Abstract
Elongation factor G (EF-G), a translational GTPase responsible for tRNA-mRNA translocation possesses a conserved histidine (H91 in Escherichia coli) at the apex of switch-II, which has been implicated in GTPase activation and GTP hydrolysis. While H91A, H91R and H91E mutants showed different degrees of defect in ribosome associated GTP hydrolysis, H91Q behaved like the WT. However, all these mutants, including H91Q, are much more defective in inorganic phosphate (Pi) release, thereby suggesting that H91 facilitates Pi release. In crystal structures of the ribosome bound EF-G•GTP a tight coupling between H91 and the γ-phosphate of GTP can be seen. Following GTP hydrolysis, H91 flips ~140° in the opposite direction, probably with Pi still coupled to it. This, we suggest, promotes Pi to detach from GDP and reach the inter-domain space of EF-G, which constitutes an exit path for the Pi. Molecular dynamics simulations are consistent with this hypothesis and demonstrate a vital role of an Mg2+ ion in the process.
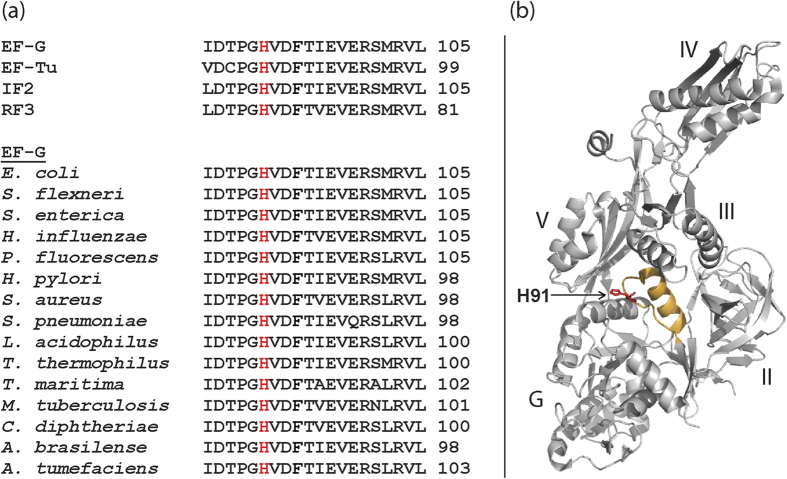
Elongation factor G (EF-G) belongs to the subfamily of translational G-proteins in the GTPase superfamily1. All G-proteins share a nucleotide binding G domain, which contains distinct and highly conserved elements (G1-G5)1,2. The G3 sequence motif, ‘switch II’, is highly flexible and contains a DXXG sequence1,2. It has been suggested that a conserved glutamine (Q) immediately after the glycine of the DXXG motif, positioned at the tip of switch II is crucial for GTP hydrolysis in Ras-like G proteins2,3. However, in all translational GTPases this glutamine is replaced by a histidine1 (Fig. 1a). In EF-G from Escherichia coli (E. coli), this histidine is H91 based on its residue number on the mature EF-G chain after removal of the starting f-Met (Fig. 1b). The corresponding histidine on EF-Tu is H84; high resolution structures of EF-G and EF-Tu on the ribosome with GTP analogues place H91 and H84 in similar conformations, pointing towards the γ-phosphate of GTP through the hydrophobic gate4,5,6.
Figure 1. Sequence conservation and location of H91 in EF-G.
(a) Comparison of the amino acid sequences of the switch II loop containing DXXG (G3) motif in four translational GTPases from E. coli, and EF-G from various bacteria (Escherichia coli, Shigella flexneri, Salmonella enterica, Haemophilus influenzae, Pseudomonas fluorescens, Helicobacter pylori, Staphylococcus aureus, Streptococcus pneumoniae, Lactobacillus acidophilus, Thermus thermophilus, Thermotoga maritime, Mycobacterium tuberculosis, Corynebacterium diphtheriae, Azospirillum brasilense and Agrobacterium tumefaciens listed from top). The conserved H91 residue is marked in red. (b) The structure of Thermus thermophilus EF-G (PDB ID: 1FNM) with H87 (corresponding to H91 in E. coli) marked in red on the apex of switch II (in gold).
The role of this His in GTP hydrolysis by EF-Tu and EF-G has been extensively studied. Several groups have reported that mutations of H84 inhibit ribosome stimulated GTP hydrolysis by EF-Tu7,8,9,10. Similarly, the H91A mutation in EF-G has been claimed to be completely11,12 or significantly13 impaired in GTP hydrolysis. Based on a crystal structure of EF-Tu•GDPNP on the ribosome it has been proposed that H84 acts as a catalytic residue that deprotonates a water molecule for in-line attack on the bond between β- and γ-phosphates of GTP6. This proposal, however, has been challenged and an alternative mechanism where H84 plays an indirect stabilizing role for the catalytic water has been suggested14,15. Thus, the current understanding of the field is that this His may not be catalytic for GTP hydrolysis, but is important for GTPase activation of the translational GTPases on the ribosome16,17.
Compared to GTP hydrolysis, fewer studies have been conducted to understand the mechanism of Pi release and its role in EF-G functions. It has been shown that, restriction of the inter-domain movements in EF-G by introducing a disulphide cross-link between domains I and V renders the factor completely inactive in translocation, but leaves both GTP hydrolysis and Pi release unaffected18. Later studies indicated that Pi release is not a pre-requisite for translocation and might occur through an independent, parallel pathway19. However, Pi release was shown to be essential for EF-G release from the ribosome20 and also, for EF-G function in ribosome disassembly21. The ribosomal stalk protein L7/L12 has been proposed to be involved in the Pi release process, but the mechanism is yet to be clarified20.
Recent high resolution structures of EF-G on the ribosome in complex with either GTP analogs (GDPCP and GDPNP)4,5, or GDP and fusidic acid (FA)22,23,24 show a dramatic change in the orientation of H91 upon GTP hydrolysis. Compared to the structures with the GTP analogue GDPCP or GDPNP, where the side chain of H91 points towards the γ-phosphate and remains tightly coupled via a salt bridge4,5, in structures with GDP and fusidic acid, it is rotated by 140° in the opposite direction. This striking observation prompted us to think that the ‘flip’ of the H91 following GTP hydrolysis might provide an efficient mechanism for Pi release. To test this idea, we created several mutant EF-Gs by replacing H91 with a series of amino acids (H91A, H91E, H91Q and H91R) and studied these mutant EF-Gs in GTP hydrolysis and Pi release with pre-steady state kinetics. As a control, we have mutated another highly conserved residue on switch II, Phe94 to Leu (F94L) and tested this mutant in parallel. F94 is known to be important for conformational changes in EF-G on and off the ribosome25,26 and mutation of F94 confers high level FA resistance to the bacteria, but also leads to loss of fitness25,27.
Our results show that the degree of defect in GTP hydrolysis varies in the H91 mutants with the exception of H91Q, which is not impaired in GTP hydrolysis. However, all the H91 mutants are defective in Pi release. Moreover, the relative decrease in the rate of Pi release is always greater than that in GTP hydrolysis. We also performed molecular dynamics (MD) simulations, which show that H91 remains coupled to Pi even after GTP hydrolysis. Combining our results with information from high resolution structures we propose that by ‘flipping’ after GTP hydrolysis, H91 guides the release of Pi by positioning it in an inter-domain space of EF-G, that constitutes an unhindered exit path for the Pi.
Results
The EF-G variants show similar affinity to (mant)-GTP
We have estimated the affinity of the EF-G variants (WT, F94L, H91A, H91E, H91Q and H91R) for mant-GTP, a fluorescent GTP analogue28. KD values were estimated by plotting the relative increase in mant fluorescence against the concentration of mant-GTP (Fig. S1). The KD values were similar for all six EF-G variants (Table 1); 8.2 ± 0.3 μM for the WT, 8.5 ± 1.9 μM for F94L, 10.6 ± 0.3 μM for H91A, 8.1 ± 0.3 μM for H91E, 13.4 ± 1.3 μM for H91Q and 14.3 ± 1.8 μM for H91R EF-G. Thus, the H91 mutants and F94L display similar proficiency in binding mant-GTP and thus, by inference native GTP.
Table 1. The rate constants of single turnover GTP hydrolysis and Pi release by the EF-G variants in the absence (−70S) and presence (+70S) of the ribosome.
| EF-G | KD GTP(μM) | kGTP (s−1) −70S | kGTP (s−1) +70S | kPi (s−1) +70S |
|---|---|---|---|---|
| WT | 8.2 ± 0.3 | 0.013 ± 0.003 | 202 ± 29 | 28 ± 4.7 |
| F94L | 8.5 ± 1.9 | 0.014 ± 0.003 | 170 ± 28 | 21 ± 3.5 |
| H91Q | 13.4 ± 1.3 | 0.02 ± 0.006 | 174 ± 12 | 1.2 ± 0.16 |
| H91R | 14.3 ± 1.8 | 0.019 ± 0.004 | 27 ± 2.5 | ------- |
| H91A | 10.6 ± 0.3 | 0.012 ± 0.001 | 28 ± 4.5 | 0.32 ± 0.04 |
| H91E | 8.1 ± 0.3 | 0.013 ± 0.003 | 1.9 ± 0.3 | 0.25 ± 0.02 |
The first column represents the affinity of the EF-Gs for (mant) GTP. Uncertainties represent standard deviations obtained from three independent experiments.
Intrinsic GTP hydrolysis by EF-G is unaffected by the H91 and F94 mutations
We studied the spontaneous GTPase activity of the EF-G variants by monitoring the time course of GTP hydrolysis in the absence of 70S ribosomes. For that, EF-G in excess was mixed with [3H]GTP at 37 °C and the reaction was quenched with 25% formic acid at different time points. The amount of GDP formed was estimated from the ratio of the peak area of [3H]GDP and [3H]GTP as separated on a MonoQ column coupled to HPLC. The rate constant for the intrinsic GTPase activity of WT EF-G was estimated as 0.013 ± 0.003 s−1, in close agreement with earlier reports13,26. Interestingly, none of the mutations affected the rate of intrinsic GTP hydrolysis. The rate constants for all EF-G variants were estimated as 0.014 ± 0.003 s−1 for F94L, 0.012 ± 0.001 s−1 for H91A, 0.014 ± 0.004 s−1 for H91E, 0.02 ± 0.006 s−1 for H91Q and 0.019 ± 0.004 s−1 for H91R (Fig. 2a, Table 1). Thus, in the absence of ribosome, GTP hydrolysis by EF-G does not involve H91. This is highly similar to a recent result with H84 mutations in EF-Tu, which did not show any defect in intrinsic GTPase activity29.
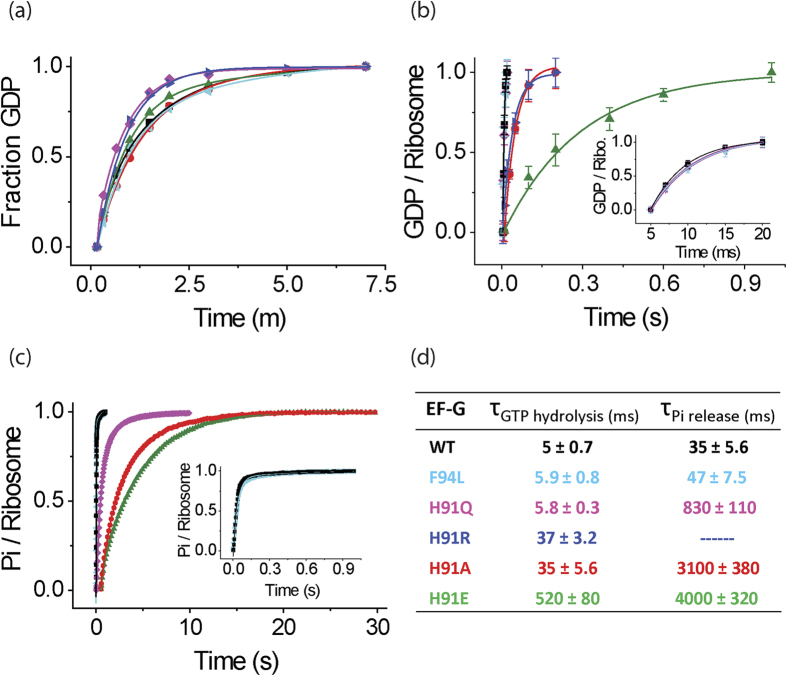
Figure 2. GTP hydrolysis and Pi release by the EF-G variants.
Time course of GTP hydrolysis by WT (black), F94L (cyan), H91A (red), H91E (green), H91Q (magenta) and H91R (blue) EF-Gs in the absence (a) and presence of 70S ribosomes (b) measured under single turnover conditions. The amount of [3H] GDP formed is plotted against time and fitted with a single exponential function to estimate the rate constant of GTP hydrolysis. Error bars represent standard deviations obtained from three independent experiments. (c) Single turnover Pi release in the presence of 70S ribosomes studied by fluorescence increase of MDCC-PBP. The traces are fitted with single exponential function. Inset in (b) shows WT, F94L and H91Q data while inset in (c) shows WT and F94L data on a shorter time scale. (d) Mean times (τ) for GTP hydrolysis and Pi release estimated from the experiments in b and c.
H91 mutated EF-Gs show different degrees of defect in ribosome-stimulated GTP hydrolysis
GTP hydrolysis experiments in the presence of 70S ribosomes were performed in a quench-flow instrument. As shown in earlier reports, the GTPase activity of EF-G was significantly stimulated by the ribosome13,26,30. The WT EF-G hydrolyzed GTP on the ribosome with a single turnover rate constant of kGTP WT = 202 ± 29 s−1, which is about 15 000 times larger than the rate constant of intrinsic GTP hydrolysis, as also reported earlier31. Mutants H91Q (kGTP H91Q = 174 ± 12 s−1) and the control mutation F94L (kGTP F94L = 170 ± 28 s−1) hydrolyzed GTP with rate constants comparable to that of WT EF-G (Fig. 2b, Table 1). In contrast, the H91A, H91E and H91R EF-Gs were defective in GTP hydrolysis, although the degree of defect varied. Mutants H91A and H91R showed a seven fold decrease in the rate constant compared to the WT (kGTP H91A = 28 ± 4.5 s−1 and kGTP H91R = 27 ± 2.5 s−1), while H91E showed a rate reduction by a factor of 100 (kGTP H91E = 1.9 ± 0.3 s−1) (Fig. 2b, Table 1). The mean time analysis (i.e. inverse of rate constants) shows that compared to 5 ± 0.7 ms for the WT, the H91A and H91R take 35 ± 5.6 ms and 37 ± 3.2 ms respectively, and H91E takes 520 ± 82 ms to hydrolyze one GTP on the ribosome (Fig. 2d). Since the H91 mutant EF-Gs show similar affinity to GTP as the WT, the defect in ribosome stimulated GTP hydrolysis must have originated from their interaction with the ribosome.
H91 mutants show larger defects in Pi release than in GTP hydrolysis
Single turnover Pi release was studied using stopped-flow, with MDCC labelled phosphate binding protein (PBP-MDCC), the fluorescence of which increases instantaneously upon Pi binding32,33. In the presence of the ribosome, WT EF-G released Pi with a rate constant of kPi WT = 28 ± 4.7 s−1 (Fig. 2c, Table 1), which is much slower than GTP hydrolysis. In comparison to the WT, except for the control mutation F94L, which did not show any defect in Pi release (kPi F94L = 21 ± 3.5 s−1), all the H91 mutants were much slower in Pi release (Fig. 2c, Table 1). The H91Q mutant, which did not show any defect in GTP hydrolysis, was over 20fold slower than WT EF-G in releasing Pi (kPi H91Q = 1.2 ± 0.16 s−1). The other two mutants, H91A and H91E released Pi with rate constants kPi H91A = 0.32 ± 0.04 s−1 and kPi H91E 0.25 ± 0.02 s−1 respectively. Pi release by H91R EF-G could not be measured. This is probably due to very slow release of Pi, which is either beyond the fluorescence life-time of PBP-MDCC or is readily removed by the phosphate mop (see Materials and Methods) present in the reaction. These results suggest that H91 plays an important role in Pi release from EF-G.
Molecular dynamics simulations are consistent with the role of H91 in Pi release
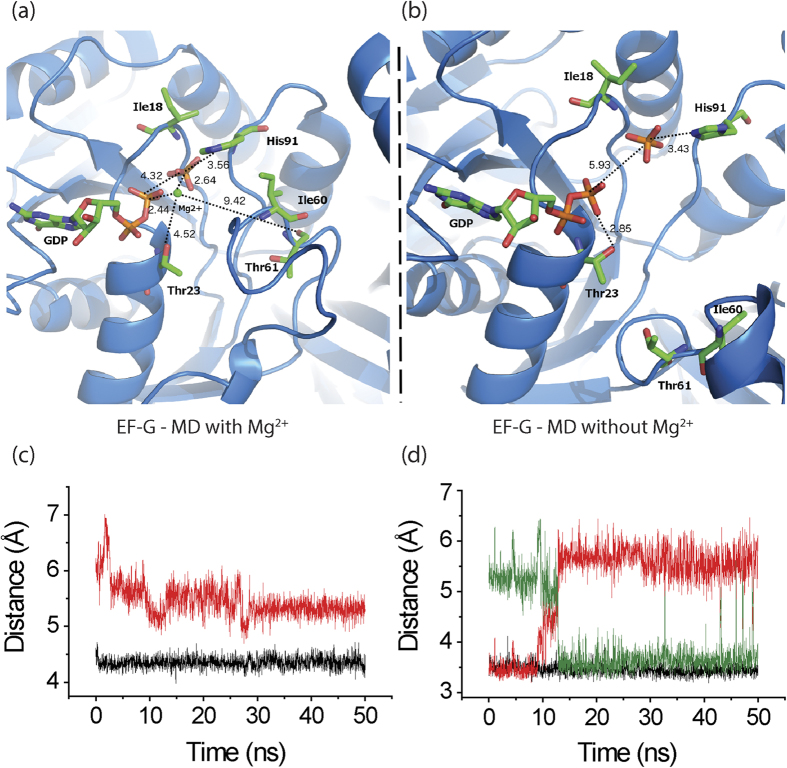
Based on the crystal structure of the ribosome bound to EF-G•GTP (PDB: 4CR1)4, we have built and equilibrated an initial model of EF-G bound to GDP and Pi using MacroMoleculeBuilder (MMB)34 and with MD simulations35. This model essentially mimics the situation after GTP hydrolysis but before Pi release. In MD, the distance between Pi and the β-phosphate was 4.3 Å (Fig. 3a,c), while between H91 (Nδ1) and Pi it was 3.6 Å (Fig. 3a,d); this did not differ significantly from what was observed with MMB. In addition, a strong coordination between Pi and Mg2+ was observed in both MMB and MD runs, suggesting that the Mg2+ stabilizes Pi even after GTP hydrolysis.
Figure 3. MD simulation results.
(a,b) Structural views of the guanine-nucleotide binding pocket and switch II of EF-G with (a) and without (b) Mg2+ ion (green sphere) from the MD simulation experiments. The initial models are built using MacroMoleculeBuilder (MMB)34, on the basis of the crystal structure of EF-G•GDPCP bound to the ribosome (PDB: 4CR1)4. Both models represent the scenario immediately after GTP hydrolysis but before Pi release. We did not observe any relevant structural differences in the replica MDs. (c,d) Time trace for the distances between Pi and β-phosphate (c) and Pi and H91 (either Nδ1 or Nε2) (d) estimated from MD structures with (black trace) or without (red/green trace) Mg2+ ion. The red and the green traces in (d) are distances involving Nδ1 or Nε2 atoms of the H91 side chain, respectively.
Close comparison of the ribosome bound EF-G•GTP structures (PDB: 4CR1, 4JUW)4,5 with the EF-G•GDP structures showed disappearance (PDB: 4KDA)24 or major repositioning of the Mg2+ with respect to β-phosphate and Pi (PDB: 2WRI)22 (Fig. S2). This suggests that in order for the Pi to be released the Mg2+ needs to be repositioned. To test this hypothesis, another model was built without the Mg2+ and equilibrated in MMB. Then the model was relaxed with MD as the former one (Fig. 3b). We saw an immediate increase in distance between β-phosphate and Pi to ~5.9 Å (Fig. 3b,c). In contrast, the distance between Pi and H91 (either Nδ1 or Nε2) remained unchanged at ~3.5 Å (Fig. 3b,d), suggesting that the Pi was strongly coupled with H91. Thus, H91 and Pi could together move towards the open GDP-bound configuration. This is consistent with the suggestion from the biochemical data that H91 facilitates Pi release.
Discussion
Our results show that GTP hydrolysis by EF-G gets strongly stimulated by the ribosome (Table 1)26, suggesting that the ribosomal components act as the GTPase activating factor for EF-G30,36. This is in agreement with the previous results and also similar to EF-Tu, where the rate of GTP hydrolysis has been reported to increase by a million fold upon binding to the ribosome29. In structural terms, the activation is probably achieved by proper positioning of the two switch regions of EF-G in relation to GTP and also by opening the so called ‘hydrophobic gate’ composed of the two hydrophobic amino acids Ile18 and Ile60 (E. coli numbers) (Fig. 4a). In crystal structures of EF-G with GTP analogues, H91 is directed towards the γ-phosphate with an average distance of 3.6 Å between them, which is typical for a salt bridge (Fig. 4a)4,5. Although the exact role of H91 in GTP hydrolysis cannot be deduced from these structures, a close coordination of H91 and the γ-phosphate is evident.
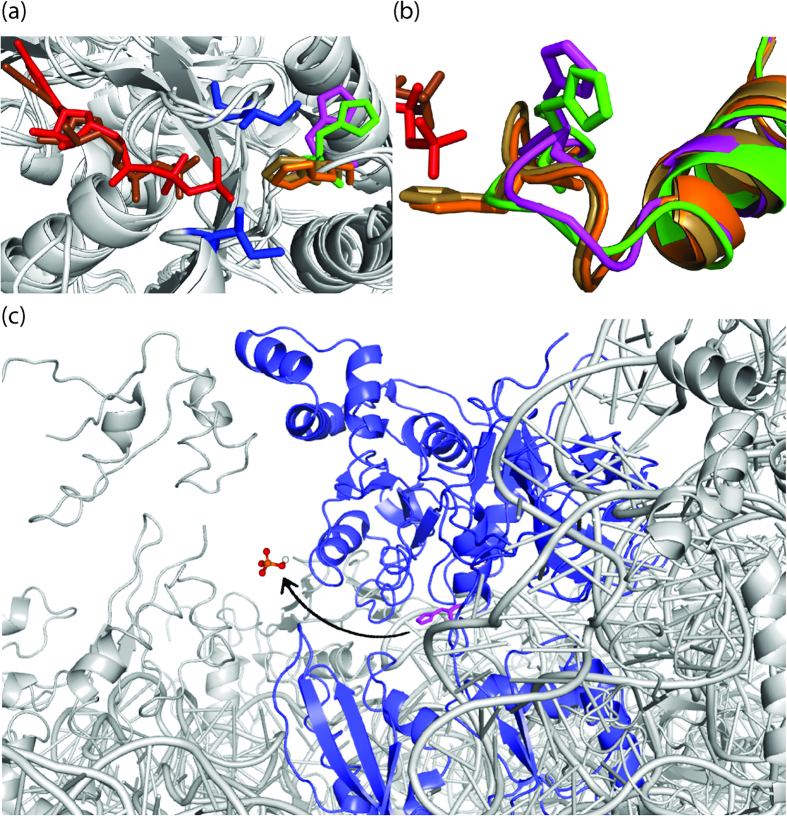
Figure 4. The orientations of the H91 side chain and the exit path for Pi.
(a) Superimposition of the crystal structures of EF-Gs (Thermus thermophilus) showing orientation of H91 side chain (H87 in T. thermophilus); with GDPCP (PDBs: 4JUW5 in orange and 4CR14 in sand) and with GDP and fusidic acid (PDBs: 4KDA24 in green and 2WRI22 in magenta). The “hydrophobic gate” formed by residues Ile20 and Ile63 are shown in blue and the bound GDP and GDPCP are shown in brown and red respectively. (b) Close-up view of the switch II loop with its conserved H91, color code as in (a) shows that the whole switch-II loop following the superimposed α helix changes its orientation in the GTP/GDP state. (c) The potential exit path of Pi (modelled in the crystal structure (PDB: 2WRI)22 through the inter-domain space of EF-G•GDP (blue) on the ribosome (grey). It can be observed that the Pi exit path (marked with an arrow) in front of H91 (magenta) is not hindered by any ribosomal components.
In this work, we have characterized four H91 mutant EF-Gs for GTP hydrolysis and Pi release and compared those with WT and F94L EF-Gs. All of these EF-G mutants have similar affinity to GTP in solution and the rate of intrinsic GTP hydrolysis is not affected by any of these mutations. This result, similar to EF-Tu suggests that the spontaneous GTP hydrolysis by EF-G does not depend on H9129. In a recent study, a monovalent cation far from H84 has been suggested as the catalytic cofactor for GTP hydrolysis in ribosome unbound EF-Tu37 and likely to be the same in EF-G. It should be noted that in the free form of EF-G, H91 points away from the γ-phosphate of GTP (PDB: 2J7K)38, which explains why H91 is not involved in intrinsic GTPase activity of EF-G. However, on the ribosome, the mutated EF-Gs display distinct behavior. The largest defect in ribosome stimulated GTP hydrolysis was identified for the H91E mutant. This result is not unexpected, since in this case the bulky positive side chain of His is replaced by a negative side chain of Glu. This swap will not only destroy the coupling between the amino acid and the γ-phosphate, but may also alter the charge distribution and the intramolecular interactions in the GTP binding pocket. In contrast, no defect was observed for the H91Q mutant. This is also expected since Gln is present instead of His in all G-proteins except the translational GTPases. When side chains are compared, the NH2 group of Gln can easily be superimposed on the NH group of His, which is actually involved in coupling with the γ-phosphate. Thus, it follows naturally that the H91Q mutant acts like the WT in GTP hydrolysis. Similar results have also been reported with the corresponding mutation in EF-Tu (H84Q), which showed only small to moderate reduction in the GTPase activity9,10. However, a recent report on H84Q mutation of E. coli EF-Tu shows about a 4000 fold decrease in the rate of GTP hydrolysis29, which contradicts the earlier results. Further investigation will be needed to solve the controversy in the rate of GTP hydrolysis by H84Q EF-Tu.
The H91A and H91R mutants are about seven fold slower than the WT in ribosome stimulated GTP hydrolysis. In these two cases, the positive side chain of His is replaced either with a small nonpolar side chain (A) or a longer basic side chain (R). In H91A, the lack of proper orientation of the β and γ-phosphate of GTP can be expected due to absence of the coupling between the γ-phosphate and the nonpolar small side chain of Ala. The H91R mutant, in contrast, might have additional coupling with multiple phosphates of GTP due to the occurrence of three N atoms in its side chain. It should be mentioned that the H91A EF-G was previously reported as completely inactive in GTP hydrolysis11. This apparent discrepancy, we think, may arise from differences in the sample preparation and experimental conditions.
Compared to the mean time of GTP hydrolysis by WT EF-G (~5 ms) Pi release takes much longer time (~35 ms) (Fig. 2d), which suggests that Pi remains trapped in the nucleotide binding pocket even after GTP is hydrolyzed. Interestingly, the Pi retention time is significantly longer than the WT for all the four H91 mutants (lowest 4000 ms), while for the F94L mutant it is similar to the WT (Fig. 2d). Moreover, comparison of the mean times of GTP hydrolysis and Pi release clearly demonstrates a much bigger defect in Pi release for all the H91 mutants than in GTP hydrolysis. Thus, our results suggest a direct involvement of H91 in Pi release.
To understand the structural basis of Pi release we compared the crystal structures of EF-G bound to GTP analogues (PDB: 4CR1, 4JUW)4,5 and GDP, both on the ribosome (PDB: 2WRI, 4KDA)22,24 and in the free state (PDB: 1FNM, 2BM0, 4M1K, 4MYU)39,40. While the most obvious change can be seen in the orientation of the H91 side chain, which ‘flipped’ almost 130°–140° (Fig. 4a) and consequently altered the conformation of the entire switch II loop (Fig. 4b), several other changes were visible. Detailed inspection of the GTP binding site dragged our attention to a Mg2+ ion coordinating with both the β- and the γ-phosphates of GTP. Interestingly, in the EF-G•GDP structures this Mg2+ was either absent (PDB: 4KDA)24 or its position varied a lot (PDB: 2WRI)22 (Fig. S2). This analysis suggested that displacement of the Mg2+ might be necessary for Pi release. Our MD simulations indeed support this hypothesis. When the EF-G•GDP•Pi model contained the Mg2+ ion, no change in the interatomic distances was observed during the MD run (Fig. 3a). In contrast, an immediate increase in the distance between the β-phosphate and Pi was noticed when the model was built without the Mg2+ (Fig. 3b). However, H91 remained constantly coupled with the Pi although the His side chain might undergo a rotation thereby switching the coupling from the Nδ1 to the Nε2 atom. Thus we propose that the tight coupling of Pi to H91 and the ‘flipping’ movement of H91are the two key determinants of Pi release.
The lack of coupling with Pi in H91A and H91E EF-Gs renders them significantly defective in Pi release. On the contrary, perhaps a too strong coupling with the Arg side chain in H91R makes Pi release so slow that it could not be measured under our experimental conditions. Most interestingly, the H91Q mutant, which is not impaired in GTP hydrolysis showed a more than 20 fold defect in Pi release. It suggests that although EF-G can attain the optimal conformation for GTP hydrolysis with Gln instead of His, the bulky side chain of His is instrumental in changing the conformation of switch II required for Pi release (Fig. 4b). This view is further supported by the observation that in the flipped conformation (GDP state), the H91 side chain opens into a tunnel which runs through the inter domain space of EF-G22,24. Since Pi remains coupled with H91 after GTP hydrolysis, most likely it follows H91 and thereby reaches this tunnel, which constitutes the exit path for the Pi (Fig. 4c). This path is free from steric obstructions even on the ribosome (Fig. 4c). Thus, we infer that the tight coordination of Pi with H91 and the ‘flipping’ of the H91 side chain are the key determinants for efficient Pi release. This mechanism may have general significance as other translational GTPases (e.g. EF-Tu) possess a His corresponding to H91, which undergoes similar structural transitions.
Materials and Methods
Mutagenesis of EF-G and protein purification
The WT fusA gene form E. coli cloned into plasmid pET24b with a C-terminal hexa-histidine tag was a kind gift from Kevin S. Wilson, Oklahoma state University, USA26. Using this construct as a template and following standard protocols for site-directed mutagenesis, the H91 residue was mutated to alanine (A), Glutamine (Q), Arginine (R) and glutamic acid (E), resulting into four mutant EF-Gs, H91A, H91Q, H91R and H91E respectively. In a similar way, residue F94 was mutated to Leucine (L), creating F94L mutant. The mutations were confirmed by DNA sequencing. Over-expression and purification of the WT and mutant EF-Gs were carried out as described earlier25, by using Ni2+ affinity chromatography followed by size exclusion. The homogeneity of the WT and mutant EF-Gs was confirmed by SDS-Page followed by mass spectrometric analysis.
Buffers and components
All experiments were performed in HEPES polymix buffer (pH 7.5) containing 5 mM NH4Cl, 5 mM Mg(OAc)2, 100 mM KCl, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 5 mM HEPES and 1 mM dithioerythritol (DTE) at 37 °C with a free Mg2+ concentration of around 1.5 mM. MRE600 70S ribosomes were purified according to the published methods33,41. Nucleotide triphosphates were from GE-Healthcare. Plasmid construct for phosphate binding protein (PBP) was a kind gift from Prof. Martin Webb, London. The purification and 7-Diethylamino-3-((((2-Maleimidyl) ethyl)amino) carbonyl)coumarin (MDCC) labelling of PBP was done according to the published protocol32.
Single-round GTP hydrolysis
Intrinsic GTP hydrolysis by EF-G was measured under single turnover condition by mixing equal volumes of [3H]GTP(15 μM) and EF-G (30 μM) manually at 37 °C; the reaction was quenched at different time points with 25% formic acid. The amount of [3H]GDP formed was estimated by separating the [3H]GTP and [3H]GDP fractions on a MonoQ column attached to a HPLC system.
The GTPase activity of the EF-Gs on the ribosome was measured in a similar way by mixing vacant 70S ribosomes (3 μM) and [3H] GTP (15 μM) (in one mix) with EF-G (15 μM) in a quench-flow device (RQF-3 KinTek Corp.) at 37 °C. In order to estimate the single turnover GTP hydrolysis rate, data points only from the initial fast phase were used. The rate constants of GTP hydrolysis (kGTP) was obtained by fitting the data with a single exponential function using ORIGIN 8.0 (Originlab Corporation). The mean time τGTP was estimated as (1/kGTP).
Single-round Pi release
The release of inorganic phosphate from EF-G after GTP hydrolysis was measured in a stopped flow instrument (Applied Photophysics SX20) by monitoring the fluorescence of MDCC-PBP that shows an immediate increase in fluorescence upon binding to Pi32,33. Mix A containing 70S ribosomes (2 μM) and GTP (100 μM) was rapidly mixed with mix B containing EF-G (20 μM) and MDCC-PBP (1 μM) after preincubating both mixes for 10 minutes at 37 °C. Both mixes also contained a phosphate mop comprising 7-methylguanosine (300 μM) and purine nucleoside phosphorylase (0.1 unit/μl). The rate constants of Pi release (kPi) was estimated by fitting the fluorescence time traces with a single exponential function using Origin 8.0 (Originlab Corp.). The mean time for Pi release (τPi) was estimated as (1/kPi).
Measurement of affinity for GTP
The equilibrium dissociation constant (KD) for GTP to EF-G was estimated using a fluorescent analog of GTP called mant-GTP ((2′, 3′)-O-N-methylanthraniloyl-GTP). When exited at 290 nm, fluorescence resonance energy transfer (FRET) occurs between the Trp residues present in the proximity of the nucleotide binding pocket in EF-G and the mant group of mant-GTP, thereby leading to increase in its fluorescence28. EF-G (2 μM) was mixed with increasing amounts of mant-GTP (5–60 μM) at 37 °C and the fluorescence change at 445 nm was recorded in a fluorimeter (HITACHI F-7000). The difference in the FRET signal with and without EF-G was plotted against the concentration of mant-GTP and the KD was estimated by fitting of a hyperbolic function using ORIGIN 8.0 (Originlab Corporation).
Modelling of EF-G•GDP – Pi and Molecular Dynamics simulations
We built and equilibrated an initial model of EF-G bound to GDP and Pi using MMB34 based on the crystal structure of the ribosome bound to EF-G•GTP (PDB: 4CR1)4, as a reduced-dimensionality prototype simulation prior to full MD. During equilibration, EF-G residues 83-91, Mg2+, β-phosphate of GDP and Pi were flexible. We placed a small water droplet restrained to a ~1.6 nm radius about H91. The PARM99 force field35 was applied to the water and all atoms within 1.2 nm of the flexible regions. A strong coordination between Pi and Mg2+ was observed, in addition to coordination between H91 and Pi. To understand the role of the Mg2+, another model without Mg2+ was built and similarly equilibrated in MMB. For MD simulation, GDP parameters (charges, dihedrals, angles, bonds, and van der Waals parameters) were based on Meagher et al.42. For Pi, the atomic point charges were calculated with the GAUSSIAN 0943 at the HF/6-31G* level, while the other parameters were obtained with general Amber force field (GAFF)35. The model with Mg2+ was solvated with 45347 single point charge (SPC) waters while the model without Mg2+ was solvated with 46144 SPC waters. Both systems were submitted to 100 steps of steep descent energy minimization to remove bad contacts between the solvent and the protein, followed by 200 ps of restrained MD simulation to relax the water molecules. Finally, for both models, two random initial velocity 50 ns MD runs were performed using a time step of 0.002 ps. Periodic boundary conditions were used in a NPT ensemble (v-rescale thermostat44 and Parrinello-Rahman barostat45 - τT = 0.1 ps, Tref = 300 K, Pref = 1 bar). The Particle Mesh Ewald (PME)46 method was applied with a cut-off of 1.0 nm. A twin range cut-off with neighbor list cut-off 1.0 was used for the van der Waals interactions. All simulations and consequent analyses were carried out using the Gromacs 4.6.5 software package conjugated with the PARM99 force field35,47.
Additional Information
How to cite this article: Koripella, R. K. et al. A conserved histidine in switch-II of EF-G moderates release of inorganic phosphate. Sci. Rep. 5, 12970; doi: 10.1038/srep12970 (2015).
Supplementary Material
Acknowledgments
We thank Kevin S. Wilson, Oklahoma state University, USA for providing us the pET24b construct for over-expression and mutagenesis of EF-G and Prof. Martin Webb, London for sharing the construct of PBP. The authors acknowledge grants from Swedish Research council (2011-6088 [NT], 2014-4423 [NT] and 2008-6593 [Linnaeus grant to Uppsala RNA Research Center]); and Knut and Alice Wallenberg Foundation (KAW 2011.0081 to RiboCORE) to SS and Swedish Foundation for International Cooperation in Research and Higher Education (led by SF). The authors gratefully acknowledge Måns Ehrenberg for critical reading of the manuscript.
Footnotes
Author Contributions S.S. conceived the idea, designed experiments and analyzed the results; R.K.K. and M.H. performed biochemical experiments and analyzed data; D.D. and S.F. conducted MD simulations; C.S.M. analyzed available structures. All the authors participated in manuscript preparation.
References
- Bourne H. R., Sanders D. A. & McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127 (1991). [DOI] [PubMed] [Google Scholar]
- Vetter I. R., Nowak C., Nishimoto T., Kuhlmann J. & Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398, 39–46 (1999). [DOI] [PubMed] [Google Scholar]
- Hilgenfeld R. Regulatory GTPases. Curr Opin Struct Biol 5, 810–817 (1995). [DOI] [PubMed] [Google Scholar]
- Chen Y., Feng S., Kumar V., Ero R. & Gao Y. G. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol 20, 1077–1084 (2013). [DOI] [PubMed] [Google Scholar]
- Tourigny D. S., Fernandez I. S., Kelley A. C. & Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340, 1235490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees R. M., Schmeing T. M., Kelley A. C. & Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool R. H. & Parmeggiani A. Substitution of histidine-84 and the GTPase mechanism of elongation factor Tu. Biochemistry 30, 362–366 (1991). [DOI] [PubMed] [Google Scholar]
- Daviter T., Wieden H. J. & Rodnina M. V. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol 332, 689–699 (2003). [DOI] [PubMed] [Google Scholar]
- Scarano G., Krab I. M., Bocchini V. & Parmeggiani A. Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: its substitution by glutamine and alanine. FEBS Lett 365, 214–218 (1995). [DOI] [PubMed] [Google Scholar]
- Zeidler W. et al. Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81 and Arg300. Eur J Biochem 229, 596–604 (1995). [DOI] [PubMed] [Google Scholar]
- Cunha C. E. et al. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation 1, e24315-1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp W. et al. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J 33, 1073–1085 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsi E., Farah E., Dann J. & Ermolenko D. N. Following movement of domain IV of elongation factor G during ribosomal translocation. Proc Natl Acad Sci USA 111, 15060–15065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljas A., Ehrenberg M. & Aqvist J. Comment on “The mechanism for activation of GTP hydrolysis on the ribosome”. Science 333, 37 (2011). [DOI] [PubMed] [Google Scholar]
- Wallin G., Kamerlin S. C. & Aqvist J. Energetics of activation of GTP hydrolysis on the ribosome. Nat Commun 4, 1733 (2013). [DOI] [PubMed] [Google Scholar]
- Aqvist J. & Kamerlin S. C. The Conformation of a Catalytic Loop Is Central to GTPase Activity on the Ribosome. Biochemistry 54, 546–556 (2015). [DOI] [PubMed] [Google Scholar]
- Li W. et al. Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G. Science Advances (2015), 10.1126/sciadv.1500169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F., Matassova N. B., Savelsbergh A., Rodnina M. V. & Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell 6, 501–505 (2000). [DOI] [PubMed] [Google Scholar]
- Savelsbergh A. et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell 11, 1517–1523 (2003). [DOI] [PubMed] [Google Scholar]
- Savelsbergh A., Mohr D., Kothe U., Wintermeyer W. & Rodnina M. V. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J 24, 4316–4323 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Rodnina M. V. & Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 15, 772–780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. G. et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H. et al. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat Rev Microbiol 12, 89–100 (2014). [DOI] [PubMed] [Google Scholar]
- Zhou J., Lancaster L., Donohue J. P. & Noller H. F. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340, 1236086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koripella R. K. et al. Mechanism of elongation factor-G-mediated fusidic acid resistance and fitness compensation in Staphylococcus aureus. J Biol Chem 287, 30257–30267 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticu C., Murataliev M., Nechifor R. & Wilson K. S. A central interdomain protein joint in elongation factor G regulates antibiotic sensitivity, GTP hydrolysis, and ribosome translocation. J Biol Chem 286, 21697–21705 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., Aevarsson A., Liljas A. & Hughes D. The dynamic structure of EF-G studied by fusidic acid resistance and internal revertants. J Mol Biol 258, 420–432 (1996). [DOI] [PubMed] [Google Scholar]
- Wilden B., Savelsbergh A., Rodnina M. V. & Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci USA 103, 13670–13675 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maracci C., Peske F., Dannies E., Pohl C. & Rodnina M. V. Ribosome-induced tuning of GTP hydrolysis by a translational GTPase. Proc Natl Acad Sci USA 111, 14418–14423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A., Mohr D., Wilden B., Wintermeyer W. & Rodnina M. V. Stimulation of the GTPase activity of translation elongation factor G by ribosomal protein L7/12. The Journal of biological chemistry 275, 890–894 (2000). [DOI] [PubMed] [Google Scholar]
- Rodnina M. V., Savelsbergh A., Katunin V. I. & Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385, 37–41 (1997). [DOI] [PubMed] [Google Scholar]
- Brune M., Hunter J. L., Corrie J. E. & Webb M. R. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33, 8262–8271 (1994). [DOI] [PubMed] [Google Scholar]
- Huang C., Mandava C. S. & Sanyal S. The ribosomal stalk plays a key role in IF2-mediated association of the ribosomal subunits. J Mol Biol 399, 145–153 (2010). [DOI] [PubMed] [Google Scholar]
- Dourado D. F. & Flores S. C. A multiscale approach to predicting affinity changes in protein-protein interfaces. Proteins 82, 2681–2690 (2014). [DOI] [PubMed] [Google Scholar]
- Wang J. M., Wolf R. M., Caldwell J. W., Kollman P. A. & Case D. A. Development and testing of a general amber force field Journal of Computational Chemistry 25, 1157–1174 (2004). [DOI] [PubMed] [Google Scholar]
- Frank J., Gao H., Sengupta J., Gao N. & Taylor D. J. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA 104, 19671–19678 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhle B. & Ficner R. A monovalent cation acts as structural and catalytic cofactor in translational GTPases. EMBO J 33, 2547–2563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson S., Singh R., Gudkov A. T., Liljas A. & Logan D. T. Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett 579, 4492–4497 (2005). [DOI] [PubMed] [Google Scholar]
- Hansson S., Singh R., Gudkov A. T., Liljas A. & Logan D. T. Structural insights into fusidic acid resistance and sensitivity in EF-G. J Mol Biol 348, 939–949 (2005). [DOI] [PubMed] [Google Scholar]
- Laurberg M. et al. Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site. J Mol Biol 303, 593–603 (2000). [DOI] [PubMed] [Google Scholar]
- Mandava C. S. et al. Bacterial ribosome requires multiple L12 dimers for efficient initiation and elongation of protein synthesis involving IF2 and EF-G. Nucleic Acids Res 40, 2054–2064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher K. L., Redman L. T. & Carlson H. A. Development of polyphosphate parameters for use with the AMBER force field. Journal of Computational Chemistry 24, 1016–1025 (2003). [DOI] [PubMed] [Google Scholar]
- Frisch M. J. et al. Gaussian 09, Revision D. 01, (ed. Gaussian , Inc., Wallingford CT, 2009). [Google Scholar]
- Bussi G., Donadio D. & Parrinello M. Canonical sampling through velocity rescaling. Journal of Chemical Physics 126, (2007); 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Parrinello M. & Rahman A. Polymorphic Transitions in Single-Crystals - a New Molecular-Dynamics Method. Journal of Applied Physics 52, 7182–7190 (1981). [Google Scholar]
- Essmann U. et al. A Smooth Particle Mesh Ewald Method. Journal of Chemical Physics 103, 8577–8593 (1995). [Google Scholar]
- Lindahl, E., Hess B. & Spoel D. V. D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. Journal of molecular modeling 7, 306–317 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.