Abstract
We evaluated effects of 9-year simulated nitrogen (N) deposition on microbial composition and diversity in the rhizosphere of two dominant temperate grassland species: grass Stipa krylovii and forb Artemisia frigida. Microbiomes in S. krylovii and A. frigida rhizosphere differed, but changed consistently along the N gradient. These changes were correlated to N-induced shifts to plant community. Hence, as plant biomass changed, so did bacterial rhizosphere communities, a result consistent with the role that N fertilizer has been shown to play in altering plant-microbial mutualisms. A total of 23 bacterial phyla were detected in the two rhizospheric soils by pyrosequencing, with Proteobacteria, Acidobacteria, and Bacteroidetes dominating the sequences of all samples. Bacterioidetes and Proteobacteria tended to increase, while Acidobacteria declined with increase in N addition rates. TM7 increased >5-fold in the high N addition rates, especially in S. krylovii rhizosphere. Nitrogen addition also decreased diversity of OTUs (operational taxonomic units), Shannon and Chao1 indices of rhizospheric microbes regardless of plant species. These results suggest that there were both similar but also specific changes in microbial communities of temperate steppes due to N deposition. These findings would contribute to our mechanistic understanding of impacts of N deposition on grassland ecosystem by linking changes in plant traits to their rhizospheric microbes-mediated processes.
Keywords: nitrogen deposition, microbial diversity, rhizosphere, Illumina Miseq, temperate steppe, Artemisia frigida, Stipa kerlovii
Introduction
Nitrogen (N), an essential mineral nutrient to plant growth, is one of the most limiting factors in many terrestrial ecosystems (Vitousek et al., 2002; Scheible et al., 2004; Asakawa and Kimura, 2008). However, elevated levels of N deposition are changing N inputs and impacting many ecological processes (Sala et al., 2000; Gilliam, 2006). Many ecosystems have developed under tight nutrient cycling and low amounts of available nutrients. In these systems, elaborate plant-microbial mutualisms have developed and are the foundation of ecosystem function (Reynolds et al., 2003). Nitrogen deposition can disrupt these plant-microbial interactions, which may feedback to alter microbial communities and ecosystem function (Sala et al., 2000; Gilliam, 2006; Martinelli et al., 2006).
Human industrial activities have led to a doubling, on average, of N deposition into terrestrial ecosystems and these inputs are expected to increase in the future (Galloway et al., 2004, 2008; Bodirsky et al., 2014). In China, N deposition has significantly increased over the last three decades, and in some regions are 5X greater than previous decades (Liu et al., 2013). Consequently, the impacts of N input due to deposition of atmospheric N on terrestrial ecosystems warrant further study (Galloway et al., 2004, 2008).
Grassland ecosystems are highly sensitive to N deposition, where long-term N addition has been shown to significantly reduce plant species richness (Stevens et al., 2004; Clark and Tilman, 2008). The semi-arid grasslands in northern China, which are a part of the Eurasian steppe, are exposed to enhanced N deposition rates (Zhang et al., 2008; Liu et al., 2013), and experienced reductions in plant species richness (Bai et al., 2010; Song et al., 2011; Fang et al., 2012; Tian et al., 2015). Several mechanisms have been proposed to explain the decline in species richness by N deposition (Suding et al., 2005; Harpole and Tilman, 2007; Bobbink et al., 2010), and among them are changes in soil microbial activity and biodiversity (Chen et al., 2014; Dean et al., 2014). Nitrogen deposition to a variety of terrestrial ecosystems has also been shown to profoundly affect soil microbial communities (Ramirez et al., 2010, 2012; He et al., 2013; Liu et al., 2014b; Zhang et al., 2014), however, these studies have not emphasized the involvement of root-zone and rhizosphere microbes; rather they have mainly focused on microbes in soils generally. The rhizosphere niche is an important interface for plant-microbes interactions and key to the success of both plants and microbes (Bakker et al., 2013). Microbe communities in the rhizosphere soils differ from those in bulk soils (Berg and Smalla, 2009), thus deserving specific attention for understanding ecosystem responses to N deposition. It is conceivable that microbial communities in the rhizosphere of different plant species may respond more strongly and perhaps, differently, to N deposition. Recently, Dean et al. (2014) found that N deposition affected host-associated plant root-associated fungi, however, there was no broader description of fungal or rhizosphere bacterial communities.
In the present study, we found that plant community shifted from co-dominance by a monocot grass, Stipa krylovii, and a dicot forb, Artemisia frigida, to exclusive dominance by a monocot grass in an Inner Mongolia steppe after 9-year of N addition. To test whether the microbial communities colonizing S. krylovii and A. frigida rhizosphere niche respond, and respond in similar or different ways to N addition, we used the high-throughput Illumina Miseq sequencing platform to characterize the rhizosphere microbial communities of the two dominant plant species under varying simulated N deposition rates. The following question was specifically addressed: Whether the rhizosphere microbial communities would be associated with plant host and its response to N deposition rate?
Materials and methods
Study site
The experiment was conducted in Duolun county (42°02′N, 116°17′E, 1324 m a.s.l.), Inner Mongolia, China. The area is located in a semiarid temperate steppe where the mean annual temperature is 2.1°C and long-term annual precipitation is 382.2 mm (Yang et al., 2011; Fang et al., 2012). The soil in the area is chestnut (Chinese classification) and Haplic Calcisols (FAO classification). The soil bulk density is 1.31 g cm−3 and pH is about 6.84 (Fang et al., 2012). The dominant species in this typical temperature steppe are A. frigida, S. krylovii, Cleistogenes squarrosa, Allium bidentatum, Potentilla acaulis, Leymus chinensiss, Salsola collina, Carex korshinskyi, Melilotoides ruthenica, and Agropyron cristatum (Niu et al., 2008; Fang et al., 2012).
Experimental design
In the experimental area, 64 plots of 15 × 10 m separated by 4-m-wide buffer strips were established in an 8 × 8 Latin square experimental design. Nitrogen was added as urea (N, 46%) at the midpoint of the growing season (July) every year since 2003. There were eight levels of N fertilization including a control, 0 (N0), 1 (N1), 2 (N2), 4 (N4), 8 (N8), 16 (N16), 32 (N32), and 64 (N64) g N m−2 year−1 with ambient N deposition of 1.6 g N m−2 year−1 (Zhang et al., 2008). In the present study, the rhizosphere soil samples were collected from 32 plots with four levels of N fertilization, ambient (N0), 2 (N2), 8 (N8), and 16 (N16) g N m−2 year−1. The rhizospheric soils from at least two individual plant roots of the two dominant species (S. krylovii, A. frigida) in each plot were sampled in August 2012 by collecting soils that were adhered to roots after vigorously shaking roots removed from field by a spade as described by Smalla et al. (2001). The rhizospheric soils were sampled from the eight plots under the four N addition levels, and the final three soil samples used for sequencing were obtained by randomly mixing the samples from 3, 3, and 2 plots, respectively. These ensure that the rhizospheric soil samples covered the eight replicates for N addition. Aboveground biomass in one quadrat (1 × 1 m) at each plot was clipped and determined since August 2004 as described by Fang et al. (2012).
Sequencing and data analysis
The total genomic DNA was extracted from 0.5 g rhizosphere soils using the SoilGen DNA Kit (CWbiotech Corporation, China) according to the manufacturer's instructions. PCR amplifications were conducted with the 515f/806r (GTGCCAGCMGCCGCGGTAA/GGA CTACHVGGGTWTCTAAT) primer set that amplified the V4 region of the 16S rRNA gene (Peiffer et al., 2013). The primer set was selected as it exhibits few biases and should yield accurate taxonomic information. The reverse primer contained a 6-bp error-correcting barcode unique to each sample. The PCR reaction was carried out in 30 μL reactions with 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs); 0.2 μM of forward and reverse primers, and about 10 ng template DNA. Thermal cycling was consisted of initial denaturation at 98°C for 1 min, then 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, elongation at 72°C for 60 s and finally 72°C for 5 min. PCR products were mixed in equal density ratios. Then, mixture PCR products were purified with GeneJET Gel Extraction Kit (Thermo Scientific). Sequencing libraries were generated using NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit @ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina MiSeq platform and 300 bp paired-end reads were generated.
Pairs of reads from the original DNA fragments were merged by using FLASH-a very fast and accurate software tool which was designed to merge pairs of reads when the original DNA fragments were shorter than twice the length of reads (Magoc and Salzberg, 2011). Sequencing reads were assigned to each sample according to the unique barcode of each sample. Sequences were analyzed with the QIIME software package (Quantitative Insights Into Microbial Ecology) and UPARSE pipeline (Caporaso et al., 2010; Edgar, 2013). First, the reads were filtered by QIIME quality filters. Default settings for Illumina processing in QIIME were used. Then we used UPARSE pipeline to pick operational taxonomic units (OTUs) by making OTU table. After removal of chimera, sequences were assigned to OTUs at 97% similarity. We picked a representative sequence for each OTU and used the version 2.2 RDP classifier to assign taxonomic data to each representative sequence with default 0.8 as confidence threshold (Wang et al., 2007). Singleton OTUs that appeared in only one sample were removed because they could be potential sequencing errors. In order to compute alpha diversity, we rarified the OTU table and calculated three metrics: Chao1 metric estimated the species richness, the observed species metric was simply the count of unique OTUs found in the sample, and Shannon index. Rarefaction curves were generated based on these three metrics. QIIME calculated unweighted unifrac, which was used to do Principal Coordinate Analysis (PCA).
Sequence accession number
The data were deposited in the National Center for Biotechnology Information Sequence Reads Archive with accession number SRS977347.
Statistical analysis
Considering that the N addition and plant host in the same plot may not be completely independent, we used the linear mixed models to analyze the effects of N addition on the rhizoshpere microbes at four levels with N treatment as the fixed effect and plant host as random effects. For each species, we conducted separate ANOVAs (Dunnett's test) to determine the difference in the aboveground biomass (AGB), microbial diversity and relative abundance of phyla between N0 and different levels of N addition (SPSS 16.0). It was regarded as significant differences when P-value was less than 0.05. Square root transformation of the OTUs data and arcsine square root transformation of relative abundance of phyla were done before proceeding with ANOVA and post-hoc test. The assumptions of normality and homogeneity of variance were checked prior to conducting the statistical tests.
Results
Nitrogen addition reduced and enhanced aboveground biomass (AGB) of forbs and grasses
Nitrogen addition for 9 years had contrasting effects on AGB of grasses and forbs, such that AGB of grasses and forbs was significantly (P < 0.05) increased and reduced by N addition, respectively (Figure 1A). The steppe community was co-dominated by grass S. krylovii and forb A. frigida in the control plot without N addition. The AGB of S. krylovii was enhanced by the N addition, while the same treatment led to a decrease in AGB of A. frigida (P < 0.05) (Figure 1B).
Figure 1.
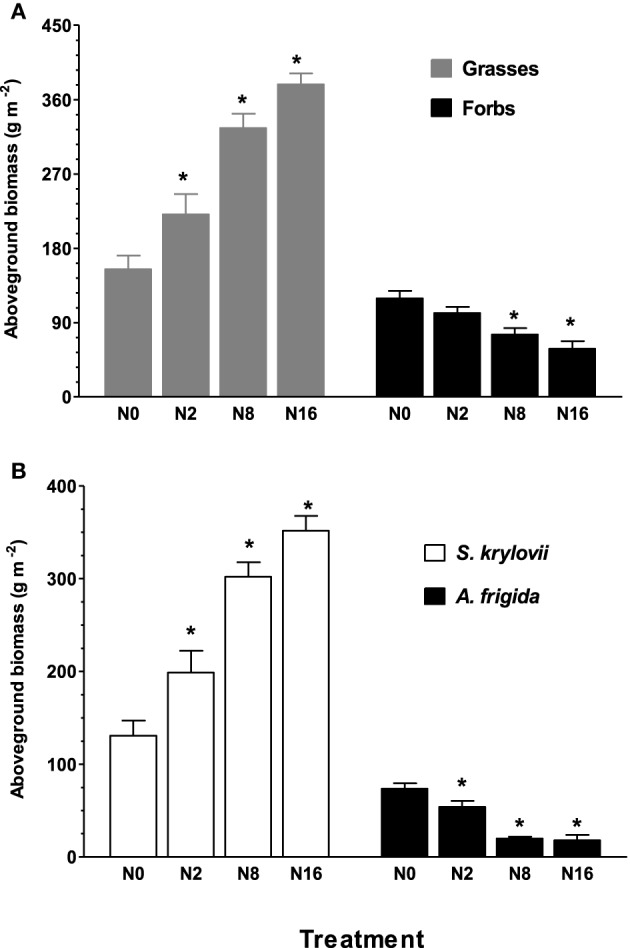
Effect of N addition on aboveground biomass of grasses and forbs (A), and S. krylovii and A. frigida (B). Aboveground biomass was determined in quadrats (1 × 1 m). N0, N2, N8, and N16 represent N addition rate of 0, 2, 8, 16 g ha−1 yr−1. Data are means ± s.e. (n = 8). Asterisks on the top of columns indicate significant difference at P < 0.05 between N0 and different rates of N addition for each species.
Sequencing and analysis of rhizospheric microbial diversity
We obtained a total of 1,931,731 clean reads after filtered by QIIME quality filters with default settings (Table A1). One sample from N0 plots was later excluded from analysis because of the low quality of the sequence reads. Rarefaction analysis was performed on each soil sample and none of the rarefaction curves reached the plateau phase, suggesting that soils were not sampled to saturation (Figure 2).
Figure 2.
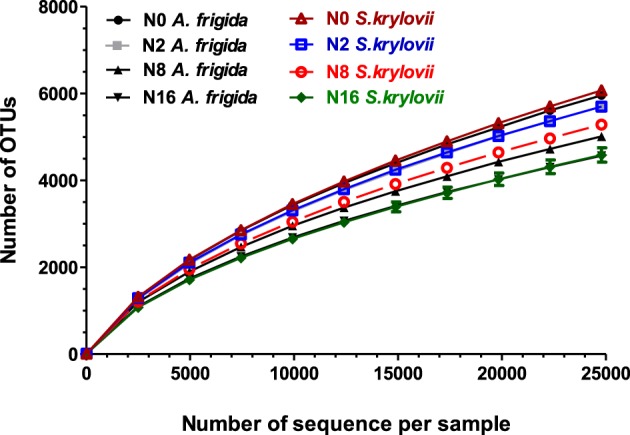
Rarefaction curves of all samples were generated for microbial OTUs which contained unique sequences and were defined at 97% sequence similarities.
Linear mixed model analysis indicated that OTU number, Chao1, and Shannon indices of rhizosphere microbes of both S. krylovii and A. frigida were significantly affected by the N addition (Figure 3 and Table 1). In control plot without N addition, the number of OTUs in the rhizosphere of S. krylovii and A. frigida was 6036 and 5957, respectively (Figure 3A). Nitrogen addition led to similar effects on microbial diversity in the rhizosphere of the two species. As N addition rate increased, the OTU number in both the S. krylovii and A. frigida rhizospheric niche decreased (Figure 3A). For example, the OTU number in the S. krylovii rhizosphere was decreased from 6036 to 5698, 5282, and 4583 in response to N addition rate of 2, 8, and 16 g N m−2 yr−1. Similarly, the OTU number in the A. frigida rhizosphere niche decreased from 5957 to 5700, 5010, and 4567 in response to the same N addition rates (Figure 3A). In addition to the number of OTUs, changes in microbial diversity using the Shannon index and the total species richness estimated by the Chao1 index in the two rhizospheric soil samples were also compared among plots treated with different rates of N addition. Shannon and Chao1 indices in the two rhizospheric soils also showed similar trends with increases in N addition rates, such that N addition reduced Shannon and Chao1 indices in the rhizosphere of S. krylovii and A. frigida (Figures 3B,C).
Figure 3.
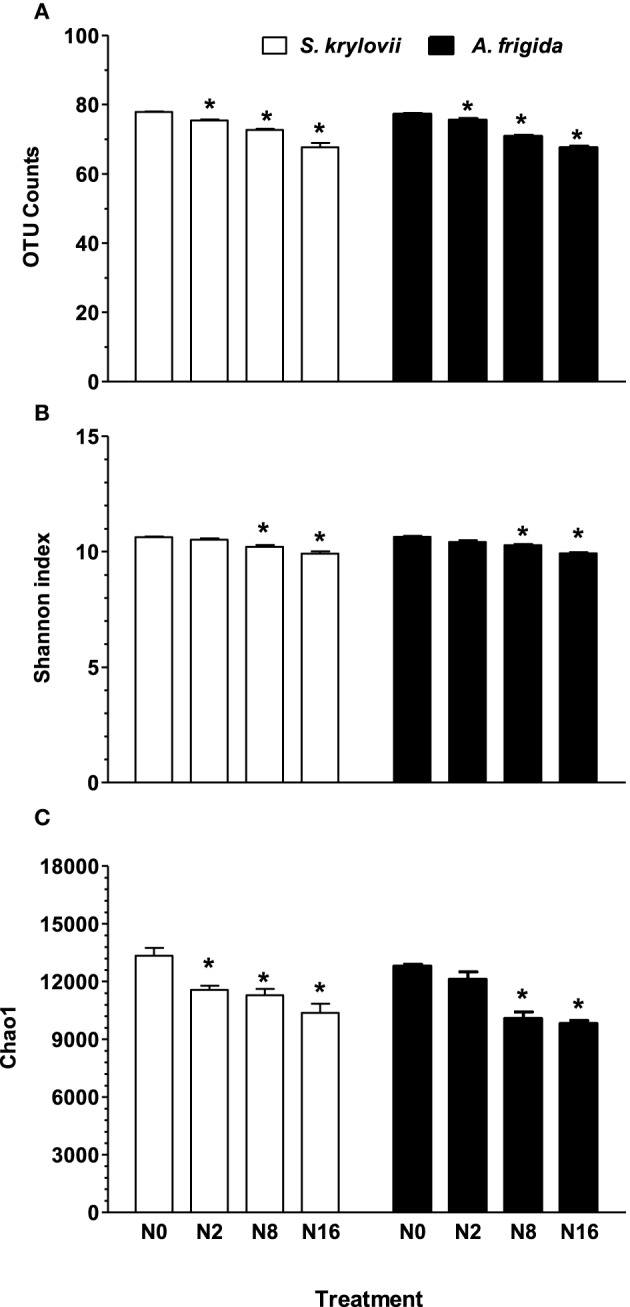
Estimated number of observed (A) OTU counts, (B) Shannon index, (C) Chao1 index of S. krylovii and A. frigida rhizosphere microbiome across all the N-supplied plots. Data are means ± s.e. (n = 3). Asterisks on the top of columns indicate significant difference at P < 0.05 between N0 and different rates of N addition for each species.
Table 1.
F-value and P-value of analysis of variance for the effects of nitrogen addition, species and their interaction (nitrogen × species) on OTU, Shannon, Chao1 indices and the relative abundance of the phyla.
| Fixed factors | Nitrogen | Species | Nitrogen × Species | |
|---|---|---|---|---|
| d.f. | 3 | 1 | 3 | |
| OTU | F | 252.74 | 7.99 | 3.18 |
| P | < 0.0001 | 0.0143 | 0.06 | |
| Shannon | F | 75.03 | 0 | 1.22 |
| P | < 0.0001 | 0.9624 | 0.3424 | |
| Chao1 | F | 49.09 | 5.20 | 4.41 |
| P | < 0.0001 | 0.0400 | 0.0239 | |
| Acidobacteria | F | 146.93 | 13.67 | 11.94 |
| P | < 0.0001 | 0.0027 | 0.0005 | |
| Proteobacteria | F | 19.53 | 9.94 | 1.04 |
| P | < 0.0001 | 0.0076 | 0.4057 | |
| Bacteroidetes | F | 30.34 | 0.09 | 9.87 |
| P | < 0.0001 | 0.7733 | 0.0012 | |
| Crenarchaeota | F | 72.80 | 0.01 | 11.96 |
| P | < 0.0001 | 0.9247 | 0.0005 | |
| Verrucomicrobia | F | 4.03 | 10.84 | 0.37 |
| P | 0.0314 | 0.0058 | 0.7754 | |
| Planctomycetes | F | 6.24 | 1.42 | 3.27 |
| P | 0.0074 | 0.2555 | 0.0559 | |
| Actinobacteria | F | 4.71 | 5.65 | 4.62 |
| P | 0.0194 | 0.0335 | 0.0207 | |
| Cyanobacteria | F | 0.62 | 0 | 0.41 |
| P | 0.6168 | 0.9601 | 0.7504 | |
| Gemmatimonadetes | F | 12.42 | 7.81 | 5.34 |
| P | 0.0004 | 0.0152 | 0.0129 | |
| Chloroflexi | F | 16.22 | 15.54 | 6.55 |
| P | 0.0001 | 0.0017 | 0.0062 | |
| Firmicutes | F | 39.78 | 21.00 | 0.28 |
| P | < 0.0001 | 0.0005 | 0.8384 | |
| WYO | F | 1.75 | 24.18 | 11.48 |
| P | 0.2057 | 0.0003 | 0.0006 | |
| TM7 | F | 195.38 | 5.60 | 7.16 |
| P | < 0.0001 | 0.0341 | 0.0044 |
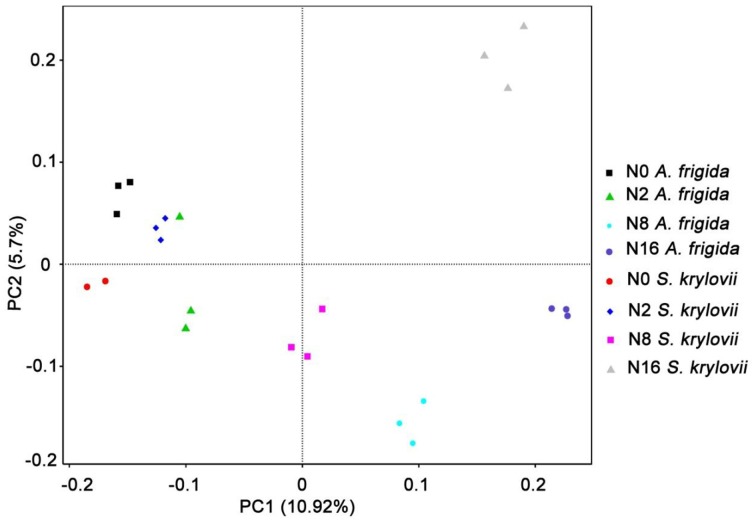
Principal component analysis was used to detect variation in the community composition. As shown in Figure 4, the two principal components accounted for 16.62% of the total microbial community variations among the individual samples. The two-dimensional figure showed that the microbial community compositions in both S. krylovii and A. frigida rhizosphere niche with different N addition rates were distributed separately among each other, exhibiting differences in the microbial community structure. These results suggest that the S. krylovii and A. frigida rhizosphere microbiome had different composition and that the composition changed along the N-gradient.
Figure 4.
Principle component analysis (PCA) of microbial communities based on OTUs for all samples from S. krylovii and A. frigida rhizosphere. The first two components were 10.68% and 5.46%, respectively.
Effect of N addition on relative abundance of rhizospheric microbe
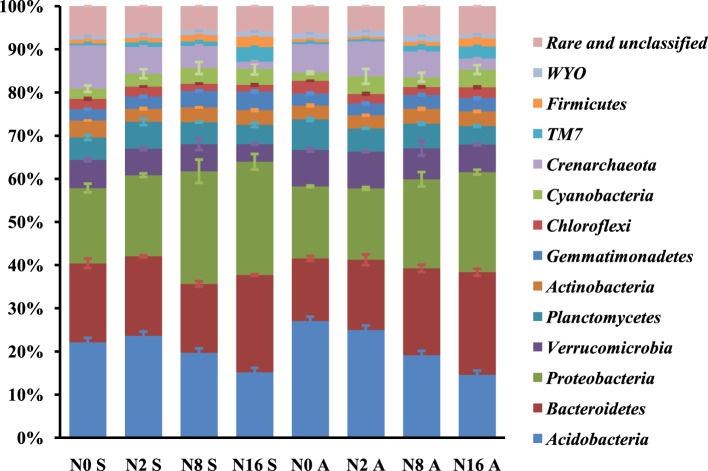
Analysis of the taxonomic groups detected in the soil samples showed that there were a total of 23 phyla in the rhizosphere of both S. krylovii and A. frigida. The most dominant phyla across all samples were Proteobacteria, Acidobacteria, and Bacteroidetes, accounting for about 60% of the bacterial sequences (Figure 5). In addition, Verrucomicrobia, Crenarchaeota, Planctomycetes, Actinobacteria, Cyanobateria, Gemmatimonadetes, Chloroflexi, TM7, Firmicutes, and WYO were detected in all the samples with low abundance, while the unclassified and rare phyla accounted for about 6.3% in the samples. In the control plots without N addition, the relative abundance of Bacteroidetes and Crenarchaeota in the rhizosphere of S. krylovii was higher than that in the rhizosphere of A. frigida (P < 0.05). In contrast, the relative abundance of Acidobacteria, Verrucomicrobia, Chloroflexi, and WYO in the rhizosphere of S. krylovii was lower than that in the rhizosphere of A. frigida (P < 0.05) (Figure 5). These results suggest that difference existed between the microbial communities in the rhizosphere of both S. krylovii and A. frigida in the control plot.
Figure 5.
The relative abundance of phyla of S. krylovii and A. frigida rhizosphere microbiome across all the N-supplied plots (% of total sequence).
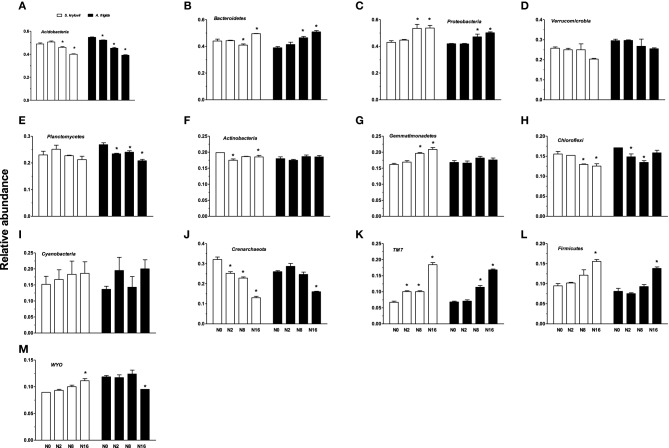
Nitrogen addition had significant impacts on the relative abundance of the phyla (Figures 5, 6). With the increases in N addition rates, the relative abundance of Bacteroidetes, Proteobacteria, Gemmatimonadetes, TM7, Firmicutes, and WYO was enhanced in the S. krylovii rhizosphere (Figures 6B,C,G,K–M). A decrease in the relative abundance of Acidobacteria, Actinobacteria, Chloroflexi, and Crenarchaeota in the S. krylovii rhizosphere niche was detected (Figures 6A,F,H,J), while the relative abundance of Verrucomicrobia, Planctomycetes, and Cyanobacteria in the S. krylovii rhizosphere niche remained relatively unchanged (Figures 6D,E,I). With the increases in N addition rates, the relative abundance of Bacteroidetes, Proteobacteria, TM7, and Firmicutes in the A. frigida rhizosphere niche was increased (Figures 6B,C,K,L). Nitrogen addition had little effects on the relative abundance of Verrucomicrobia, Actinobacteria, Gemmatimonadetes, and Cyanobacteria in the A. frigida rhizosphere niche (Figures 6D,F,G,I). By contrast, the relative abundance of Acidobacteria, Planctomycetes, Chloroflexi, Crenarchaeota, and WYO in the A. frigida rhizosphere niche was reduced by N addition (Figures 6A,E,H,J,M).
Figure 6.
Effect of N addition on the relative abundance of phyla A, of S. krylovii and A. frigida rhizosphere microbiome across all the N-supplied plots. (A) Acidobacteria, (B) Bacteroidetes, (C) Proteobacteria, (D) Verrucomicrobia, (E) Planctomycetes, (F) Actinobacteria, (G) Gemmatimonadetes, (H) Chloroflexi, (I) Cyanobacteria, (J) Crenarchaeota, (K) TM7, (L) Firmicutes, (M) WYO. Data are means ± s.e. (n = 3). Asterisks on the top of columns indicate significant difference at P < 0.05 between N0 and different rates of N addition for each species.
Discussion
In this study, we found that 9-year N addition shifted plant community structure from co-dominance by a monocot grass, S. krylovii, and a dicot forb, A. frigida, to exclusive dominance by a monocot grass in an Inner Mongolia steppe (Figure 1). These results concurred with other studies which showed that N enrichment favors more for grass growth than for forb growth (Song et al., 2011; Fang et al., 2012; Zhang et al., 2014). The differential growth responses of the two plants to N addition prompted us to test whether N addition may also have different impacts on the microbial communities in the rhizosphere of the two species. We thus characterized microbial communities in the rhizosphere of the two dominant species, S. krylovii and A. frigida, under varying rates of N addition by high-throughput sequencing technique. Our results showed that the relative abundance of most microbial phyla in the rhizosphere of S. krylovii differed from that of A. frigida under control, ambient N conditions, suggesting that the microbiomes in the rhizosphere of the two species differ intrinsically. Bakker et al. (2013) have suggested that root exudates can explain the differences in the rhizospheric microbiomes of various plant species. In this context, our previous studies showed that S. krylovii and A. frigida differed in their rhizospheric processes, such that S. krylovii roots can exude greater amount of organic anions (malate and citrate) than A. frigida roots under ambient N conditions (Liu et al., 2014a). In addition, Acidobacteria, Bacteroidetes, and Proteobacteria were dominant microbial species in the two rhizosphere microbiomes, accounting for about 60% of the total microbiomes, implying that the three phyla may play important roles in the rhizosphere of the two species.
Several studies have reported that elevated N deposition profoundly impacts the soil microbial communities across different terrestrial ecosystems (Ramirez et al., 2010, 2012; He et al., 2013; Zhang et al., 2014). The microbes in the bulk soils have often been used to evaluate the effect of N addition on composition and biodiversity of bacterial community. However, in contrast to our studies, few studies have specifically investigated the effect of N addition on rhizospheric microbial communities. In a recent study, Zhu et al. (2015) reported that short-term N addition has minimal influence on rhizosphere effect of smooth crabgrass and bermudagrass by T-RFLP and analysis of enzyme activities. In the present study, we examined the rhizosphere microbiomes of S. krylovii and A. frigida across different N addition rates. We found that the OTU number, Shannon and Chao1 indices in the rhizospheric soils of the two dominant species showed a similar trend in response to N addition. Nitrogen addition reduced Shannon index in the rhizosphere of S. krylovii and A. frigida, indicating that the rhizospheric microbiomes of the two species became less diverse with increases in N-addition rates. In a similar study, Zhang et al. (2014) reported that long-term N addition had no effects on the overall OTU number of bulk soils in the Inner Mongolia steppe. Given the high heterogeneity of soils and co-existence of multiple plant species in a natural ecosystem, results obtained from monitoring microbes in the bulk soils may not truly reflect the changes in rhizospheric microbial communities. Roots are high in C, so the microbial community closer to the root may be copiotrophic relative to those in bulk soils (Nguyen, 2003; Badri et al., 2009; Gottel et al., 2011). We also found that the relative abundance of most phyla was altered in response to the long-term N addition. The relative abundance of all phyla except Verrucomicrobia, Planctomycetes, and Cyanobacteria in the rhizospheric microbiomes of S. krylovii was altered in response to the N addition. In the rhizospheric microbiomes of A. frigida, we found that the relative abundance of Verrucomicrobia, Actinobacteria, Gemmatimonadetes, and Cyanobacteria remained relatively unchanged by the same N addition. Furthermore, our results showed that the direction of the response of individual phyla to N addition was dependent upon host identity. These findings are in contrast to those previously reported results, where no comparable increases and decreases in response to N addition were observed in the same bacterial phylum of grasslands (Ramirez et al., 2010, 2012; He et al., 2013; Liu et al., 2014b). The differences in soil sampling methods (rhizospheric soils vs. bulk soils) between our studies and others may partly account for the different results. It is more likely that microbes in the rhizosphere simply behave differently from those in bulk soils.
It has been established that interaction can occur between plant communities and soil microbial communities (Zak et al., 2003; Dean et al., 2014). Many studies have attempted to link the above-ground plant diversity and productivity to below-ground bacterial diversities by characterizing soil bacterial communities (Zak et al., 2003; Dean et al., 2014). Rooney et al. (2006) showed that in agricultural grasslands the response of plants and bacterial communities to sheep urine deposition is dependent on both the concentration of synthetic sheep urine applied and the grass species. The rhizospheric microbial community associated with plant roots is highly diverse, and it is conceivable that the complex plant-associated microbial community is important for plant health (Kyselková et al., 2009; Berendsen et al., 2012). The rhizospheric microbial community may contribute to maintaining plant health directly by releasing pathogen inhibitors, or indirectly by promoting plant growth (Kyselková et al., 2009; Berendsen et al., 2012). More specifically, the phylum Proteobacteria is involved in cycling of essential mineral nutrients (Lesaulnier et al., 2008; Chaudhry et al., 2012). For example, Chaudhry et al. (2012) showed that the higher abundance of Proteobacteria may contribute to improved soil fertility and plant growth. It has been reported that the phylum Verrucomicrobia acts as inhabitants of paddy soil (Asakawa and Kimura, 2008; Do Thi et al., 2012). However, little is known about the function of Verrucomicrobia in the semi-arid grasslands. Although its specific physiological functions in the soil remain to be clarified, the wide occurrence of this predominant phylum Acidobacteria across all samples may indicate a key role in soil ecosystem functioning. Firmicutes is related to plant health and the high abundance of Firmicutes can threaten plant fitness (Berendsen et al., 2012; Zhang et al., 2014). These results may indicate that these phyla in rhizosphere soil may be associated with the shift of plant community structure from co-dominance by a grass, S. krylovii, and a forb species, A. frigida, to exclusive dominance by the grass.
The arbuscular mycorrhiza (AM) is the mutualistic symbiosis between terrestrial plants and fungi. Fungi may play an important role in driving changes in plant biomass. Goomaral et al. (2013) reported that that S. krylovii increased in biomass as soil N availability increased, and this was associated with increased mycorrhizal colonization. Furthermore, altered fungal communities in response to environmental stressors in other systems have been linked to changing plant communities (Deslippe et al., 2012; Semenova et al., 2015). Therefore, further studies to investigate the involvement of fungi diversity and composition in N-induced shifts in species composition in the temperate steppe are warranted.
In the present study, to simulate the effects of N deposition on temperate grassland ecosystems, long-term N addition experiment was conducted in Inner Mongolia steppes by application of urea. The applied urea is first hydrolyzed to ammonia/ammonium by the enzyme urease in soils, and ammonium is further converted into nitrate by ammonia oxidizing bacteria and ammonia oxidizing archaea, leading to increases in inorganic N in soils (Zhang et al., 2012). Previous studies showed that application of urea in the Inner Mongolia steppes led to significant increases in soil nitrate concentrations (Fang et al., 2012). In addition, a similar reduction in plant species richness by addition of inorganic NH4NO3 in the Inner Mongolia steppes has been observed (Lan and Bai, 2012; Yang et al., 2012). Therefore, our N addition experiments with urea can simulate the natural N deposition.
In summary, our results demonstrated that long-term N addition had a profound influence on productivity of the steppe and composition of the rhizospheric microbial community of S. krylovii and A. frigida. These results highlight the importance of rhizospheric microbial communities in control and/or feedback of N addition-invoked steppe communities in response to N deposition. Our findings would contribute to our mechanistic understanding of impacts of N deposition on grassland ecosystem by linking changes in plant traits to their rhizospheric microbes-mediated processes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank the staff at the Duolun Restoration Ecology Research Station, Institute of Botany, Chinese Academy of Sciences, for their help in maintaining the field facilities and collecting the aboveground biomass data. This work was supported by the National Natural Science Foundation of China (41273090, 31470466 and 31470006).
Appendix
Table A1.
The number of reads before and after quality filtering.
| Sample | Before quality filtering | After quality filtering | |
|---|---|---|---|
| N0 S. krylovii | 1 | 151330 | 147221 |
| 2 | 62531 | 60743 | |
| N2 S. krylovii | 1 | 37876 | 36842 |
| 2 | 88768 | 86471 | |
| 3 | 71252 | 69278 | |
| N8 S. krylovii | 1 | 68660 | 66952 |
| 2 | 51083 | 49787 | |
| 3 | 122252 | 118084 | |
| N16 S. krylovii | 1 | 100960 | 97848 |
| 2 | 118645 | 115232 | |
| 3 | 54686 | 53313 | |
| N0 A. frigida | 1 | 83187 | 80841 |
| 2 | 159845 | 155362 | |
| 3 | 80904 | 78636 | |
| N2 A. frigida | 1 | 102194 | 99208 |
| 2 | 77534 | 75623 | |
| 3 | 80417 | 78241 | |
| N8 A. frigida | 1 | 72041 | 70274 |
| 2 | 72393 | 70461 | |
| 3 | 72187 | 70604 | |
| N16 A. frigida | 1 | 91416 | 89042 |
| 2 | 81153 | 79019 | |
| 3 | 84889 | 82649 | |
References
- Asakawa S., Kimura M. (2008). Comparison of bacterial community structures at main habitats in paddy field ecosystem based on DGGE analysis. Soil Biol. Biochem. 40, 1322–1329. 10.1016/j.soilbio.2007.09.024 [DOI] [Google Scholar]
- Badri D. V., Weir T. L., van der Lelie D., Vivanco J. M. (2009). Rhizosphere chemical dialogues: plant-microbe interactions. Curr. Opin. Biotechnol. 20, 642–650. 10.1016/j.copbio.2009.09.014 [DOI] [PubMed] [Google Scholar]
- Bai Y. F., Wu J. G., Clark C. M., Naeem S., Pan Q. M., Huang J. H., et al. (2010). Tradeoffs and thresholds in the effects of nitrogen addition on biodiversity and ecosystem functioning: evidence from inner Mongolia Grasslands. Glob. Chang. Biol. 16, 358–372. 10.1111/j.1365-2486.2009.01950.x [DOI] [Google Scholar]
- Bakker P. A., Berendsen R. L., Doornbos R. F., Wintermans P. C., Pieterse C. M. (2013). The rhizosphere revisited: root microbiomics. Front. Plant Sci. 4:165. 10.3389/fpls.2013.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen R. L., Pieterse C. M., Bakker P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Berg G., Smalla K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. 10.1111/j.1574-6941.2009.00654.x [DOI] [PubMed] [Google Scholar]
- Bobbink R., Hicks K., Galloway J., Spranger T., Alkemade R., Ashmore M., et al. (2010). Global assessment of nitrogen deposition effects on terrestrial plant diversity: a synthesis. Ecol. Appl. 20, 30–59. 10.1890/08-1140.1 [DOI] [PubMed] [Google Scholar]
- Bodirsky B. L., Popp A., Lotze-Campen H., Dietrich J. P., Rolinski S., Weindl I., et al. (2014). Reactive nitrogen requirements to feed the world in 2050 and potential to mitigate nitrogen pollution. Nat. Commun. 5, 3858. 10.1038/ncomms4858 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry V., Rehman A., Mishra A., Chauhan P. S., Nautiyal C. S. (2012). Changes in bacterial community structure of agricultural Land due to long-term organic and chemical amendments. Microb. Ecol. 64, 450–460. 10.1007/s00248-012-0025-y [DOI] [PubMed] [Google Scholar]
- Chen D. M., Mi J., Chu P. F., Zhang L. X., Pan Q. M., Xie Y. C., et al. (2014). Patterns and drivers of soil microbial communities along a precipitation gradient on the Mongolian Plateau. Landsc. Ecol. 1–14. 10.1007/s10980-014-9996-z [DOI] [Google Scholar]
- Clark C. M., Tilman D. (2008). Loss of plant species after chronic low-level nitrogen deposition to prairie grasslands. Nature 451, 712–715. 10.1038/nature06503 [DOI] [PubMed] [Google Scholar]
- Dean S. L., Farrer E. C., Taylor D. L., Porras−Alfaro A., Suding K. N., Sinsabaugh R. L. (2014). Nitrogen deposition alters plant–fungal relationships: linking belowground dynamics to aboveground vegetation change. Mol. Ecol. 23, 1364–1378. 10.1111/mec.12541 [DOI] [PubMed] [Google Scholar]
- Deslippe J. R., Hartmann M., Simard S. W., Mohn W. W. (2012). Long-term warming alters the composition of Arctic soil microbial communities. FEMS Microbiol. Ecol. 82, 303–315. 10.1111/j.1574-6941.2012.01350.x [DOI] [PubMed] [Google Scholar]
- Do Thi X., Vo Thi G., Rosling A., Alstrom S., Chai B., Hogberg N. (2012). Different crop rotation systems as drivers of change in soil bacterial community structure and yield of rice, Oryza sativa. Biol. Fertil. Soils 48, 217–225. 10.1007/s00374-011-0618-5 [DOI] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Fang Y., Xun F., Bai W., Zhang W., Li L. (2012). Long-term nitrogen addition leads to loss of species richness due to litter accumulation and soil acidification in a temperate steppe. PLoS ONE 7:e47369. 10.1371/journal.pone.0047369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway J. N., Dentener F. J., Capone D. G., Boyer E. W., Howarth R. W., Seitzinger S. P., et al. (2004). Nitrogen cycles: past, present, and future. Biogeochemistry 70, 153–226. 10.1007/s10533-004-0370-0 [DOI] [Google Scholar]
- Galloway J. N., Townsend A. R., Erisman J. W., Bekunda M., Cai Z. C., Freney J. R., et al. (2008). Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320, 889–892. 10.1126/science.1136674 [DOI] [PubMed] [Google Scholar]
- Gilliam F. S. (2006). Response of the herbaceous layer of forest ecosystems to excess nitrogen deposition. J. Ecol. 94, 1176–1191. 10.1111/j.1365-2745.2006.01155.x [DOI] [Google Scholar]
- Goomaral A., Iwase K., Undarmaa J., Matsumoto T., Yamato M. (2013). Communities of arbuscular mycorrhizal fungi in Stipa krylovii (Poaceae) in the Mongolian steppe. Mycoscience 54, 122–129. 10.1016/j.myc.2012.09.006 [DOI] [Google Scholar]
- Gottel N. R., Castro H. F., Kerley M., Yang Z., Pelletier D. A., Podar M., et al. (2011). Distinct microbial communities within the endosphere and rhizosphere of populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 77, 5934–5944. 10.1128/AEM.05255-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpole W. S., Tilman D. (2007). Grassland species loss resulting from reduced niche dimension. Nature 446, 791–793. 10.1038/nature05684 [DOI] [PubMed] [Google Scholar]
- He Y. T., Qi Y. C., Dong Y. S., Xiao S. S., Peng Q., Liu X. C., et al. (2013). Effects of nitrogen fertilization on soil microbial biomass and community functional diversity in temperate grassland in inner Mongolia, China. Clean Soil Air Water 41, 1216–1221. 10.1002/clen.201200021 [DOI] [Google Scholar]
- Kyselková M., Kopecký J., Frapolli M., Défago G., Ságová-Mareckova M., Grundmann G. L., et al. (2009). Comparison of rhizobacterial community composition in soil suppressive or conducive to tobacco black root rot disease. ISME J. 3, 1127–1138. 10.1038/ismej.2009.61 [DOI] [PubMed] [Google Scholar]
- Lan Z., Bai Y. (2012). Testing mechanisms of N-enrichment-induced species loss in a semiarid Inner Mongolia grassland: critical thresholds and implications for long-term ecosystem responses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 3125–3134. 10.1098/rstb.2011.0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesaulnier C., Papamichail D., McCorkle S., Ollivier B., Skiena S., Taghavi S., et al. (2008). Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ. Microbiol. 10, 926–941. 10.1111/j.1462-2920.2007.01512.x [DOI] [PubMed] [Google Scholar]
- Liu N. N., Tian Q. Y., Zhang W. H. (2014a). Comparison of adaptive strategies to phosphorus-deficient soil between dominant species Artemisia frigida and Stipa krylovii in typical steppe of Nei Mongol. Chin. J. Plant. Ecol. 38, 905–915. 10.3724/SP.J.1258.2014.00085 [DOI] [Google Scholar]
- Liu W. X., Jiang L., Hu S. J., Li L. H., Liu L. L., Wan S. Q. (2014b). Decoupling of soil microbes and plants with increasing anthropogenic nitrogen inputs in a temperate steppe. Soil Biol. Biochem. 72, 116–122. 10.1016/j.soilbio.2014.01.022 [DOI] [Google Scholar]
- Liu X. J., Zhang Y., Han W. X., Tang A. H., Shen J. L., Cui Z. L., et al. (2013). Enhanced nitrogen deposition over China. Nature 494, 459–462. 10.1038/nature11917 [DOI] [PubMed] [Google Scholar]
- Magoc T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli L. A., Howarth R. W., Cuevas E., Filoso S., Austin A. T., Donoso L., et al. (2006). Sources of reactive nitrogen affecting ecosystems in Latin America and the Caribbean: current trends and future perspectives. Biogeochemistry 79, 3–24. 10.1007/s10533-006-9000-3 [DOI] [Google Scholar]
- Nguyen C. (2003). Rhizodeposition of organic C by plants: mechanisms and controls. Agronomie 23, 375–396. 10.1051/agro:2003011 [DOI] [Google Scholar]
- Niu S., Wu M., Han Y., Xia J., Li L., Wan S. (2008). Water-mediated responses of ecosystem carbon fluxes to climatic change in a temperate steppe. New Phytol. 177, 209–219. 10.1111/j.1469-8137.2007.02237.x [DOI] [PubMed] [Google Scholar]
- Peiffer J. A., Spor A., Koren O., Jin Z., Tringe S. G., Dangl J. L., et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A. 110, 6548–6553. 10.1073/pnas.1302837110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez K. S., Craine J. M., Fierer N. (2012). Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Chang. Biol. 18, 1918–1927. 10.1111/j.1365-2486.2012.02639.x [DOI] [Google Scholar]
- Ramirez K. S., Lauber C. L., Knight R., Bradford M. A., Fierer N. (2010). Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology 91, 3463–3470. 10.1890/10-0426.1 [DOI] [PubMed] [Google Scholar]
- Reynolds H. L., Packer A., Bever J. D., Clay K. (2003). Grassroots ecology: plant-microbe-soil interactions as drivers of plant community structure and dynamics. Ecology 84, 2281–2291. 10.1890/02-0298 [DOI] [Google Scholar]
- Rooney D., Kennedy N., Deering L., Gleeson D., Clipson N. (2006). Effect of sheep urine deposition on the bacterial community structure in an acidic upland grassland soil. Appl. Environ. Microbiol. 72, 7231–7237. 10.1128/AEM.00926-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala O. E., Chapin F. S., Armesto J. J., Berlow E., Bloomfield J., Dirzo R., et al. (2000). Biodiversity-Global biodiversity scenarios for the year 2100. Science 287, 1770–1774. 10.1126/science.287.5459.1770 [DOI] [PubMed] [Google Scholar]
- Scheible W. R., Morcuende R., Czechowski T., Fritz C., Osuna D., Palacios-Rajas N., et al. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136, 2483–2499. 10.1104/pp.104.047019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova T. A., Morgado L. N., Welker J. M., Walker M. D., Smets E., Geml J. (2015). Long-term experimental warming alters community composition of ascomycetes in Alaskan moist and dry arctic tundra. Mol. Ecol. 24, 424–437. 10.1111/mec.13045 [DOI] [PubMed] [Google Scholar]
- Smalla K., Wieland G., Buchner A., Zock A., Parzy J., Kaiser S., et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67, 4742–4751. 10.1128/AEM.67.10.4742-4751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Bao X., Liu X. J., Zhang Y., Christie P., Fangmeier A., et al. (2011). Nitrogen enrichment enhances the dominance of grasses over forbs in a temperate steppe ecosystem. Biogeosciences 8, 2341–2350. 10.5194/bg-8-2341-2011 [DOI] [Google Scholar]
- Stevens C. J., Dise N. B., Mountford J. O., Gowing D. J. (2004). Impact of nitrogen deposition on the species richness of grasslands. Science 303, 1876–1879. 10.1126/science.1094678 [DOI] [PubMed] [Google Scholar]
- Suding K. N., Collins S. L., Gough L., Clark C., Cleland E. E., Gross K. L., et al. (2005). Functional- and abundance-based mechanisms explain diversity loss due to N fertilization. Proc. Natl. Acad. Sci. U.S.A. 102, 4387–4392. 10.1073/pnas.0408648102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q. Y., Liu N. N., Bai W. M., Li L. H., Zhang W. H. (2015). Disruption of metal ion homeostasis insoils is associated with nitrogendeposition-induced species loss in anInner Mongolia steppe. Biogeosciences 12, 3499–3512. 10.5194/bg-12-3499-2015 [DOI] [Google Scholar]
- Vitousek P. M., Hättenschwiler S., Olander L., Allison S. (2002). Nitrogen and nature. Ambio 31, 97–101. 10.1579/0044-7447-31.2.97 [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Jiang L., Li L., Li A., Wu M., Wan S. (2012). Diversity-dependent stability under mowing and nutrient addition: evidence from a 7-year grassland experiment. Ecol. Lett. 15, 619–626. 10.1111/j.1461-0248.2012.01778.x [DOI] [PubMed] [Google Scholar]
- Yang H. J., Wu M. Y., Liu W. X., Zhang Z., Zhang N. L., Wan S. Q. (2011). Community structure and composition in response to climate change in a temperate steppe. Glob. Chang. Biol. 17, 452–465. 10.1111/j.1365-2486.2010.02253.x [DOI] [Google Scholar]
- Zak D. R., Holmes W. E., White D. C., Peacock A. D., Tilman D. (2003). Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecology 84, 2042–2050. 10.1890/02-0433 [DOI] [Google Scholar]
- Zhang X., Wang Q., Gilliam F. S., Bai W., Han X., Li L. (2012). Effect of nitrogen fertilization on net nitrogen mineralization in a grassland soil, northern China. Grass Forage Sci. 67, 219–230. 10.1111/j.1365-2494.2011.00836.x [DOI] [Google Scholar]
- Zhang X. M., Wei H. W., Chen Q. S., Han X. G. (2014). The counteractive effects of nitrogen addition and watering on soil bacterial communities in a steppe ecosystem. Soil Biol. Biochem. 72, 26–34. 10.1016/j.soilbio.2014.01.034 [DOI] [Google Scholar]
- Zhang Y., Zheng L., Liu X. J., Jickells T., Cape J. N., Goulding K., et al. (2008). Evidence for organic N deposition and its anthroponic sources in China. Atmos. Environ. 42, 1035–1041. 10.1016/j.atmosenv.2007.12.015 [DOI] [Google Scholar]
- Zhu B., Panke-Buisse K., Kao-Kniffin J. (2015). Nitrogen fertilization has minimal influence on rhizosphere effects of smooth crabgrass (Digitaria ischaemum) and bermudagrass (Cynodon dactylon). J. Plant Ecol. 8, 390–400. 10.1093/jpe/rtu034 [DOI] [Google Scholar]