion at the plasma membrane. Dis Model Mech. 2014;7:1335–1340. doi: 10.1242/dmm.017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald ML, Lindquist S. Impaired ERAD and ER stress are early and specific events in polyglutamine toxicity. Genes Dev. 2008;22:3308–3319. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Zhang X-D, Wu J-C, Lin F, Wang J, DiFiglia M, et al. The role of chaperone-mediated autophagy in Huntingtin degradation. PLoS ONE. 2012;7:e46834. doi: 10.1371/journal.pone.0046834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Martin DDO, Ladha S, Ehrnhoefer DE, Hayden MR. Autophagy in Huntington disease and huntingtin in autophagy. Trends Neurosci. 2015;38:26–35. doi: 10.1016/j.tins.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Orhon I, Dupont N, Pampliega O, Cuervo AM, Codogno P. Autophagy and regulation of cilia function and assembly. Cell Death Differ. 2015;22:389–397. doi: 10.1038/cdd.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Lee H, Noh J-Y, Oh Y, Kim Y, Chang J-W, Chung C-W, et al. IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux. Hum Mol Genet. 2012;21:101–114. doi: 10.1093/hmg/ddr445. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, et al. IKK phosphorylates huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, et al. SUMO modification of huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia Jr, TJ, Mazzulli JR, Cui L, Savas JN, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DDO, Heit RJ, Yap MC, Davidson MW, Hayden MR, Berthiaume LG. Identification of a post-translationally myristoylated autophagy-inducing domain released by caspase cleavage of huntingtin. Hum Mol Genet. 2014;23:3166–3179. doi: 10.1093/hmg/ddu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkar AU, Chu K, Raj L, Bouley R, Lee S-H, Kim Y-B, et al. Identification of ROCK1 kinase as a critical regulator of Beclin1-mediated autophagy during metabolic stress. Nat Commun. 2013;4:2189. doi: 10.1038/ncomms3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, et al. Regulation of intracellular accumulation of mutant huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- Wong YC, Holzbaur ELF. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci. 2014;34:1293–1305. doi: 10.1523/JNEUROSCI.1870-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Zheng S, Clabough EBD, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6:e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y-N, Xu Z, Patel B, Chen Z, Chen D, Tito A, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015;17:262–275. doi: 10.1038/ncb3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates Hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, et al. PDGFRα signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Clement CA, Ajbro KD, Koefoed K, Vestergaard ML, Veland IR, Henriques de Jesus MPR, et al. TGF-β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep. 2013;3:1806–1814. doi: 10.1016/j.celrep.2013.05.020. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Shyer AE, Dowdle WE, Gaulden J, Singla V, Reiter JF. Kif3a constrains β-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- Guemez-Gamboa A, Coufal NG, Gleeson JG. Primary cilia in the developing and mature brain. Neuron. 2014;82:511–521. doi: 10.1016/j.neuron.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y-G, Spassky N, Romaguera-Ros M, Garcia-Verdugo J-M, Aguilar A, Schneider-Maunoury S, et al. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- Sotthibundhu A, Li Q-X, Thangnipon W, Coulson EJ. Aβ1–42 stimulates adult SVZ neurogenesis through the p75 neurotrophin receptor. Neurobiol Aging. 2009;30:1975–1985. doi: 10.1016/j.neurobiolaging.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, Benstaali C, et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest. 2011;121:4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, et al. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–439. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Wei Q, Zhang Y, Li Y, Zhang Q, Ling K, Hu J. The BBSome controls IFT assembly and turnaround in cilia. Nat Cell Biol. 2012;14:950–957. doi: 10.1038/ncb2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT): role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Ke Y-N, Yang W-X. Primary cilium: an elaborate structure that blocks cell division. Gene. 2014;547:175–185. doi: 10.1016/j.gene.2014.06.050. [DOI] [PubMed] [Google Scholar]
- Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA, HEF1-Dependent Aurora A. Activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123:511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baly C, Aioun J, Badonnel K, Lacroix M-C, Durieux D, Schlegel C, et al. Leptin and its receptors are present in the rat olfactory mucosa and modulated by the nutritional status. Brain Res. 2007;1129:130–141. doi: 10.1016/j.brainres.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Händel M, Schulz S, Stanarius A, Schreff M, Erdtmann-Vourliotis M, Schmidt H, et al. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 1999;89:909–926. doi: 10.1016/s0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- Boehlke C, Kotsis F, Patel V, Braeg S, Voelker H, Bredt S, et al. Primary cilia regulate mTORC1 activity and cell size through Lkb1. Nat Cell Biol. 2010;12:1115–1122. doi: 10.1038/ncb2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiprilov EN, Awan A, Desprat R, Velho M, Clement CA, Byskov AG, et al. Human embryonic stem cells in culture possess primary cilia with hedgehog signaling machinery. J Cell Biol. 2008;180:897–904. doi: 10.1083/jcb.200706028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu Z-X, Hennessy B, Ding Z, Larrea M, et al. The energy sensing LKB1–AMPK pathway regulates p27kip1 phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampliega O, Orhon I, Patel B, Sridhar S, Díaz-Carretero A, Beau I, et al. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boada-Romero E, Letek M, Fleischer A, Pallauf K, Ramón-Barros C, Pimentel-Muiños FX. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 2013;32:566–582. doi: 10.1038/emboj.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, et al. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502:254–257. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante MI, Zullo A, Barra A, Bimonte S, Messaddeq N, Studer M, et al. Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat Genet. 2006;38:112–117. doi: 10.1038/ng1684. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur ELF. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci USA. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, et al. Huntingtin-associated Protein 1 (HAP1) interacts with the p150Glued Bubunit of Dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- Li X-J, Li S-H, Sharp AH, Nucifora FC, Schilling G, Lanahan A, et al. A huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- Wu LL, Fan Y, Li S, Li X-J, Zhou X-F. Huntingtin-associated protein-1 interacts with pro-brain-derived neurotrophic factor and mediates its transport and release. J Biol Chem. 2010;285:5614–5623. doi: 10.1074/jbc.M109.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Wueseke O, Hyman AA. Pericentriolar material structure and dynamics. Philos Trans R Soc B Biol Sci. 2014;369:20130459. doi: 10.1098/rstb.2013.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krishnaswami SR, Gleeson JG. CEP290 interacts with the centriolar satellite component PCM-1 and is required for Rab8 localization to the primary cilium. Hum Mol Genet. 2008;17:3796–3805. doi: 10.1093/hmg/ddn277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- Plotkin JL, Day M, Peterson JD, Xie Z, Kress GJ, Rafalovich I, et al. Impaired TrkB receptor signaling underlies corticostriatal dysfunction in Huntington's disease. Neuron. 2014;83:178–188. doi: 10.1016/j.neuron.2014.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liot G, Zala D, Pla P, Mottet G, Piel M, Saudou F. Mutant huntingtin alters retrograde transport of TrkB receptors in striatal dendrites. J Neurosci Off J Soc Neurosci. 2013;33:6298–6309. doi: 10.1523/JNEUROSCI.2033-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito V, Puigdellívol M, Giralt A, del Toro D, Alberch J, Ginés S. Imbalance of p75(NTR)/TrkB protein expression in Huntington's disease: implication for neuroprotective therapies. Cell Death Dis. 2013;4:e595. doi: 10.1038/cddis.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginés S, Paoletti P, Alberch J. Impaired TrkB-mediated ERK1/2 activation in huntington disease knock-in striatal cells involves reduced p52/p46 Shc expression. J Biol Chem. 2010;285:21537–21548. doi: 10.1074/jbc.M109.084202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch CC, Zaghloul NA. BBS4 is necessary for ciliary localization of TrkB receptor and activation by BDNF. PLoS ONE. 2014;9:e98687. doi: 10.1371/journal.pone.0098687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy B, Gaudet C, Ménard M, Atkinson T, Chiarini A, Dal Prà I, et al. The p75 neurotrophin receptor is localized to primary cilia in adult murine hippocampal dentate gyrus granule cells. Biochem Biophys Res Commun. 2010;401:458–462. doi: 10.1016/j.bbrc.2010.09.081. [DOI] [PubMed] [Google Scholar]
- Maiuri T, Woloshansky T, Xia J, Truant R. The huntingtin N17 domain is a multifunctional CRM1 and Ran-dependent nuclear and cilial export signal. Hum Mol Genet. 2013;22:1383–1394. doi: 10.1093/hmg/dds554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal RS, Desmond CR, Caron N, Maiuri T, Xia J, Sipione S, et al. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat Chem Biol. 2011;7:453–460. doi: 10.1038/nchembio.582. [DOI] [PubMed] [Google Scholar]
- Finetti F, Paccani SR, Riparbelli MG, Giacomello E, Perinetti G, Pazour GJ, et al. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse. Nat Cell Biol. 2009;11:1332–1339. doi: 10.1038/ncb1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldari CT, Rosenbaum J. Intraflagellar transport: it's not just for cilia anymore. Curr Opin Cell Biol. 2010;22:75–80. doi: 10.1016/j.ceb.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P, Marshall W. Ciliary secretion: switching the cellular antenna to “transmit.”. Curr Biol. 2013;23:R471–R473. doi: 10.1016/j.cub.2013.04.056. [DOI] [PubMed] [Google Scholar]
- Wang J, Silva M, Haas LA, Morsci NS, Nguyen KCQ, Hall DH, et al. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil V, Marzesco A-M, Corbeil D, Huttner WB, Wilsch-Bräuninger M. Midbody and primary cilium of neural progenitors release extracellular membrane particles enriched in the stem cell marker prominin-1. J Cell Biol. 2007;176:483–495. doi: 10.1083/jcb.200608137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon-Heszele MF, Choi SY, Zuo X, Baek J-I, Ward C, Lipschutz JH. The exocyst and regulatory GTPases in urinary exosomes. Physiol Rep. 2014;2:e12116–e12116. doi: 10.14814/phy2.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11:155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YP, Albin RL, Penney JB, Young AB, Anderson KD, Reiner A. Differential loss of striatal projection systems in Huntington's disease: a quantitative immunohistochemical study. J Chem Neuroanat. 2004;27:143–164. doi: 10.1016/j.jchemneu.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11:301–307. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Lee VMY. Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat Med. 2014;20:130–138. doi: 10.1038/nm.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, et al. Alzheimer's disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 2006;103:11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats P, Lee H-J, Bae E-J, Patrick C, Rockenstein E, Crews L, et al. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of alpha-synuclein. Proc Natl Acad Sci USA. 2009;106:13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfoury N, Holmes BB, Jiang H, Holtzman DM, Diamond MI. Trans-cellular propagation of Tau aggregation by fibrillar species. J Biol Chem. 2012;287:19440–19451. doi: 10.1074/jbc.M112.346072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Cleveland DW. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia R, Tattum MH, Jones S, Collinge J, Fisher EMC, Jackson GS. Superoxide dismutase 1 and tgSOD1 mouse spinal cord seed fibrils, suggesting a propagative cell death mechanism in amyotrophic lateral sclerosis. PLoS ONE. 2010;5:e10627. doi: 10.1371/journal.pone.0010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem. 2011;286:18664–18672. doi: 10.1074/jbc.M111.231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad LI, Yerbury JJ, Turner BJ, Guest WC, Pokrishevsky E, O'Neill MA, et al. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 2014;111:3620–3625. doi: 10.1073/pnas.1312245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P-H, Lauckner JE, Kachirskaia I, Heuser JE, Melki R, Kopito RR. Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat Cell Biol. 2009;11:219–225. doi: 10.1038/ncb1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Abounit S, Marzo L, Danckaert A, Chamoun Z, Roux P, et al. Transfer of polyglutamine aggregates in neuronal cells occurs in tunneling nanotubes. J Cell Sci. 2013;126:3678–3685. doi: 10.1242/jcs.126086. [DOI] [PubMed] [Google Scholar]
- Pecho-Vrieseling E, Rieker C, Fuchs S, Bleckmann D, Esposito MS, Botta P, et al. Transneuronal propagation of mutant huntingtin contributes to non-cell autonomous pathology in neurons. Nat Neurosci. 2014;17:1064–1072. doi: 10.1038/nn.3761. [DOI] [PubMed] [Google Scholar]
- Luk KC, Kehm V, Carroll J, Zhang B, O'Brien P, Trojanowski JQ, et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338:949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Z, Cooper J, Murray TK, Garn K, McNaughton E, Clarke H, et al. A novel in vivo model of tau propagation with rapid and progressive neurofibrillary tangle pathology: the pattern of spread is determined by connectivity, not proximity. Acta Neuropathol (Berl) 2014;127:667–683. doi: 10.1007/s00401-014-1254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Calignon A, Polydoro M, Suárez-Calvet M, William C, Adamowicz DH, Kopeikina KJ, et al. Propagation of tau pathology in a model of early Alzheimer's disease. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, et al. Trans-synaptic spread of tau pathology in vivo. PLoS ONE. 2012;7:e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M, Walker LC. Pathogenic protein seeding in alzheimer disease and other neurodegenerative disorders. Ann Neurol. 2011;70:532–540. doi: 10.1002/ana.22615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Spada AR. Something wicked this way comes: huntingtin. Nat Neurosci. 2014;17:1014–1015. doi: 10.1038/nn.3770. [DOI] [PubMed] [Google Scholar]
- Pearce MMP, Spartz EJ, Hong W, Luo L, Kopito RR. Prion-like transmission of neuronal huntingtin aggregates to phagocytic glia in the Drosophila brain. Nat Commun. 2015;6:6768. doi: 10.1038/ncomms7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-Y, Englund E, Holton JL, Soulet D, Hagell P, Lees AJ, et al. Lewy bodies in grafted neurons in subjects with Parkinson's disease suggest host-to-graft disease propagation. Nat Med. 2008;14:501–503. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]
- Li J-Y, Englund E, Widner H, Rehncrona S, Björklund A, Lindvall O, et al. Characterization of Lewy body pathology in 12- and 16-year-old intrastriatal mesencephalic grafts surviving in a patient with Parkinson's disease. Mov Disord Off J Mov Disord Soc. 2010;25:1091–1096. doi: 10.1002/mds.23012. [DOI] [PubMed] [Google Scholar]
- Kordower JH, Chu Y, Hauser RA, Olanow CW, Freeman TB. Transplanted dopaminergic neurons develop PD pathologic changes: a second case report. Mov Disord Off J Mov Disord Soc. 2008;23:2303–2306. doi: 10.1002/mds.22369. [DOI] [PubMed] [Google Scholar]
- Kordower JH, Chu Y, Hauser RA, Freeman TB, Olanow CW. Lewy body-like pathology in long-term embryonic nigral transplants in Parkinson's disease. Nat Med. 2008;14:504–506. doi: 10.1038/nm1747. [DOI] [PubMed] [Google Scholar]
- Cicchetti F, Lacroix S, Cisbani G, Vallières N, Saint-Pierre M, St-Amour I, et al. Mutant huntingtin is present in neuronal grafts in Huntington disease patients. Ann Neurol. 2014;76:31–42. doi: 10.1002/ana.24174. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang J, Yang H, Zhao T, Sun M, Xu X, Zhou X-F, et al. Huntingtin-associated protein 1 regulates postnatal neurogenesis and neurotrophin receptor sorting. J Clin Invest. 2014;124:85–98. doi: 10.1172/JCI69206. [DOI] [PMC free article] [PubMed] [Google Scholar]