Abstract
In the mammalian olfactory epithelium (OE), olfactory receptor neurons (ORNs) are continuously regenerated throughout the animal's lifetime. Horizontal basal cells (HBCs) in the OE express the epithelial marker keratin 5 (K5) and the stem cell marker Pax6 and are considered relatively quiescent tissue stem cells in the OE. Pax6 is a key regulator of several developmental processes in the central nervous system and in sensory organs. Although Pax6 is expressed in the OE, its precise role remains unknown, particularly with respect to stem cell-like HBCs. To investigate the function of Pax6 in the developmental and regenerative processes in the OE, we generated conditional Pax6-knockout mice carrying a loxP-floxed Pax6 gene. Homozygous Pax6-floxed mice were crossed with K5-Cre transgenic mice to generate HBC-specific Pax6-knockout (Pax6-cKO) mice. We confirmed that the deletion of Pax6 expression in HBCs was sufficiently achieved in zone 1 of the OE in Pax6-cKO mice 3 days after methimazole-induced severe damage. In this condition, regeneration of the OE was dramatically impaired; both OE thickness and the number of ORNs were significantly decreased in the regenerated OE of Pax6-cKO mice. These results suggest that Pax6 expression is essential for HBCs to differentiate into neuronal cells during the regeneration process following severe injury.
Introduction
Olfaction (ie, the ability to sense odorants) is one of the most evolutionarily conserved senses among the animal kingdom. The vertebrate olfactory system comprises the olfactory epithelium (OE), the olfactory bulb, and the olfactory cortex [1]. In the mammalian OE, olfactory receptor neurons (ORNs) are continuously regenerated by physiological turnover and are replaced following damage to the OE [2,3]; these processes continue throughout the animal's lifetime. Therefore, the OE is an attractive model for studying adult neurogenesis [4]. In the OE, two subpopulations of basal cells have been proposed to function as tissue stem cells; globose basal cells (GBCs) are the major proliferating cell population within the adult OE, and horizontal basal cells (HBCs) are relatively quiescent cells that lie just above the basal lamina. HBCs express the epithelial markers keratin 5 (K5) and CD54 (also known as ICAM-1), and HBCs express the stem cell markers Pax6 and Sox2 [5–8]. Although the evidence collected to date is far from conclusive, it has been suggested that HBCs are the tissue stem cells in the OE because HBCs can self-renew and generate neuronal and non-neuronal cells, including the GBCs, in vivo (both during normal turnover and following OE injury) as well as in vitro [4,9–11]. HBCs express the transcription factor p63, which is essential for the proliferation of epithelial stem cells that give rise to the thymus and epidermis [12]. Recently, two interesting studies clarified the role of p63 in the regulation of HBC's cellular dynamics [13,14]; however, despite these and other studies, the detailed mechanisms that underlie HBC differentiation and proliferation remain unclear.
The transcription factor Pax6 is highly conserved among vertebrates and is essential for a wide variety of developmental processes within the central nervous system (CNS), sensory organs, and the endocrine system [15–19]. Homozygous Pax6 mutant mice (Pax6Sey/Sey) and rats (Pax6rSey/rSey) lack eyes and nasal structures, and these animals die immediately after birth with the same pathology as the laboratory-generated Pax6-knockout mouse (Pax6−/−) [20–23]. In the adult OE, Pax6 is expressed in several cell types, including HBCs, GBCs, sustentacular cells (SUSs), and Bowman's gland cells (BGs) [5,6]. Thus, understanding the role of Pax6 in tissue stem cell-like HBCs is clearly important, even though the OE of heterozygous Pax6Sey/+ mice is normal in appearance [24].
In this study, we investigated the role of Pax6 in HBCs during normal development and following severe OE damage in adult animals. We generated Pax6-floxed (Pax6flox) mice by homologous recombination in embryonic stem (ES) cells from the C57BL/6 background. Homozygous mice (Pax6flox/flox) were obtained by crossing male and female Pax6flox/+ mice, and these homozygotes were crossed with Keratin5-Cre transgenic mice (K5-Cre+/−) [25] to generate HBC-specific Pax6-knockout mice (K5-Cre+/−::Pax6flox/flox; these mice are referred to here as “Pax6-cKO” mice). We could not evaluate the role of Pax6 in HBCs during the normal OE development because Pax6 deletion in HBCs might not perfectly be induced (see below). However, we successfully deleted Pax6 expression in HBCs in zone 1 of the OE in Pax6-cKO mice 3 days after methimazole-induced severe damage. In this condition, we observed severe impairment in regeneration of the OE with the significantly reduced number of ORNs. These results suggest that Pax6 plays a necessary role in HBCs for their proper differentiation into olfactory cells, particularly into neuronal cells during the OE regeneration process. Thus, these findings reveal a novel essential role for Pax6 in tissue stem cells.
Materials and Methods
Animals
Pax6flox mice were generated by homologous recombination using the ES cell line RENKA, which was established from the C57BL/6N mouse strain [26]. The targeting vector was constructed as follows (Fig. 1A). First, three fragments (the 5′ arm, the floxed-out region, and the 3′ arm) were subcloned using polymerase chain reaction (PCR) from the genomic DNA of the C57BL/6 mouse. A 0.85-kb region containing exon 5 that contains a part of the DNA-binding paired domain was used for the floxed-out region. The two homologous genomic DNA fragments (the 5′ and 3′ arms) were 3.3 and 7.3 kb in size, respectively. These three PCR products were inserted into a vector containing the neomycin resistance (neo) cassette flanked by two Flp recognition target (frt) sites and two loxP sites. The floxed-out exon 5 fragment was inserted between the second frt site and the second loxP site.
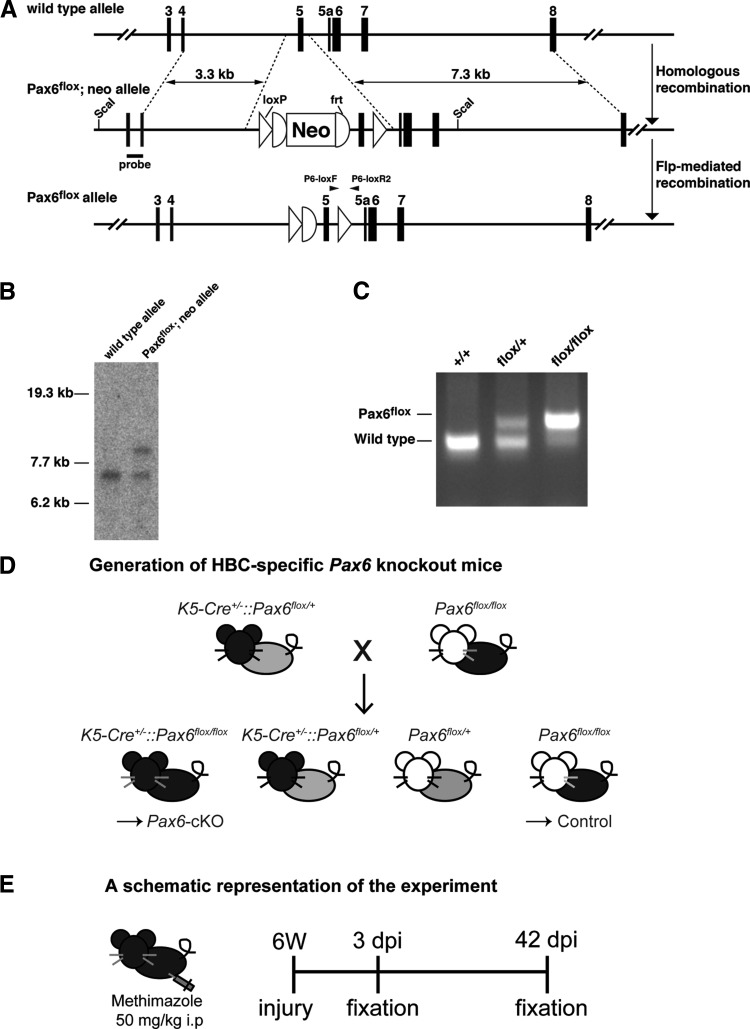
FIG. 1.
Generation of Pax6flox mice and HBC-specific Pax6-knockout mice. (A) Schematic of the Pax6 allele (wild type), the floxed neo-containing allele (Pax6flox/neo), and the floxed (neo-excised) allele (Pax6flox). The Pax6flox; neo allele contains two loxP sites flanking exon 5 in Pax6, and the neo gene is flanked by two frt sequences. The neo gene was removed by crossing Pax6flox; neo mice with FLP66 mice, which express Flp recombinase. The location and orientation of the P6-loxF and P6-loxR2 PCR primers are indicated as closed triangles; the location of the 5′ probe used for Southern blot analysis is also indicated. (B) Southern blot analysis of genomic DNA from wild-type and Pax6flox; neo ES clones. Genomic DNA was digested with ScaI and probed with the 5′ probe, as shown in (A). (C) PCR analysis of the genomic DNA isolated from the progeny, using the P6-loxF and P6-loxR2 primers. The wild-type and mutant alleles produce 269- and 392-bp amplification products, respectively. (D) Breeding scheme for the generation of HBC-specific Pax6-knockout mice. K5-Cre+/−::Pax6flox/+ mice were crossed with Pax6flox/flox mice to obtain K5-Cre+/−::Pax6flox/flox (Pax6-cKO) offspring. Littermates of the Pax6flox/flox mice were used as control mice. (E) Schematic representation of the experimental paradigm. Six-week-old (6W) mice were injected with methimazole (50 mg/kg body weight). The mice were then sacrificed and analyzed at 3 or 42 days postinjury (dpi). HBC, horizontal basal cell; PCR, polymerase chain reaction.
Homologous recombination in the ES cells and production of chimeric founder mice were performed as described previously [26]. Recombinant clones were confirmed by Southern blot analysis (Fig. 1B). The resulting chimeric mice were mated to FLP66 transgenic mice on the C57BL/6 strain [27] to remove the neo cassette.
The Pax6flox mutant allele was detected using PCR (Fig. 1C) with the following primers: P6-loxF 5′-TGGTAACAGTGTACAAACTG-3′ and P6-loxR2 5′-CTGACCTTGCCTAAAGTAG-3′. Amplification of the wild-type and mutant alleles generates 269- and 392-bp fragments, respectively.
To delete Pax6 expression selectively in HBCs, we generated HBC-specific conditional Pax6 knockout mice using K5-Cre mice (a generous gift from Dr. Junji Takeda, Osaka University) [25]. Heterozygous K5-Cre+/− mice were mated with Pax6flox/flox mice to obtain heterozygous K5-Cre+/−::Pax6flox/+ mice, which were then crossed with Pax6flox/flox mice to obtain homozygous HBC-specific Pax6-knockout mice (K5-Cre+/−::Pax6flox/flox, referred to hereafter as Pax6-cKO mice). Littermates of the Pax6flox/flox mice were used as control animals (Fig. 1D). Six-week-old mice were used in this study.
All of the experimental procedures used in this study were approved by the Ethics Committee for Animal Experiments of Tohoku University Graduate School of Medicine (No. 2013-201), and all animals were treated in accordance with the National Institutes of Health's guidelines for the care and use of laboratory animals.
Induction of OE lesions
OE lesions were induced as described previously [7]. In brief, methimazole (63760 Fluka; Sigma-Aldrich) was diluted to 5 mg/mL in 0.9% NaCl and injected intraperitoneally into the mice at 50 mg/kg body weight. The mice were then sacrificed 3 or 42 days postinjury (dpi).
Tissue preparation
The tissues were prepared essentially as described previously [7]. Deeply anesthetized mice were transcardially perfused with cold phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA, P6148; Sigma-Aldrich) in PBS. The nose was removed and postfixed in 4% PFA in PBS overnight at 4°C and then decalcified in 10% ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA, 345-01865; Dojindo Laboratories) for 4 days at 4°C. After sequential incubation in 10% and 30% sucrose in PBS (w/v), the tissues were embedded in the optimum cutting temperature compound (Tissue-Tek O.C.T. Compound, Sakura Finetek) and snap-frozen on dry ice. Coronal sections (5 μm in thickness) were cut using a model CM3050 cryostat (Leica Instruments), mounted on MAS-coated glass slides (Superfrost; Matsunami), and stored at −80°C for subsequent analysis.
Histological analysis
The sections were stained with hematoxylin and eosin (H&E) and visualized using a light microscope (BZ-9000; Keyence). Immunohistochemistry was performed as previously described, with slight modifications [7]. The sections were first washed in 0.1% Triton X-100/Tris-buffered saline (TBS) to remove the O.C.T. compound. For staining with the Pax6, Ki-67, Sox2, and p63 antibodies, the sections were boiled in 0.01 M citrate buffer (pH 6.0) for 15 min. The sections were then blocked with 3% bovine serum albumin/0.3% Triton X-100/TBS for 30 min at room temperature. The following primary antibodies were used: rabbit anti-Pax6 (1:1,000 [28]), rabbit anti-Keratin5 (1:500, PRB-160P; Covance), rabbit anti-Ki-67 (1:500, NCL-Ki67p; Novocastra), goat anti-Sox2 (1:500, AF2018; R&D systems), goat anti-Sox10 (1:200, AF2864; R&D systems), goat anti-NQO1 (1:400, ab2346; Abcam), goat anti-OMP (1:1,000, 019-22291; Wako), armenian hamster anti-ICAM-1 (CD54) (1:200; BD Biosciences), and mouse anti-p63 (1:200, sc8431; Santa Cruz Biotechnology). Following an overnight incubation in the primary antibodies at 4°C, the sections were washed and then incubated in the following secondary antibodies (where appropriate) for 1 h at room temperature: Cy3-conjugated donkey anti-rabbit (1:400; Jackson ImmunoResearch), Cy3-conjugated goat anti-armenian hamster (1:400; Jackson ImmunoResearch), FITC-conjugated donkey anti-rabbit (1:400; Jackson ImmunoResearch), Alexa Fluor 488 donkey anti-mouse (1:400; Invitrogen), Biotin-SP donkey anti-rabbit (1:1,000; Jackson ImmunoResearch), and/or Biotin-SP donkey anti-goat (1:1,000; Jackson ImmunoResearch). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1:2,000; Sigma-Aldrich), and the slides were mounted with coverslips using a mounting medium (VECTASHIELD; Vector Labs). For the enzyme-based antibody visualization technique, the signal was visualized using the avidin–biotin complex (VECTASTAIN Elite ABC Kit; Vector Labs) and 3,3′-diaminobenzidine (DAB); the nuclei were counterstained with hematoxylin, and the slides were mounted with the mounting medium (Entellan New; Merck). Light images of the H&E-stained and enzyme antibody-stained sections were obtained using an HS All-in-One fluorescence microscope (BZ-9000; Keyence) with BZ-H1C analysis software (Keyence). Immunofluorescence images were obtained using a laser-scanning confocal microscope (LSM 5-PASCAL; Zeiss) and LSM Image Browser analysis software (Zeiss). The images were compiled using Adobe Photoshop software (Adobe Systems, Inc.).
OE thickness measurement and cell counting
To measure OE thickness, we used ≥3 sections per animal from three to five animals; total basement membrane (BM) length was >2,800 μm. Each OE was divided into two regions (zone 1, the dorsomedial region; zones 2–4, the ventrolateral region) using NQO1 (NAD(P)H dehydrogenase [quinone] 1) staining; NQO1-positive fields were defined as zone 1 [29]. Using BZ-H1C analysis software (Keyence), we calculated the OE area (from the basal lamina to the nasal lumen) of zone 1 or zones 2–4 at the nasal septum, and we measured the total length of the BM. OE thickness (in microns) was calculated by dividing the OE area (in microns2) by total BM length (in microns).
The mean number of HBCs in the area of zone 1 (or zones 2–4) at the nasal septum was estimated by counting the mean number of p63-positive cells/field in 9–14 randomly selected fields at 63×magnification (for normal OE at 6 weeks of age, n=3) or 100×magnification (for regenerating OE at 3 dpi, n=3).
To measure the percentage of Pax6-positive HBCs (ie, p63-positive cells that were also Pax6 positive), the number of cells that were positive for both Pax6 and p63 was divided by the total number of p63-positive cells (at least 80 cells per animal were counted) lining the nasal septum at zone 1 or zones 2–4 (n=3 animals); this value was then multiplied by 100. Because Pax6 expression in HBCs is relatively weak in zone 1 compared to the background signal, we defined any cells that lacked or had considerably low Pax6 expression as Pax6 negative. Images were analyzed using the Keyence BZ-H1C or LSM Image Browser software program to merge channels and measure the percentage of double-positive cells.
To analyze the proliferative ability of HBCs in zone 1 at 3 dpi, the percentage of Ki-67-positive basal cells against the total number of basal cells was calculated in the area just above the BM lining the nasal septum (150 cells per animal, n=2–3 animals). We used the BZ-9000 at 20×magnification and analyzed the images using the BZ-H1C software program.
The mean number of NQO1-positive cells in zone 1 in the nasal septum was calculated by dividing the total number of NQO1-positive cells by total BM length (in mm); a minimum of 3 sections per animal (n=3–5 animals with total BM length >3,600 μm) was used. The mean number of Sox10-positive cells in zone 1 in the nasal septum was calculated by dividing the total number of Sox10-positive cells by total BM length (in mm); a minimum of 3 sections per animal (n=3–5 animals with total BM length>3,600 μm) was used. We obtained the images using the BZ-9000 at 20×magnification with the BZ-H1C software program.
Statistical analysis
All data are presented as the mean±the standard error of the mean. A two-sample Student's t-test or Welch's t-test was used to compare the data from different groups; P<0.05 was considered to be statistically significant.
Results
Generation of HBC-specific Pax6-knockout mice
To examine the specific function of Pax6 in the development and regeneration of the OE, we first created conditional Pax6-knockout mice using the Cre/loxP conditional gene deletion system. We generated Pax6flox mice in which exon 5 is flanked by loxP sites (Fig. 1A–C). Excision of the floxed exon 5 in Cre recombinase-expressing cells creates a frameshift mutation that introduces a premature stop codon at codon 11. The mammalian Pax6 gene also encodes a spliced isoform (Pax6-5a) that encodes an additional 14 amino acids. Thus, the excised exon 5 also generates a premature stop codon at codon 16 in Pax6 mRNA containing exon 5a.
To knockout Pax6 expression specifically in HBCs, we used K5-Cre mice [25]. Expression of the K5 promoter in HBCs begins at the late embryonic stage [7,30]. Using conventional reporter mice (CAG-CAT-EGFP mice) [31], we confirmed the expression and activity of K5-Cre in nearly all HBCs in both zone 1 and zones 2–4 (data not shown).
Normal OE development in HBC-specific Pax6-knockout mice
To analyze the effect of deleting Pax6 expression in HBCs on OE development, we examined the OE in 6-week-old Pax6flox/flox and Pax6-cKO mice. Low-magnification H&E images of coronal nasal sections from Pax6flox/flox and Pax6-cKO mice are shown in Figure 2A and B, respectively. Representative high-magnification H&E images of zone 1 and zones 2–4 are shown in Figure 2A1 and B2. Zone 1 was distinguished from zones 2–4 using the zone 1-specific anti-NQO1 antibody (Fig. 2C–F) [29]. Based on this analysis, we found no obvious anatomical differences in any region of the OE between the two genotypes (Fig. 2A–F).
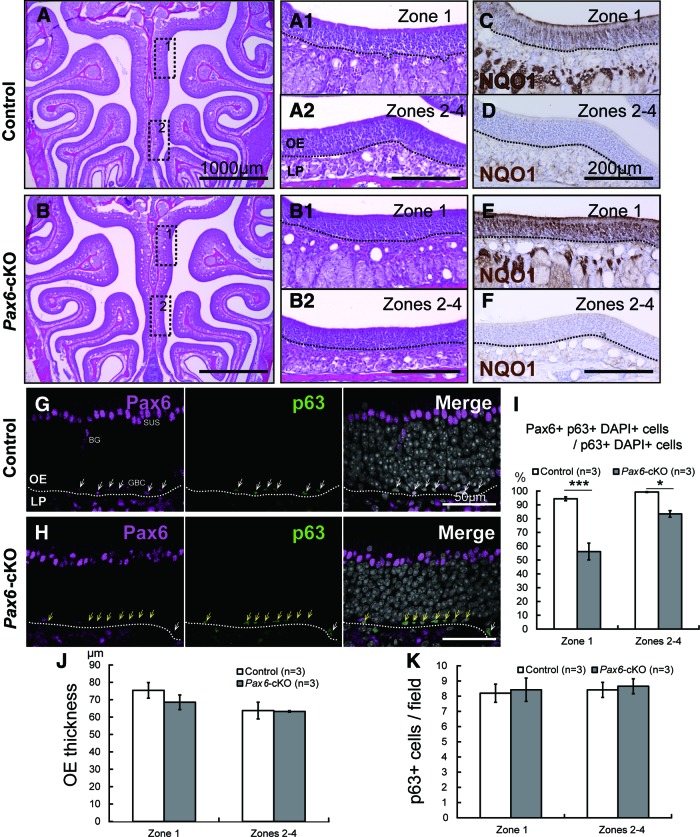
FIG. 2.
The OE of HBC-specific Pax6-knockout mice. (A–F) Images of hematoxylin and eosin-stained coronal sections from the OE of 6-week-old Pax6flox/flox mice (control; A) and K5-Cre+/−::Pax6flox/flox mice (Pax6-cKO; B). (C–F) NQO1 was stained as a marker for zone 1. (G, H) Pax6 and p63 expression in the OE of control and Pax6-cKO mice. In the OE of control mice, Pax6 was observed in HBCs, GBCs, SUSs, and BG cells (G). The majority of HBCs coexpressed p63 and Pax6 (G, white arrows). In the Pax6-cKO OE, many of the HBCs lacked Pax6 expression (H, yellow arrows), and the percentage of Pax6-positive HBCs was significantly lower in zone 1 compared to control mice (I, left); the percentage of Pax6-positive HBCs was also significantly reduced in zones 2–4 in the Pax6-cKO mice, although to a lesser extent than in zone 1 (I, right). (J) OE thickness did not differ significantly between control and Pax6-cKO mice in zone 1 or zones 2–4. (K) The absolute number of p63-positive HBCs did not differ significantly between control and Pax6-cKO mice in either zone 1 or zones 2–4. The dotted line in (A–H) indicates the basal lamina. Scale bars represent 1,000 μm (A–B), 200 μm (A2, B2, D, F), and 50 μm (G–H). *P<0.05; ***P<0.001. BG, Bowman's gland; GBCs, globose basal cells; LP, lamina propria; OE, olfactory epithelium; NQO1, NAD(P)H dehydrogenase [quinone] 1; SUSs, sustentacular cells. Color images available online at www.liebertpub.com/scd
Next, we measured the percentage of p63-positive HBCs that also expressed Pax6 to determine the specificity of our HBC-specific Pax6-knockout strategy. In the OE of control mice, Pax6 expression was observed in HBCs, GBCs, SUSs, and BGs (Fig. 2G); the majority of HBCs expressed both p63 and Pax6 (Fig. 2G, arrows). In the Pax6-cKO OE (Fig. 2H), the percentage of Pax6-positive HBCs was reduced to 56.2% in zone 1 (Fig. 2I, left). We found no apparent change in Pax6 expression in any other cell types. We also observed a significant reduction in Pax6-positive HBCs in zones 2–4 in the Pax6-cKO mice (Fig. 2I, right).
Despite the significant loss of Pax6 expression in the OE of Pax6-cKO mice, OE thickness did not differ significantly between the Pax6flox/flox and Pax6-cKO mice in either zone 1 or zones 2–4 (Fig. 2J). Moreover, the number of p63-positive HBCs did not differ significantly between the Pax6flox/flox and Pax6-cKO mice in either zone 1 or zones 2–4 (Fig. 2K). These results suggest that the decrease of Pax6-expressing HBCs does not seem to affect the normal development of the OE. This may be caused by the compensation of HBC population that escaped from the Pax6 deletion due to the limitation of the Cre-loxP system; there may be an insufficiency of Cre expression or there may be HBC progenitors (possibly Pax6 positive) that already exist before K5-Cre activity initiates [14].
HBCs in the OE of Pax6-cKO mice have self-renewal capacity
To evaluate the role of Pax6 in HBCs with respect to regeneration of the OE following injury, we induced severe damage to the OE of Pax6flox/flox and Pax6-cKO mice and monitored regeneration of the OE (Fig. 1E). We used the standard approach of injecting methimazole (a drug used to treat hyperthyroidism) intraperitoneally; in mice, methimazole triggers the degeneration and subsequent regeneration of the OE from HBCs [4,7]. Methimazole treatment causes death of a large percentage of differentiated OE cells within 24 h, but generally spares the HBCs [13].
Using this experimental paradigm, we first examined the percentage of p63-positive and Pax6-positive HBCs at 3 days postinjury (3 dpi). We stained OE sections of Pax6flox/flox (Fig. 3A, B) and Pax6-cKO (Fig. 3C, D) with anti-p63 and anti-Pax6 antibodies; the nuclei were counterstained with DAPI. In the Pax6flox/flox mice, p63-positive cells were detected in the basal layer just above the basal lamina (Fig. 3A, B), and Pax6-positive cells were observed in various cell types within the layers of the OE (Fig. 3A, B). Nearly all p63-positive cells expressed Pax6 at 3 dpi (white arrowheads). In contrast, in the Pax6-cKO mice, the majority of p63-positive cells in zone 1 were negative for Pax6 (Fig. 3C, yellow arrowheads), although there seemed similar numbers of p63-positive cells both in Pax6flox/flox and Pax6-cKO mice (see below). As we expected for Pax6-cKO mice, more p63-positive HBCs in zones 2–4 were Pax6-positive compared to zone 1 (Fig. 3D). A quantitative analyses revealed that the percentage of HBCs coexpressing p63 and Pax6 in zone 1 was significantly lower in the Pax6-cKO mice (16.9%±5.7%) comparing to the Pax6flox/flox mice (99.8%±0.2%) (Fig. 3E, left). In contrast, we found no difference between the Pax6flox/flox and Pax6-cKO mice with respect to zones 2–4 (Fig. 3E, right).
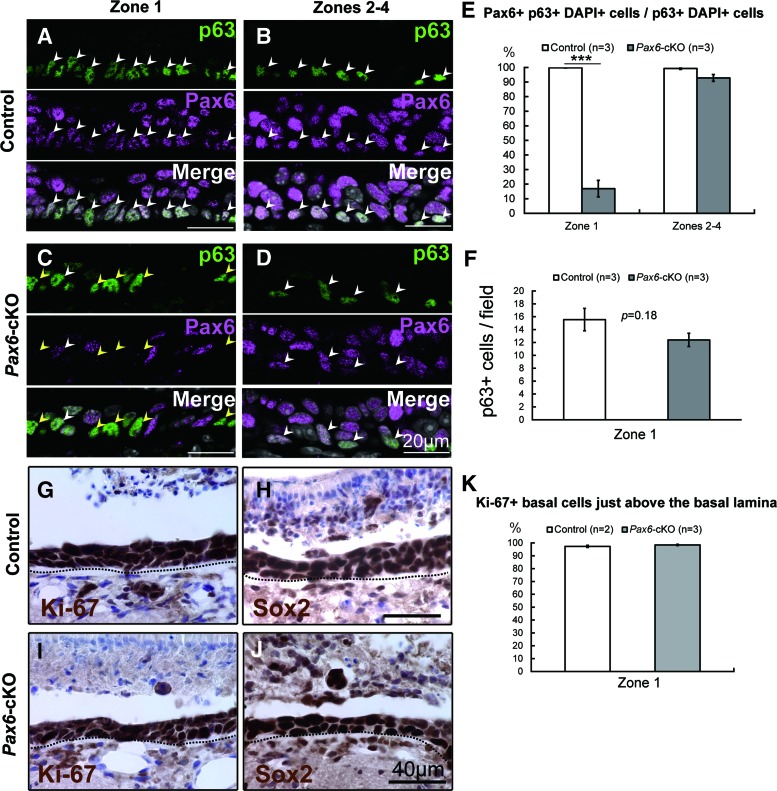
FIG. 3.
HBCs in the OE of Pax6-cKO mice have self-renewal capacity. (A–D) Pax6 and p63 expression in the OE of Pax6flox/flox mice (control) (zone 1, A; zones 2–4, B) and K5-Cre+/−::Pax6flox/flox mice (Pax6-cKO) (zone 1, C; zones 2–4, D) at 3 days postinjury (3 dpi). In the control OE, p63-positive cells were present in the basal layer just above the basal lamina, and Pax6-positive cells were present in various cell types within the OE. Nearly all p63-positive cells also expressed Pax6 (white arrowheads). In contrast, in the Pax6-cKO mice, Pax6 expression was nearly undetectable in the majority of p63-positive cells in zone 1 (C, yellow arrowheads). (E) Summary of the percentage of p63-postive cells that were also Pax6-positive in zone 1 (left) and zones 2–4 (right). (F) Summary of the number of p63-positive HBCs in zone 1. (G–J) Immunohistochemical analysis of the proliferation marker Ki-67 and the stem cell marker Sox2 in the OE of control (G, H) and Pax6-cKO (I, J) mice at 3 dpi. Note that nearly all OE cells were positive for both Ki-67 and Sox2 in both the control and Pax6-cKO mice. (K) The percentages of Ki-67-positive HBCs were almost 100% both in the Pax6flox/flox and Pax6-cKO mice. The dotted line in (G–J) indicates the basal lamina. Scale bars represent 20 μm (A–D) and 40 μm (H, J). ***P<0.001. Color images available online at www.liebertpub.com/scd
To examine the self-renewal capacity of HBCs, we carefully counted the absolute number of p63-positive HBCs at 3 dpi. Our analysis shows that the number of p63-positive HBCs in zone 1 did not differ significantly between the Pax6flox/flox and Pax6-cKO mice (Fig. 3F). These results suggest that in the injured OE of Pax6-cKO mice, the self-renewal capacity of HBCs is almost intact.
We next examined the expression of the proliferation marker Ki-67 and the stem cell marker Sox2 in the OE of Pax6flox/flox and Pax6-cKO mice at 3 dpi (Fig. 3G–J). Most surviving basal cells (ie, HBCs and GBCs) are proliferative and express the stem cell markers Pax6 and Sox2 3 days after injury [6]. We found that nearly all of the cells in the OE were positive for a proliferation marker Ki-67 and a neural stem cell marker Sox2 in both Pax6flox/flox (Fig. 3G, H) and Pax6-cKO (Fig. 3I, J) mice at 3 dpi. To quantify the percentage of proliferating HBCs in zone 1, we counted the number of Ki-67-positive basal cells just above the BM. The percentages of Ki-67-positive HBCs were almost 100% both in the Pax6flox/flox (97.3%±0.8%) and Pax6-cKO mice (98.5%±0.5%) (Fig. 3K). Indeed, we found no apparent difference between the two genotypes with respect to the expression of either Ki-67 or Sox2. Taken together, our results show that the HBCs retain their proliferative capacity at 3 dpi in zone 1 where Pax6 expression in HBCs was specifically deleted.
HBC-specific Pax6-knockout mice have severely impaired OE regeneration
Finally, we assessed the long-term regenerative capacity of the OE in Pax6flox/flox (Fig. 4A, D–F) and Pax6-cKO (Fig. 4B, G–I) mice by examining the OE at 42 dpi. Consistent with our previous results [7], in Pax6flox/flox mice, the OE had recovered nearly fully in both zone 1 and zones 2–4 within 42 days of injury (Fig. 4A). In contrast, the regenerative capacity of the OE was severely impaired in zone 1 of the Pax6-cKO mice (Fig. 4B, G–I). A quantitative analysis revealed that the OE in zone 1 was significantly thinner in Pax6-cKO mice (26.3±1.9 μm) compared to the Pax6flox/flox mice (50.5±3.7 μm) (Fig. 4C, left); in contrast, we found no significant difference in OE thickness in zones 2–4 (Fig. 4C, right). It is known that the OE lesion using methimazole disrupts the GBC population and produces multiple types of cells derived from HBCs [4]. Thus, our deletion of Pax6 function in HBCs is considered to severely impair the capacity of the OE to regenerate after severe injury.
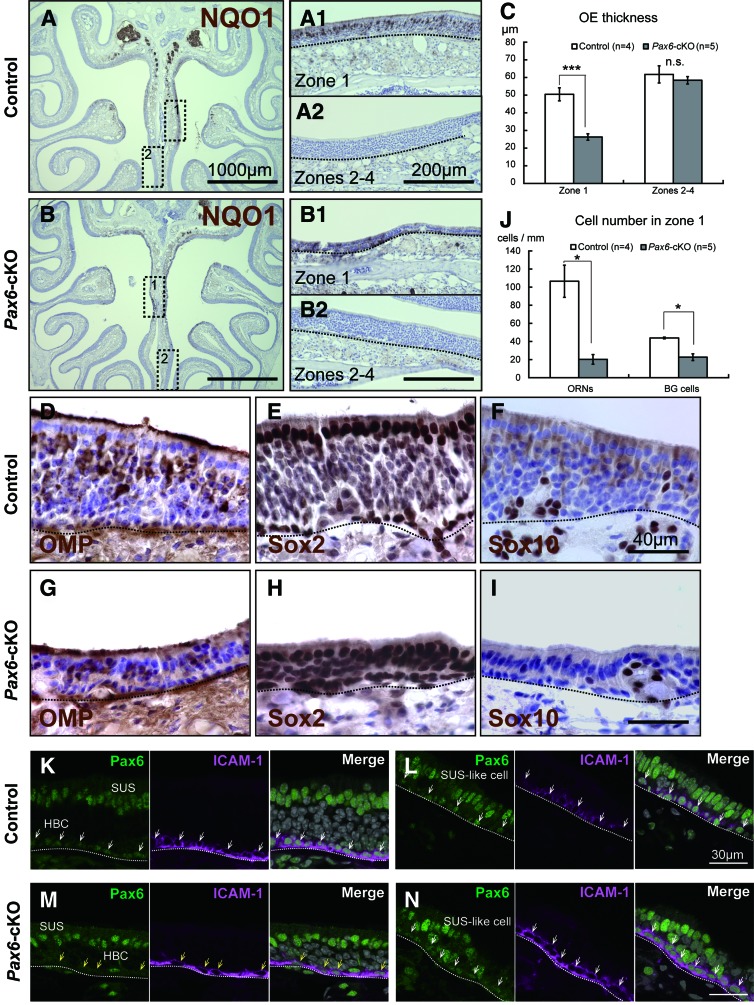
FIG. 4.
HBC-specific Pax6-knockout severely impairs the long-term regeneration of the OE. (A, B) Images of NQO1-immunostained coronal sections of the OE from Pax6flox/flox mice (control; A) and K5-Cre+/−::Pax6flox/flox mice (Pax6-cKO; B) at 42 days postinjury (42 dpi). In the control mice, the OE had recovered nearly completely in both zone 1 and zones 2–4 (A). In contrast, regeneration of the OE was severely impaired in zone 1 in the Pax6-cKO mice (B). (C) Summary of OE thickness in zone 1 and zones 2–4. (D–I) Images of immunostained sections from zone 1 of control (D–F) and Pax6-cKO (G–I) mice. The sections were stained using antibodies against OMP (D, G), Sox2 (E, H), and Sox10 (F, I). The number of OMP-positive mORNs and Sox10-positive BG cells was reduced in Pax6-cKO mice (G, I) compared to control mice (D, F). Note the abnormal expression of Sox2 in cells in the middle layer of the OE in the Pax6-cKO mice (H). (J) Summary of the number of NQO1-positive ORNs (left panel) and Sox10-positive BG cells (right panel) in zone 1. (K–N) Expression of Pax6 (green) and ICAM-1 (magenta) in zone 1 of control (K, L) and Pax6-cKO (M, N) mice at 42 dpi. In the control OE, ICAM-1-positive HBCs express Pax6 (white arrows), while there are Pax6-negative HBCs in the Pax6-cKO OE (yellow arrows). Pax6-positive SUSs and SUS-like cells (green) localize at the apical area of the OE in the control and Pax6-cKO mice. The dotted line in (A, B, D–I, and K–N) indicates the basal lamina. Scale bars represent 1,000 μm (A, B), 200 μm (A2, B2), 40 μm (F, I), and 30 μm (L, N). *P<0.05; ***P<0.001; n.s., not significant. mORNs, mature olfactory receptor neurons. Color images available online at www.liebertpub.com/scd
To investigate the regeneration process of the OE in Pax6-cKO mice in further detail, we performed immunohistochemical staining using various cell type-specific markers. We found fewer OMP-positive mature ORNs (mORNs) in zone 1 of Pax6-cKO mice (Fig. 4G) compared to Pax6flox/flox mice (Fig. 4D). To quantify the number of ORNs, we used the zone 1-specific marker NQO1 [32]. The number of NQO1-positive ORNs in zone 1 was significantly lower in the Pax6-cKO mice (20.2±5.3/mm) compared to the Pax6flox/flox mice (106.4±11.7/mm) (Fig. 4J, left). We also measured the number of BG cells. Since we found a specific expression of Sox10, a transcription factor that regulates the development of neural crest cells [33] in BGs in the OE of Pax6flox/flox mice (Fig. 4F), we measured the number of Sox-10-positive cells as the number of BG cells. In zone 1, the number of Sox10-positive BG cells was significantly lower in the Pax6-cKO mice (22.3±3.7/mm) compared to the Pax6flox/flox mice (43.9±0.9/mm) (Fig. 4J, right). Finally, basal cells (ie, HBCs and GBCs) and SUSs in the regenerated OE express Sox2 [6]. Although the background signal was relatively high in the regenerated OE, we observed strong Sox2 expression in all three cell types in the Pax6flox/flox mice (Fig. 4E). Interestingly, in the Pax6-cKO mice, in addition to normal Sox2 expression in the basal cells and SUSs, Sox2 was also expressed in cells located in the middle layer of the OE (Fig. 4H). This expression pattern has never been reported in wild-type mice; in normal OE, only Sox2-negative immature and mature ORNs are localized in the middle layer [6]. Taken together, our data suggest that deleting Pax6 in HBCs inhibits the differentiation of stem cell-like HBCs to mature OE cells (ie, mORNs and BG cells). These results also suggest that deleting Pax6 in HBCs severely impairs the ability of HBCs to differentiate during the regenerative process following severe OE damage. We also observed the difference of Pax6-positive SUSs in Pax6flox/flox (Fig. 4K, L) and Pax6-cKO (Fig. 4M, N) mice at 42 dpi. In Pax6flox/flox mice, Pax6-positive SUSs were abundantly localized at the apical area of the OE in the normaly reconstructed region (Fig. 4K), and Pax6-positive SUS-like cells were localized at the apical area of the OE in the severely damaged region (Fig. 4L). Similary, Pax6-positive SUSs and Pax6-positive SUS-like cells were also localized at the apical area of the OE in Pax6-cKO (Fig. 4M, N). These results suggest that HBC-specific deletion of Pax6 does not seem to affect the regeneration of Pax6-positive SUSs in the apical area of the OE.
Discussion
In this study, we report that deleting Pax6 specifically in HBCs in zone 1 of the OE impairs the ability of HBCs (ie, the tissue stem-like cells in the OE) to differentiate into mature cells during the regenerative process following severe OE damage. Although we could not evaluate the role of Pax6 in HBCs in zones 2–4 due to insufficient deletion of Pax6 in these areas, our findings are instrumental to understand how HBCs regenerate after injury.
Pax6 is a multifunctional protein that regulates the proliferation and differentiation of neural stem/progenitor cells (NSCs) in the CNS by regulating the expression of several downstream genes, including Fabp7, Ngn2, and Wnt7b [16,34–37]. Pax6 is also expressed in astrocytes in late embryogenesis and postnatally, and deleting Pax6 causes cells to retain their NSC-like properties and inhibits astrocyte maturation [28]. Expression of Pax6 in NSCs continues both in the developing CNS to mature astrocytes and in the adult CNS [28], which suggests that Pax6 is required for cells in the CNS to retain their stem cell-like properties and differentiate into glial lineage cells. In contrast, overexpressing Pax6 gives rise to spiking neuroblasts in non-neurogenic regions in an adult mouse model of mild transient brain ischemia [38]. Thus, the essential role of Pax6 in the CNS is highly context dependent. Pax6 also plays an important role as a multilevel regulator of ocular development [17]. In the retina, Pax6 directly controls the transcriptional activation of retinogenic bHLH factors and mediates the full retinogenic potential of retinal progenitor cells [39]. As discussed above, Pax6 regulates the proliferation and differentiation of stem/progenitor cells depending on the context within the CNS (including the retina), which is consistent with our finding that Pax6 is important in the olfactory system, in which Pax6 regulates the ability of tissue stem cells in the OE to differentiate into neuronal lineage cells.
Under normal conditions, HBCs are quiescent tissue stem cells in the OE, and a dual origin of HBCs—that is, from the olfactory placode and neural crest—has recently been reported [7,40,41]. Our group has reported that the main population of HBCs gradually change from placode-derived cells to neural crest-derived ones during postnatal maturation both in zone 1 and zones 2–4 [7,41]. However, distinctive characteristics of these HBCs are still unknown. Future studies on the dual origin of HBCs could contribute to understand the regeneration capacity of HBCs. The stem cell dynamics of HBCs are regulated by the transcription factor p63 [13,14]. Although p63-knockout mice lack HBC differentiation in the OE, other cell types in the OE—including neurons—are essentially normal [14]. This finding suggests that HBCs are not required for the formation of the majority of the OE during development, which is consistent with our observation that the OE of HBC-specific Pax6-cKO mice (even though their knockout is not thorough) has no obvious structural abnormalities at 6 weeks of age. In a previous study of regenerated OE in K5-CrePR::Rosa26YFP mice on the p63lox/lox background, YFP-positive cells that differentiated from HBCs were observed in both neuronal and non-neuronal cells, including ORNs and GBCs; however, HBC markers were strikingly absent from these YFP-lineage cells [13]. The authors concluded that p63 is required for maintaining HBC fate, but is not required for HBC differentiation in the OE during regeneration. In contrast, although the self-renewal capacity of HBCs was not affected in our Pax6-cKO mice (Fig. 3F), we found that HBC differentiation in the OE during regeneration was severely impaired following damage to the OE. Taken together, these finding suggest that Pax6 and p63 play distinct roles in the self-renewal and differentiation of HBCs.
The transcription factor Sox2 is important for maintaining ES cells and for inducting pluripotent stem cells [42,43]. Deleting Sox2 in the brain causes neurodegeneration and impaired neurogenesis in adult mice [44]. On the other hand, constitutively expressing SOX2 inhibits neuronal differentiation and causes cells to retain their neural progenitor properties [45]. In addition, overexpressing Sox2 drives the differentiation of neural progenitors into astroglia, but inhibits neurogenesis in the developing neocortex [46]. Therefore, Sox2 also plays an essential context-dependent role in the CNS. Sox2 forms a complex with its partners to function, and Pax6 is one of the factors that interact with Sox2 in both stem/progenitor cells (eg, NSCs in the CNS and optic cup progenitor cells) and terminally differentiated cells (eg, lens cells) [16,47–49]. In the OE, Pax6 and Sox2 are coexpressed in HBCs, GBCs, and SUSs, and they are believed to suppress neuronal differentiation [6]. In our Pax6-cKO mice, we found that Sox2 was expressed ectopically in many cells in the OE (cells in which neurogenesis was severely impaired during OE regeneration). This finding is consistent with previous studies that reported that high Sox2 expression blocks neuronal differentiation and promotes glial differentiation [45,46]. Taken together, we hypothesize that appropriate Pax6 expression in HBCs is required for the cells to differentiate into neuronal lineage cells by suppressing Sox2 expression during OE regeneration.
In recent decades, regeneration in sensory organs such as the eyes, ears, and nose has received considerable attention, and the reprogramming process used in the regeneration of sensory receptor cells appears to be similar among these organs [50]. A recent study found that Pax6+/Sox2+/Nestin+ multipotent retinal stem cells (RSCs) in the adult mouse retina are capable of producing functional photoreceptor cells [51]. Moreover, in humans, PAX6 and p63 are coexpressed in K5-positive limbal stem/progenitor cells (LSCs), which can differentiate into corneal epithelial cells, and WNT7A acts as an upstream regulator of PAX6, regulating corneal epithelium differentiation [52]. Finally, transfecting PAX6 into cultured skin epithelial stem cells differentiates the cells into corneal epithelial-like cells; conversely, deleting PAX6/WNT7A in LSCs causes abnormal skin epidermis-like differentiation [52]. These results obtained from research in the ocular system have several features in common with our findings. First, Pax6 and Sox2 are coexpressed in tissue stem cells (RSCs, HBCs). Second, Pax6, p63, and K5 are coexpressed in tissue stem cells (LSCs, HBCs). Third, deleting Pax6 expression causes abnormal differentiation of tissue stem cells (LSCs, HBCs).
Based on these findings, we hypothesize that Pax6 functions as a master regulator in the differentiation of a variety of tissue stem cells in several sensory organs. Future studies will be designed to identify the specific molecular mechanisms that underlie the balance between maintaining tissue stem cells and inducing their appropriate differentiation.
Acknowledgments
We are grateful to Dr. M. Mishina for providing FLP66 transgenic mice and Dr. J. Takeda, Dr. H. Motohashi, and Dr. K. Taguchi for K5-Cre transgenic mice. We thank Dr. F. Suto for providing technical advice with immunohistochemistry, Ms. S. Mochizuki for assistance with the H&E staining, and Ms. A. Ogasawara and Ms. S. Makino for animal care. We also thank all the members of the Osumi laboratory for their valuable comments. This work was supported by the Tohoku Neuroscience Global COE program “Basic and Translational Research Center for Global Brain Science” from the Japanese Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Treloar HB, Miller AM, Ray A. and Greer CA. (2010). Development of the olfactory system. In: The Neurobiology of Olfaction. Menini A, ed. Boca Raton, FL: [PubMed] [Google Scholar]
- 2.Graziadei PP. and Graziadei GA. (1979). Neurogenesis and neuron regeneration in the olfactory system of mammals. I. Morphological aspects of differentiation and structural organization of the olfactory sensory neurons. J Neurocytol 8:1–18 [DOI] [PubMed] [Google Scholar]
- 3.Schwob JE. (2002). Neural regeneration and the peripheral olfactory system. Anat Rec 269:33–49 [DOI] [PubMed] [Google Scholar]
- 4.Leung CT, Coulombe PA. and Reed RR. (2007). Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci 10:720–726 [DOI] [PubMed] [Google Scholar]
- 5.Davis JA. and Reed RR. (1996). Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci 16:5082–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Z, Packard A, Krolewski RC, Harris MT, Manglapus GL. and Schwob JE. (2010). Expression of pax6 and sox2 in adult olfactory epithelium. J Comp Neurol 518:4395–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki J, Yoshizaki K, Kobayashi T. and Osumi N. (2013). Neural crest-derived horizontal basal cells as tissue stem cells in the adult olfactory epithelium. Neurosci Res 75:112–120 [DOI] [PubMed] [Google Scholar]
- 8.Murdoch B. and Roskams AJ. (2007). Olfactory epithelium progenitors: insights from transgenic mice and in vitro biology. J Mol Histol 38:581–599 [DOI] [PubMed] [Google Scholar]
- 9.Carter LA, MacDonald JL. and Roskams AJ. (2004). Olfactory horizontal basal cells demonstrate a conserved multipotent progenitor phenotype. J Neurosci 24:5670–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwai N, Zhou Z, Roop DR. and Behringer RR. (2008). Horizontal basal cells are multipotent progenitors in normal and injured adult olfactory epithelium. Stem Cells 26:1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackay-Sim A. (2010). Stem cells and their niche in the adult olfactory mucosa. Arch Ital Biol 148:47–58 [PubMed] [Google Scholar]
- 12.Senoo M, Pinto F, Crum CP. and McKeon F. (2007). p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129:523–536 [DOI] [PubMed] [Google Scholar]
- 13.Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG. and Ngai J. (2011). p63 Regulates olfactory stem cell self-renewal and differentiation. Neuron 72:748–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Packard A, Schnittke N, Romano RA, Sinha S. and Schwob JE. (2011). DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J Neurosci 31:8748–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dohrmann C, Gruss P. and Lemaire L. (2000). Pax genes and the differentiation of hormone-producing endocrine cells in the pancreas. Mech Dev 92:47–54 [DOI] [PubMed] [Google Scholar]
- 16.Osumi N, Shinohara H, Numayama-Tsuruta K. and Maekawa M. (2008). Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells 26:1663–1672 [DOI] [PubMed] [Google Scholar]
- 17.Shaham O, Menuchin Y, Farhy C. and Ashery-Padan R. (2012). Pax6: a multi-level regulator of ocular development. Prog Retin Eye Res 31:351–376 [DOI] [PubMed] [Google Scholar]
- 18.Nomura T, Haba H. and Osumi N. (2007). Role of a transcription factor Pax6 in the developing vertebrate olfactory system. Dev Growth Differ 49:683–690 [DOI] [PubMed] [Google Scholar]
- 19.Georgala PA, Carr CB. and Price DJ. (2011). The role of Pax6 in forebrain development. Dev Neurobiol 71:690–709 [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara M, Uchida T, Osumi-Yamashita N. and Eto K. (1994). Uchida rat (rSey): a new mutant rat with craniofacial abnormalities resembling those of the mouse Sey mutant. Differentiation 57:31–38 [DOI] [PubMed] [Google Scholar]
- 21.Hogan BL, Horsburgh G, Cohen J, Hetherington CM, Fisher G. and Lyon MF. (1986). Small eyes (Sey): a homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J Embryol Exp Morphol 97:95–110 [PubMed] [Google Scholar]
- 22.Matsuo T, Osumi-Yamashita N, Noji S, Ohuchi H, Koyama E, Myokai F, Matsuo N, Taniguchi S, Doi H, et al. (1993). A mutation in the Pax-6 gene in rat small eye is associated with impaired migration of midbrain crest cells. Nat Genet 3:299–304 [DOI] [PubMed] [Google Scholar]
- 23.Pratt T, Quinn JC, Simpson TI, West JD, Mason JO. and Price DJ. (2002). Disruption of early events in thalamocortical tract formation in mice lacking the transcription factors Pax6 or Foxg1. J Neurosci 22:8523–8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dellovade TL, Pfaff DW. and Schwanzel-Fukuda M. (1998). Olfactory bulb development is altered in small-eye (Sey) mice. J Comp Neurol 402:402–418 [PubMed] [Google Scholar]
- 25.Tarutani M, Itami S, Okabe M, Ikawa M, Tezuka T, Yoshikawa K, Kinoshita T. and Takeda J. (1997). Tissue-specific knockout of the mouse Pig-a gene reveals important roles for GPI-anchored proteins in skin development. Proc Natl Acad Sci U S A 94:7400–7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishina M. and Sakimura K. (2007). Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci Res 58:105–112 [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi T, Nomura T, Tsujita M, Suzuki M, Fuse T, Mori H. and Mishina M. (2002). Flp recombinase transgenic mice of C57BL/6 strain for conditional gene targeting. Biochem Biophys Res Commun 293:953–957 [DOI] [PubMed] [Google Scholar]
- 28.Sakurai K. and Osumi N. (2008). The neurogenesis-controlling factor, Pax6, inhibits proliferation and promotes maturation in murine astrocytes. J Neurosci 28:4604–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vedin V, Molander M, Bohm S. and Berghard A. (2009). Regional differences in olfactory epithelial homeostasis in the adult mouse. J Comp Neurol 513:375–384 [DOI] [PubMed] [Google Scholar]
- 30.Holbrook EH, Szumowski KE. and Schwob JE. (1995). An immunochemical, ultrastructural, and developmental characterization of the horizontal basal cells of rat olfactory epithelium. J Comp Neurol 363:129–146 [DOI] [PubMed] [Google Scholar]
- 31.Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K. and Miyazaki J. (2000). A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett 470:263–268 [DOI] [PubMed] [Google Scholar]
- 32.Gussing F. and Bohm S. (2004). NQO1 activity in the main and the accessory olfactory systems correlates with the zonal topography of projection maps. Eur J Neurosci 19:2511–2518 [DOI] [PubMed] [Google Scholar]
- 33.Haldin CE. and LaBonne C. (2010). SoxE factors as multifunctional neural crest regulatory factors. Int J Biochem Cell Biol 42:441–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arai Y, Funatsu N, Numayama-Tsuruta K, Nomura T, Nakamura S. and Osumi N. (2005). Role of Fabp7, a downstream gene of Pax6, in the maintenance of neuroepithelial cells during early embryonic development of the rat cortex. J Neurosci 25:9752–9761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maekawa M, Takashima N, Matsumata M, Ikegami S, Kontani M, Hara Y, Kawashima H, Owada Y, Kiso Y, et al. (2009). Arachidonic acid drives postnatal neurogenesis and elicits a beneficial effect on prepulse inhibition, a biological trait of psychiatric illnesses. PLoS One 4:e5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scardigli R, Baumer N, Gruss P, Guillemot F. and Le Roux I. (2003). Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 130:3269–3281 [DOI] [PubMed] [Google Scholar]
- 37.Osumi N, Hirota A, Ohuchi H, Nakafuku M, Iimura T, Kuratani S, Fujiwara M, Noji S. and Eto K. (1997). Pax-6 is involved in the specification of hindbrain motor neuron subtype. Development 124:2961–2972 [DOI] [PubMed] [Google Scholar]
- 38.Kronenberg G, Gertz K, Cheung G, Buffo A, Kettenmann H, Gotz M. and Endres M. (2010). Modulation of fate determinants Olig2 and Pax6 in resident glia evokes spiking neuroblasts in a model of mild brain ischemia. Stroke 41:2944–2949 [DOI] [PubMed] [Google Scholar]
- 39.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F. and Gruss P. (2001). Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105:43–55 [DOI] [PubMed] [Google Scholar]
- 40.Katoh H, Shibata S, Fukuda K, Sato M, Satoh E, Nagoshi N, Minematsu T, Matsuzaki Y, Akazawa C, et al. (2011). The dual origin of the peripheral olfactory system: placode and neural crest. Mol Brain 4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki J. and Osumi N. (2015). Neural crest and placode contributions to olfactory development. Curr Top Dev Biol 111:351–374 [DOI] [PubMed] [Google Scholar]
- 42.Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676 [DOI] [PubMed] [Google Scholar]
- 43.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, et al. (2007). Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 9:625–635 [DOI] [PubMed] [Google Scholar]
- 44.Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S. and Nicolis SK. (2004). Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131:3805–3819 [DOI] [PubMed] [Google Scholar]
- 45.Graham V, Khudyakov J, Ellis P. and Pevny L. (2003). SOX2 functions to maintain neural progenitor identity. Neuron 39:749–765 [DOI] [PubMed] [Google Scholar]
- 46.Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR. and Sikorska M. (2006). Role of Sox2 in the development of the mouse neocortex. Dev Biol 295:52–66 [DOI] [PubMed] [Google Scholar]
- 47.Shimozaki K. (2014). Sox2 transcription network acts as a molecular switch to regulate properties of neural stem cells. World J Stem Cells 6:485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsushima D, Heavner W. and Pevny LH. (2011). Combinatorial regulation of optic cup progenitor cell fate by SOX2 and PAX6. Development 138:443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kondoh H, Uchikawa M. and Kamachi Y. (2004). Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation. Int J Dev Biol 48:819–827 [DOI] [PubMed] [Google Scholar]
- 50.Bermingham-McDonogh O. and Reh TA. (2011). Regulated reprogramming in the regeneration of sensory receptor cells. Neuron 71:389–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T, Lewallen M, Chen S, Yu W, Zhang N. and Xie T. (2013). Multipotent stem cells isolated from the adult mouse retina are capable of producing functional photoreceptor cells. Cell Res 23:788–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ouyang H, Xue Y, Lin Y, Zhang X, Xi L, Patel S, Cai H, Luo J, Zhang M, et al. (2014). WNT7A and PAX6 define corneal epithelium homeostasis and pathogenesis. Nature 511:358–361 [DOI] [PMC free article] [PubMed] [Google Scholar]