Abstract
Synthetic regulatory circuits encoded on RNA rather than DNA could provide a means to control cell behavior while avoiding potentially harmful genomic integration in therapeutic applications. We create post-transcriptional circuits using RNA-binding proteins, which can be wired in a plug-and-play fashion to create networks of higher complexity. We show that the circuits function in mammalian cells when encoded on modified mRNA or self-replicating RNA.
Gene delivery using messenger RNA (mRNA) rather than plasmid DNA (pDNA) may be safer owing to a reduced risk of genomic integration1. Advances in chemical mRNA modification technology have made it possible to use stable in vitro synthesized mRNA with low immunogenicity for gene therapy2. Self-replicating RNAs that couple RNA-only delivery with prolonged gene expression are of interest for biomedical applications including vaccination and stem cell reprogramming2. Synthetic biology, however, has so far relied exclusively or partially on transcriptional regulation, which requires introduction of foreign DNA3,4. RNA-based regulatory parts, such as aptamers or riboswitches5–7 cannot currently be interconnected to build complex RNA-encoded circuits. RNA strand displacement reactions, used to date only in bacteria8,9 could be combined into logic circuits10. However, such multi-layered RNA circuits have not yet been successfully implemented. We propose that RNA-binding proteins (RBPs)11 can function as both the input and the output of RNA regulatory devices and be wired to regulate production of each other towards the construction of complex circuits. The synthetic circuits containing RBPs reported to date have not shown that one RBP can regulate another and have depended on both translational and transcriptional regulation, requiring the use of pDNA for circuit delivery12. Additionally, general mechanisms to regulate expression from synthetic mRNA or RNA replicons have not yet been implemented. In this article we report that RBP regulatory devices can be wired together and interconnected with cellular and synthetic signaling pathways to build complex circuits that can be delivered to mammalian cells as RNA. We characterize and optimize a set of RBP devices and then use them to engineer diverse regulatory circuits including a multi-input cell type classifier, a cascade and a switch (Supplementary Fig. 1). These circuits carry out signal processing operations that detect intracellular biomarker levels, transmit information between cascaded regulatory devices and conserve circuit state through feedback regulation. We also show that the classifier can be used for selective induction of apoptosis in a targeted cell type (HeLa cancer cells) using RNA-only delivery.
As a first step toward creating RNA-encoded circuits, we optimized and characterized a set of RNA repressor devices comprising RBPs and their binding motifs (Supplementary Fig. 2,3, Supplementary Note 1). Of the tested devices, L7Ae:K-turn system11 and MS2-CNOT7:MS2 binding motif13 were the most potent and used for further circuit construction.
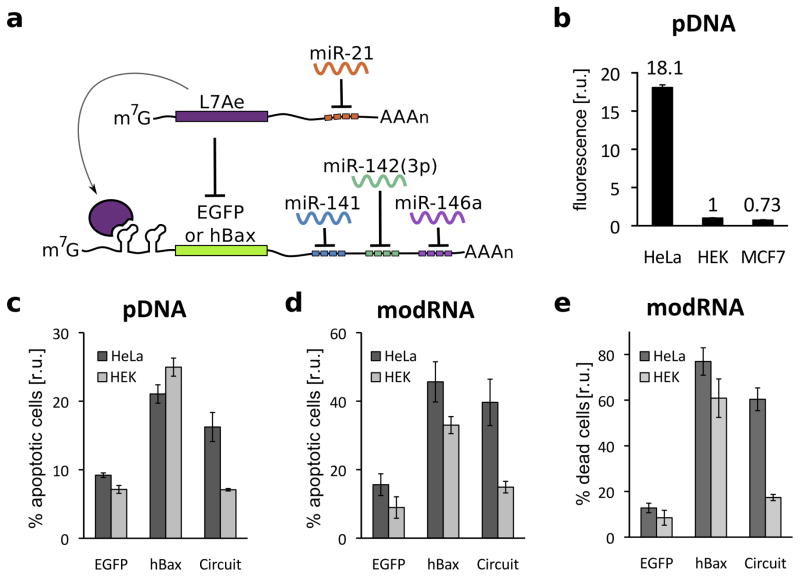
To show that these RBP-based repressors can be used as a platform for composite RNA-encoded circuits, we engineered a multi-input microRNA sensing circuit that is a simplified post-transcriptional only version of our previously reported HeLa cell classifier14. The circuit recognizes whether the cell has a microRNA expression profile indicative of HeLa cells (high miR-21, low miR141, 142(3p) and 146a) and triggers a response only if the profile is matched (Fig. 1, Supplementary Fig. 1a). The circuit topology consists of two basic sensory modules, one for specific microRNAs that are highly expressed in the cancer phenotype (HeLa-high) and one for the microRNAs that are expressed at low levels (HeLa-low). HeLa-high microRNAs affect circuit output via double inversion by repressing L7Ae, which allows expression of an output protein. HeLa-low microRNAs directly repress translation of the output. As shown in Fig. 1b and Supplementary Fig. 4, the L7Ae-based classifier is able to distinguish HeLa cells from HEK 293 and MCF7 in a fluorescence assay. While single microRNAs are often sufficient to differentiate between pairs of cell types (Supplementary Fig. 5), a multi-input circuit is needed to distinguish HeLa cells from many other cell types simultaneously14. When a pro-apoptotic gene hBax is incorporated as circuit output, the classifier selectively kills HeLa cells and does not strongly affect viability of HEK cells (Fig. 1c,d, Supplementary Fig. 6–7). Specific induction of apoptosis was achieved using both pDNA and modified mRNA (modRNA) to deliver circuits. Furthermore, the modRNA circuit specifically killed HeLa cells in a mixed HeLa/HEK cell population (Fig. 2e, Supplementary Fig. 8). The performance of our new classifier coupled with the RNA-only delivery provides a safer means for using such classifier synthetic network for a range of applications, including selective stem cell reprogramming or vaccination.
Figure 1.
RNA-only multi-input microRNA classifier circuit differentiates between HeLa, HEK 293 and MCF7 cells.
(a) An L7Ae-based multi-input microRNA classifier specifically recognizes HeLa cells based on a unique microRNA profile (highly expressed miR21 and low levels of miR141, 142(3p) and 146a). (b) Differential expression of output protein EGFP in HeLa, HEK and MCF7 cells with transient pDNA transfection. EGFP expression from the classifier circuit results in 18-fold and 25-fold higher output in HeLa cells in comparison to HEK and MCF7 cells, respectively (HEK fluorescence was normalized to 1 and circuit-regulated EGFP fluorescence was normalized to mKate expressed constitutively from the same promoter, to account for different expression levels across cell types). (c–d) Specific induction of apoptosis in HeLa cells by expression of circuit-controlled hBax protein compared with constitutive hBax expression: Annexin V positive cells in pDNA (c) and modRNA (d) transfected cells. (e) Cell death assay in a mixed HEK/HeLa-EBFP2 culture with modRNA delivery. The graphs indicate percent of dead cells as measured with AADvanced staining, with HEK and HeLa cells distinguished by EBFP fluorescence. EGFP-only transfection was used as a control in all apoptotic/cell death assays.
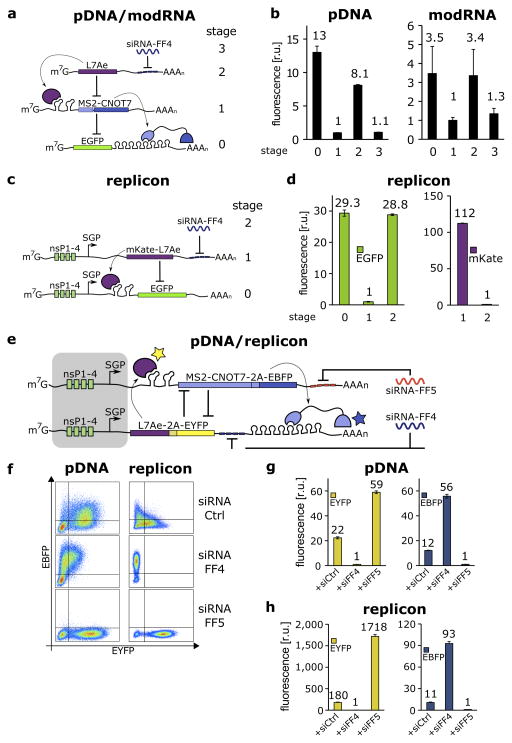
Figure 2.
Post-transcriptional cascades and two-state switch.
(a) Cascade design for the pDNA and modRNA experiments.(b) Normalized mean EGFP fluorescence for the indicated cascade stages encoded either on pDNA or modRNA. Each stage n involves co-transfection of constructs 0 to n. (c) Replicon encoded two-stage cascade. L7Ae was fused to red fluorescent protein mKate. Each replicon additionally encodes four non-structural proteins (nsP1-4) and a subgenomic promoter (SGP) driving expression of circuit components. (d) Normalized mean EGFP and mKate fluorescence for cascade encoded on self-replicating RNA. (e) Switch design; shaded: replicon components that include nsP1-4 and SGP. (f) Corresponding representative two-dimensional flow cytometry plots for pDNA and replicon transfections. (g,h) Normalized mean fluorescence of the two reporters in the different states of the switch encoded on pDNA (g) or replicon (h). Fluorescence was normalized to the lowest level in each chart.
We next connected RBP devices to produce a scalable RNA-only circuit design platform. To generate a one-way information transmitter, we designed a post-transcriptional cascade with three repression stages (Fig. 2a–d, Supplementary Fig. 1b). The input to the cascade (Fig. 2a) is a synthetic siRNA-FF4, which modulates expression of L7Ae through four repeats of the FF4 target site in the 3′UTR. L7Ae then binds the K-turn motifs in the 5′UTR of RNA that encodes a second repressor, MS2-CNOT7, which regulates expression of output EGFP containing eight repeats of the MS2 binding site in its 3′UTR. We tested the behavior of the circuit with pDNA and modRNA transfections (Fig. 2b, Supplementary Fig. 9a,b) and quantified cascade operation for a range of input concentrations and times (Supplementary Fig. 10, 11, 12).
A two-stage version of the cascade was encoded on self-replicating RNA derived from Sindbis virus15 (Fig. 2c–d, Supplementary Fig. 9c). Replication is mediated by the viral RNA-dependent RNA polymerase (RdRp; comprised of nonstructural proteins nsP1-nsP4) and enables long-term gene expression (Supplementary Note 2, Supplementary Fig. 13–16). Production of exogenous genes is driven by a subgenomic promoter of the replicon (SGP). In our replicon-encoded cascade circuit, siRNA-FF4 (input) regulates expression of L7Ae. The repressor is under the control of the replicon SGP and additionally contains four repeats of the FF4 target site in its 3′UTR. A separate co-transfected replicon encodes output EGFP with two repeats of the K-turn motif in the 5′UTR. As shown in Figure 2d and Supplementary Figure 17, L7Ae expression results in 29-fold repression of EGFP (stage 1), and knockdown of L7Ae by synthetic siRNA-FF4 fully restores the output (stage 2). The cascade also functions with combined replicon/pDNA co-electroporation, albeit with reduced repression efficiency (Supplementary Fig. 18).
Finally, we created an RNA-based switch circuit in which two RBPs cross-repress each other to demonstrate two-way signal transmission and feedback regulation (Fig. 2e–h, Supplementary Fig. 1c, 19). The general topology of our switch is similar to previously described bacterial and mammalian transcriptional toggle switches16,17. The switch components include MS2-CNOT7 with two 5′UTR K-turn motifs and L7Ae with eight repeats of the MS2 binding site in the 3′UTR. To monitor switch behavior we additionally co-express (via 2A tags) a blue fluorescent protein (EBFP2) with MS2-CNOT7 and EYFP with L7Ae. The state of the system can be set with transient introduction of exogenous siRNA, or alternatively, endogenously expressed miRNA. We use two artificial and orthogonal siRNAs (FF4 and FF5). For pDNA transfection, when no specific siRNA is present, both repressors and associated reporters remain at intermediate levels after 48 hours (Fig. 2f, g, Supplementary Fig. 20). siRNA-FF4 sets the state to high MS2-CNOT7 (blue) and low L7Ae (yellow), while siRNA-FF5 transfection results in the opposite state. The ON/OFF ratio between the two states is 56-fold for EBFP2 and 59-fold for EYFP. Since many potential applications of the switch will require longer-term expression, we also encoded the circuit on self-replicating RNA (Figure 2f, h, Supplementary Fig. 20–22). Similar to pDNA, siRNA-FF4 and FF5 set the state effectively, with ON/OFF ratios of 93-fold for EBFP2 and 1718-fold for EYFP. In the absence of specific siRNA, the replicon-encoded circuit had stronger bimodality than pDNA. We further explored this observation using a computational model of the pDNA and replicon-encoded switch circuits (Supplementary Note 3, Supplementary Fig. 23–27). Based on literature18 and our computational model we hypothesize that in the absence of specific siRNA, initial pDNA expression of the two switch branches (each encoding an RBP) is simultaneous (multiple plasmids delivered to the nucleus at the same time) and results in production of stable proteins. These remain in the cell at relatively high levels for the duration of the transfection experiment. In contrast, initial replicon RNA and replicon-encoded RBP production is more stochastic as single replicon species are rapidly amplified, typically leading to one of the two possible states.
To our knowledge no previous study has shown that complex cellular logic can be encoded exclusively at the post-transcriptional level in mammalian cells, offering potentially significant benefits for in vivo applications. This is made possible through the use of RBPs, which can act as both the input and the output of a regulatory device, and are promising candidates for creating scalable and modular control and information processing circuits. Our engineered circuits are functional when encoded either on modified mRNA (transient response) or self-replicating RNA (prolonged circuit operation). The inherently transient nature of RNA makes it an appealing platform for applications where safety is a primary concern, as RNA circuits could be programmed to act for a defined period of time and do not leave a long-term genetic footprint. Additionally, the different expression dynamics, lifetime (Supplementary Fig. 11–12, 14–15) and possibly noise properties (Supplementary Notes 2, 3, Supplementary Fig. 28) of modRNA and replicon delivery provide further potential for circuit design. Finally, the application of our RNA-only multi-input cell classifier circuit for specific induction of apoptosis, potentially concise formulation (two RNA species in case of the classifier) and its safety characteristics (transient expression and no chromosomal integration) make this a promising framework for future in vivo applications.
ONLINE METHODS
Cell culture
HEK293FT and HEK293 (293-H) cell lines were purchased from Invitrogen. HeLa (CCL.2) and MCF7 (HTB-22) cell lines were originally obtained from ATCC. The performance of DNA-encoded miRNA sensors in these cell lines had been characterized previously14. HEK293FT were freshly purchased from the supplier. HeLa, MCF7 and BHK21, although not recently authenticated, were tested for mycoplasma. All cell lines used in this study were maintained in Dulbecco’s modified Eagle medium (DMEM, Cellgro) supplemented with 10% FBS (Atlanta BIO), 1% penicillin/streptomycin/L-Glutamine (Sigma-Aldrich) and 1% non-essential amino acids (HyClone) at 37 °C and 5% CO2. In the case of MCF7 cells, DMEM without phenol red was used. BHK21 cells were maintained in Eagle’s Minimum Essential Medium (EMEM, ATCC) supplemented with 10% FBS.
DNA preparation and transfection
All transfections were carried out in 24-well format. Parallel transfections in HEK293, HeLa and MCF7 cells (4-input sensor, Fig. 1b, c, Supplementary Fig. 3–6) were performed with Lipofectamine LTX (Life Technologies) according to manufacturer’s protocol. Lipofectamine LTX was used as it provides the best transfection efficiencies across the 3 cell lines among tested reagents. Total of 400 ng DNA was mixed with Opti-MEM I reduced serum medium (Life Technologies) to a final volume of 100ul followed by addition of 0.5ul PLUS reagent. After 5 minutes 1.5 ul Lipofectamine LTX was added, the samples were briefly vortexed and incubated for 30 min at room temperature. During the incubation time, cells were harvested by trypsinization and seeded in 500ul of complete culture medium in 24-well plate (HEK: 2×10^5, HeLa: 1.2×10^5 and MCF7: 1.5×10^5 cells per well). Transfection complexes were added dropwise to the freshly seeded cells followed by gentle mixing. Cells were supplemented with 1 ml of fresh growth medium 5 hours post transfection and analyzed by flow cytometry after 48 hours (after 24 hours for apoptosis assay). Plasmid DNA and siRNA co-transfections (cascade and switch circuits, Fig. 2b, g, Supplementary Fig. 9a, 10, 19, 20) were carried out with Lipofectamine 2000 according to the manufacturer’s protocol. Lipofectamine 2000 was used as it provides the best DNA/siRNA co-transfection efficiencies among tested reagents. A total of up to 300ng DNA and 1–5pmol siRNA were mixed with Opti-MEM I reduced serum medium (Life Technologies) to a final volume of 50ul. Separately, 2ul of Lipofectamine 2000 was mixed with 50ul of Opti-MEM. After 5 min incubation, lipofectamine and DNA/siRNA dilutions were combined and briefly vortexed. Cells were prepared, transfected and analyzed as described above. All the remaining transfections (repressor optimization in Supplementary Fig. 2 and time lapse in Supplementary Fig. 11) were carried out with Attractene (Qiagen). Up to 300ng total DNA was mixed with DMEM base medium (Cellgro) without supplements to a final volume of 60ul. 1.5ul attractene was added to the dilutions and the samples were promptly vortexed to mix. The complexes were incubated for 10–15 min and subsequently added to cells prepared as described above. 500ul of fresh medium was added to each well the next day (media change was not necessary in the case of attractene transfections). Transfection details for each experiment are shown in Supplemenatry Table 1. The list of all plasmids used in this study is shown in Supplementary Table 2.
Modified RNA preparation and mRNA transfection
A template DNA for in vitro transcription was generated via PCR, using a forward primer containing T7 promoter and a reverse primer containing 120-nucleotide-long Poly(T) tract transcribed into a Poly(A) tail. PCR products amplified from plasmids were subjected to digestion by Dpn I restriction enzyme and purified. Reactions of in vitro transcription were performed using MegaScript T7 kit (Life Technologies) under a modified condition, in which GTP, CTP and UTP was replaced by GTP mixed with Anti Reverse Cap Analog (New England Biolabs) at the ratio of 1 to 4, 5-methylcytosine-triphosphate and pseudouridine-triphosphate (TriLink BioTechnologies), respectively. Transcripts were treated with Turbo DNase (Life Technologies) for 30 min at 37 °C and purified using RNeasy MiniElute Cleanup Kit (QIAGEN). Resulting mRNAs were incubated with Antarctic Phosphatase (New England Biolabs) for 30 min at 37 °C and purified again. Modified mRNAs were transfected into the cells using TransIT-mRNA transfection kit (Mirus Bio) according to manufacturer’s protocol. StemFect (Stemgent) was used to perform co-transfections of modified mRNAs with siRNAs, according to manufacturer’s instruction. The medium was exchanged 4 hours after the transfection, and transfected cells were subjected to the analysis after 24 hours. Transfection details for each experiment are shown in Supplemenatry Table 1. Detailed configurations for modified mRNA and sequences of mRNA used in this study are shown in Supplementary Tables 3 and 4.
Self-replicating RNA preparation and electroporation
All replicon experiments were performed in BHK21 cells (a kind gift from Dr. Odisse Azizgolshani19) using an alphaviral replicon derived from the genome of the Sindbis virus TE12 strain20 containing a P726S mutation in nsP221 as described previously22 or an alphaviral replicon derived from the Venezuelan equine encephalitis (VEE) TC-83 strain containing a A3G mutation in the 5′UTR and a Q739L mutation in nsP223 constructed in this study. Briefly, BHK21 cells cultured at 37 degrees C and 5% CO2 in EMEM (ATCC) medium containing 10% FBS (PAA) were electroporated using the Neon® Transfection System (Life Technologies) per the manufacturer’s instructions with ~1–6 ug of replicon RNA per ~100,000 cells and plated in 24 well plates (Corning). Transfection details for all experiments are provided in Supplementary Table 1. Sindbis replicon RNA was produced by run-off in vitro transcription (IVT) of SacI-HF (NEB)-digested replicon plasmid DNA using the mMESSAGE mMACHINE® SP6 Kit (Life Technologies) and purified using the RNeasy® Mini Kit (Qiagen). VEE replicon RNA was produced by run-off in vitro transcription (IVT) of I-SceI (NEB)-digested replicon plasmid DNA using the MEGAscript® T7 Transcription Kit, followed by purification using the RNeasy® Mini Kit (Qiagen), denaturation of the RNA at 65 degrees C, enzymatic (cap1) capping of the RNA using the ScriptCap™ 2′-O-Methyltransferase Kit (Cellscript) and ScriptCap™ m7G Capping System (Cellscript), and a final purification using the RNeasy® Mini Kit (Qiagen) following the manufacturers’ protocols. siRNAs (IDT) were co-electroporated (0 - 10 nM final concentration) along with replicon RNA. Cells were analyzed by flow cytometry 24h post electroporation. Replicon encoding plasmids used as templates for IVT are listed in Supplementary Table 5.
qRT-PCR
In the case of pDNA and modRNA, total RNA was reverse transcribed with High-Capacity cDNA Reverse Transcription Kit (Life Technologies). Resulting cDNA was subjected to qPCR on StepOnePlus (Life Technologies) for modRNA using Power SYBR Green PCR Master Mix (Life Technologies). Same Master Mix and Mastercycler ep Realplex (Eppendorf) was used for pDNA experiments. For qRT-PCR of RNA replicons, total RNA was purified from BHK21 cells using the RNeasy® Mini Kit (Qiagen). RNA was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen) and qPCR was performed on a Mastercycler ep Realplex (Eppendorf) using the KAPA SYBR® FAST Universal 2X qPCR Master Mix (Kapa Biosystems) or the KAPA PROBE FAST Universal 2X qPCR Master Mix (Kapa Biosystems) following the manufacturer’s recommended protocol. Primers unique to the genomic RNA regions were used to calculate the absolute copy number of genomic and antigenomic RNA using a standard curve of synthetic DNA. Subgenomic RNA copy numbers were calculated by subtracting the copy numbers of genomic and antigenomic RNA from the absolute copy numbers of all replicon RNA (i.e. genomic, antigenomic, and subgenomic RNA) using primers spanning the regions downstream of the SGP. Genomic and subgenomic RNA quantities were then normalized to 18S rRNA (internal control) levels quantified using QuantumRNA™ Universal 18S Internal Standard (Life Technologies) or Eukaryotic 18S rRNA Endogenous Control (FAM™/MGB probe, non-primer limited; Life Technologies).
Primer sequences
| EGFP-qPCR-F | AAGGGCATCGACTTCAAGG |
| EGFP-qPCR-R | TGCTTGTCGGCCATGATATAG |
| VEE-nsP1-qPCR-F | CTGACCTGGAAACTGAGACTATG |
| VEE-nsP1-qPCR-R | GGCGACTCTAACTCCCTTATTG |
| VEE-nsP4-EGFP-qPCR-F | CCCTATAACTCTCTACGGCTAAC |
| VEE-nsP4-EGFP-qPCR-R | AGAAGTCGTGCTGCTTCA |
| SIN-nsP4-L7Ae-qPCR-F | GGCGTGGTTTAGAGTAGGTATAA |
| SIN-nsP4-L7Ae-qPCR-R | TCGTCTCGTTGGTACCTTTC |
| MS2-Taqman-F1 | GCTGAATGGATCAGCTCTAACT |
| MS2-Taqman-R1 | CAGTCTGGGTTGCCACTTTA |
| MS2-Taqman-P1-2 | ACCTGTAGCGTTCGTCAGTCCTCT |
Flow cytometry and data analysis
Cells were analyzed with LSR Fortessa or FACSAria flow cytometer, equipped with 405, 488 and 561 nm lasers (BD Biosciences). We collected 30,000–100,000 events per sample and fluorescence data were acquired with the following cytometer settings: 488 nm laser and 530/30 nm bandpass filter for EYFP/EGFP, 561 nm laser and 610/20 nm filter for mKate, and 405 nm laser, 450/50 filter for EBFP. In detecting mKate by FACSAria, a 780/60 nm bandpass filter was used. Data analysis was performed with FACSDiva software (BD Biosciences) and FlowJo (http://www.flowjo.com). For all fluorescence assays, populations containing live, single cells were first determined based on forward and side scatter. Red fluorescent protein (mKate) was used in all pDNA experiments as a transfection marker. Reported fluorescence values of pDNA experiments present normalized mean output fluorescence (EYFP, EGFP or EBFP) for all mKate positive cells. Non-transfected cells were used to set the gate determining mKate positive cells. For replicon electroporations and modRNA transfections the efficiency of nucleic acid delivery usually exceeds 90% and therefore all live, single cells were taken into account for calculating mean output fluorescence.
In Fig. 1b, output (EGFP) fluorescence level may depend both on circuit function and overall expression in a particular cell line (different promoter activities and transfection efficiencies). To account for cell type specific expression we therefore applied here normalization to mKate. Mean EGFP fluorescence for each sample was divided by mean mKate fluorescence and the ratio was normalized to the HEK293 level (HEK293 relative fluorescence set to 1).
Microscope measurements and image processing
Fluorescence microscopy images of live cells were taken in 24-well plates using Zeiss Axiovert 200 microscope and Plan-Neofluar 10x/0.30 Ph1 objective. The filters used were 390/22 (excitation) and 460/50 (emission) for EBFP2, 500/20 (excitation) and 535/50 (emission) for EYFP and 565/30 (excitation) and 620/60 (emission) for mKate. Data collection and processing were performed using AxioVision software (Zeiss).
Apoptosis and cell death assays
Sample cells including those in supernatant were collected 24h post-transfection, washed with PBS and stained with Pacific Blue conjugated 1 μL of Annexin V (Life Technologies) or 0.5 μL of SYTOX AADvanced (Life Technologies) in 50 μL of binding buffer for 30 min at room temperature. The cells were analyzed by flow cytometry. Percentage of apoptosis induction was defined as the percentage of Annexin V positive cells. In the case of HEK/HeLa co-culture assay, HeLa cells were labeled with stable expression of EBFP2 fluorescent protein (excitation/emission maxima of 383 nm and 448 nm) and therefore SYTOX AADvanced (excitation/emission maxima of 546 nm and 647 nm) was used instead Pacific Blue Annexin V (excitation/emission maxima of 415 nm and 455 nm). HEK293 and HeLa-EBFP2 cells were mixed in 1:1 ratio, cultured together and the cell mixture was transfected with modRNA-encoded circuit or controls. Cells were stained with SYTOX AADvanced and analyzed by flow cytometry 24h post-transfection. % of cell death was calculated as follows: (number of HEK (or HeLa) AAdvanced positive cells / total number of HEK (or HeLa) cells) *100%.
Generation of HeLa-EBFP2 cells for co-culture cell death assay
HeLa-EBFP2 cells were generated through lentiviral infection and antibiotic selection. First, HEK293FT packaging cells (Invitrogen) were used for virus production. 2×106 cells were seeded in a 60mm dish (to ~80% confluency), approximately 3h later supplemented with 3 ml of fresh complete medium and co-transfected with the following plasmids:
0.5 μg pLV-hEF1a-EBFP2-P2A-Bla (hEF1a – human elongation factor 1alpha promoter, P2A – ribosomal skipping 2A sequence from porcine teschovirus-124, Bla – blasticidin resistance gene)
1.1 μg pCMV-dR8.2 dvpr helper plasmid25 (Addgene plasmid 8455)
0.55 μg pCMV-VSV-G helper plasmid25 (Addgene plasmid 8454)
Transfections were performed using attractene (Qiagen) and standard manufacturer’s protocol. Transfection complexes were added dropwise to the adhered cells without additional media change. 2 days later, media from virus producing cells were collected into 3ml syringe, and pressed through a low protein binding 0.45μm sterile filter. 1ml of the filtered virus containing media was mixed with 4×103 HeLa cells in 0.5 ml fresh culture media and placed in a 12-well dish. Cells were supplemented with fresh media the next day and 10 μg/ml blasticidin (Invivogen) was added to the media on days 3–8 post-infection. Selected cells were over 99% BFP positive throughout the course of experiments as determined by flow cytometry. We additionally performed a fluorescent assay using our classifier circuit (as described in Fig. 2b) with HEK293, HeLa and HeLa-EBFP2 cells and we verified that the HeLa-EBFP2 cells behave as the parent cell line (data not shown).
Supplementary Material
Acknowledgments
We thank K. Hayashi and N. Nishimura (Kyoto University) for supporting modified mRNA experiments. We also thank Prof. A.C. Goldstrohm and C. Weidmann (University of Michigan Medical School) for sharing Drosophila constructs containing MS2-fused repressors and the corresponding reporter, X. Zhang (MIT) and O. Andries (Ghent University) for assisting in VEE replicon construction, T.E. Wagner, Prof. D. Densmore (Boston University), and S. Payne (MIT) for communicating results before publication, Prof. W.M. Gelbart, Prof. C.M. Knobler, Prof. A. Berk, O. Azizgolshani and J.M. Parker (University of California, Los Angeles) for sharing replicon constructs and expertise, as well as A. Graziano (MIT) for supporting pDNA experiments and H. Chung (Harvard University) for designing the intronic mKate construct. This work was supported by DARPA-BAA-11-23, NIH Grants No. P50-GM098792, 5-R01-CA155320-03, NSF GRFP Grant No. 1122374 (R.W.) and JSPS Grant Number 23681042, Research Center Network for Realization of Regenerative Medicine from the Japan Science and Technology Agency (H.S.).
Footnotes
AUTHOR CONTRIBUTIONS
LW and RW conceived the ideas, LW designed and performed pDNA experiments, KE designed and performed modRNA experiments, TK designed and performed replicon experiments, VS designed and performed pDNA apoptotic assay and 3’UTR repressor test in HeLa cells. BS created computational model of pDNA and replicon-based switch. LW, RW, KE and HS wrote the manuscript with input from all other authors.
COMPETING FINANCIAL INTERESTS
HS, KE and RW, LW, TK and VS are co-inventors on patent applications covering the RNA circuits described here.
References
- 1.Tavernier G, et al. mRNA as gene therapeutic: how to control protein expression. Journal of controlled release : official journal of the Controlled Release Society. 2011;150:238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat Rev Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 3.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nature reviews Genetics. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubel D, Fussenegger M. Mammalian synthetic biology--from tools to therapies. BioEssays : news and reviews in molecular, cellular and developmental biology. 2010;32:332–345. doi: 10.1002/bies.200900149. [DOI] [PubMed] [Google Scholar]
- 5.An CI. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausländer S, et al. A general design strategy for protein-responsive riboswitches in mammalian cells. Nature Methods. 2014;11:1154–1160. doi: 10.1038/nmeth.3136. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigo G, Landrain TE, Jaramillo A. De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells. Proc Natl Acad Sci USA. 2012;109:15271–15276. doi: 10.1073/pnas.1203831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 11.Saito H, et al. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nature chemical biology. 2010;6:71–78. doi: 10.1038/nchembio.273. [DOI] [PubMed] [Google Scholar]
- 12.Ausländer S, Ausländer D, Müller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 13.Van Etten J, et al. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem. 2012;287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 15.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 17.Kramer BP, et al. An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 18.Mortimer I, et al. Cationic lipid-mediated transfection of cells in culture requires mitotic activity. Gene Ther. 1999;6:403–411. doi: 10.1038/sj.gt.3300837. [DOI] [PubMed] [Google Scholar]
- 19.Azizgolshani O, Garmann RF, Cadena-Nava R, Knobler CM, Gelbart WM. Reconstituted plant viral capsids can release genes to mammalian cells. Virology. 2013;441:12–17. doi: 10.1016/j.virol.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Lustig S, et al. Molecular basis of Sindbis virus neurovirulence in mice. J Virol. 1988;62:2329–2336. doi: 10.1128/jvi.62.7.2329-2336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frolov I, et al. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol. 1999;73:3854–3865. doi: 10.1128/jvi.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beal J, et al. Model-driven engineering of gene expression from RNA replicons. ACS Synth Biol. 2015;4:48–56. doi: 10.1021/sb500173f. [DOI] [PubMed] [Google Scholar]
- 23.Petrakova O, et al. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in Mammalian cells. J Virol. 2005;79:7597–7608. doi: 10.1128/JVI.79.12.7597-7608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 25.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.