Abstract

The central dogma of the action of current anticancer drugs is that the drug tightly binds to its molecular target for inhibition. The reliance on tight ligand–receptor binding, however, is also the major root of drug resistance in cancer therapy. In this article, we highlight enzyme-instructed self-assembly (EISA)—the integration of enzymatic transformation and molecular self-assembly—as a multistep process for the development of cancer therapy. Using apoptosis as an example, we illustrate that the combination of enzymatic transformation and self-assembly, in fact, is an inherent feature of apoptosis. After the introduction of EISA of small molecules in the context of supramolecular hydrogelation, we describe several key studies to underscore the promises of EISA for developing cancer therapy. Particularly, we will highlight that EISA allows one to develop approaches to target “undruggable” targets or “untargetable” features of cancer cells and provides the opportunity for simultaneously interacting with multiple targets. We envision that EISA, used separately or in combination with current anticancer therapeutics, will ultimately lead to a paradigm shift for developing anticancer medicine that inhibit multiple hallmark capabilities of cancer.
Introduction
Cancer remains a major challenge to public health. The estimated new cases and deaths from cancer in the United States in 2014 were 1,665,540 and 585,720, respectively.1 Conventional molecular therapy or chemotherapy, based on tight ligand–receptor interactions or modification of nucleic acids, has been largely unable to meet the challenges posed by the great complexity of cancer cells2,3 that causes cancer drug resistance2,4 and metastasis.2,5 Despite the recent success in cancer immunotherapy,6 only a fraction of cancer patients are responsive to immunotherapy.7 Thus, there has always been an urgent need to develop innovative approaches for cancer therapy. Here we introduce an emerging approach that promises new directions in anticancer therapy by highlighting enzyme-instructed self-assembly (EISA)—the integration of enzymatic transformation (ET) and self-assembly (SA)—as a paradigm shift for the development of cancer therapy.
This perspective starts with a brief description of the major challenges in current cancer therapy; then, apoptosis is used as an example to illustrate that EISA, as a common theme conserved during evolution of life, constitutes the inherent mechanisms of programmed cell death; and after that, the development of EISA of small molecules is introduced followed by the discussion of several key studies to illustrate the concept of EISA for cancer therapy. A particular highlight is that EISA allows one to develop approaches to target “undruggable” targets or “untargetable” features of cancer cells and provides opportunities for simultaneously interacting with multiple targets. Finally, we suggest that EISA, used separately or in combination with current anticancer therapeutics, will ultimately provide a paradigm shift for developing anticancer medicines to target multiple hallmark capabilities of cancer that are the major challenges in current cancer therapy.
Ligand–Receptor Interactions in Drug Resistance
Anticancer drug resistance has been a major challenge in cancer therapy. Considerable efforts have focused on overcoming drug resistance, and the approaches largely fall into three categories: inhibiting new targets (including multidrug-resistant (MDR) transporters), improving drug specificity, or using combined therapeutics to reduce the odds of resistance.8−11 The outcome of these strategies, so far, has been disappointing.12,13 These approaches aim to inhibit tumors by interrupting one or two specific essential cellular processes or functions (e.g., DNA synthesis, RNA synthesis, protein synthesis, or protein function),14 which are insufficient due to a daunting range of resistance mechanisms.4 For example, multiple inherent cellular mechanisms, such as up-regulating growth factors or efflux transporters, the mutations of drug targets, and increasing metabolic drug degradation,15−18 work against the drugs that function via ligand–receptor binding.19 In addition, tumor microenvironment,20 genomic instability,21 intratumoral heterogeneity,22 and the up-regulation of cell survival pathways further evolve the great complexity of cancer. As pointed out by Weinberg et al.2 and illustrated in Figure 1, the cancer drugs aimed at a specific molecular target (e.g., based on tight ligand–receptor interactions) only result in a transitory clinical response that is (almost) always followed by relapses. Thus, a new paradigm of anticancer therapy is urgently needed.
Figure 1.
Representative mechanisms of cancer drug resistance: (I) plasma proteins bind the drug to reduce its effectiveness; (II) efflux pump decreases intracellular concentration of the drug; (III) mutations in the binding site abolish the inhibitory effect of the drug; (IV) redundant pathways alleviate the dependence of the cancer cell on the original target; (V) genomic instability accelerates mutation; (VI) tumor microenvironment provides prosurvival signals.
EISA in Apoptosis
As one of the most promising directions in cancer therapy in the past decade, immunotherapy utilizes the immune system to treat certain cancers and is able to achieve complete tumor regression in some cases.23 Regardless of its subtypes (i.e., cell-based therapies, antibody therapies, and cytokine therapies), immunotherapy eliminates cancer cells based on (i) generic difference between cancer and normal cells, that is, tumor cells carry cancer antigens, but normal cells do not; (ii) immune system killing the tumor cells largely by inducing apoptosis, that is, programmed cell death. While most of the attention centers on the discovery of cancer specific antigens and the development of the corresponding antibodies, an overlooked fact is that EISA, as a multistep process, constitutes an inherent feature of apoptosis (Figure 2). As part of the intrinsic pathway of apoptosis, enzymatic transformation changes the conformation of Apaf-1 to accommodate its interaction with cytochrome c,24 and the subsequent protein complex self-assembles (a.k.a., aggregates) to form the apoptosome25 as the necessary scaffold to result in cascade events of cell death; during the extrinsically induced cell death,26,27 enzymatic transformation generates certain ligands (e.g., TRAIL, TNF, and CD95L),28−30 which self-assemble (a.k.a., oligomerize) the cell death receptors (e.g., TRAIL-R1/R2, TNFR1, and CD95) and initiate the downstream signaling of apoptosis. These fundamental features of apoptosis are not a simple “on or off” (or “live or dead”) switch, but assume quantitative aspects of signaling transduction, such as location, duration, thresholds, and amplitude. These multistep processes not only are necessary for the precise and effective killing of the targeted cells without causing side effects, but also imply that EISA should be one of the strategies for developing anticancer therapeutics that selectively kill cancer cells without harming normal cells.
Figure 2.
EISA—the integration of enzymatic transformation (ET) and self-assembly (SA)—as the inherent feature of apoptosis. That is, enzymatic transformation generates (I) TRAIL to self-assemble TRAIL-R1/R2 or (II) CD95L to self-assemble CD95 and initiate the downstream signaling, including apoptosis; (III) enzymatic transformation changes the conformation of Apaf-1 to bind with cytochrome c, and the protein complex self-assembles to form the apoptosome, which results in cell death.
EISA of Small Molecules
As a ubiquitous process in nature, self-assembly (or aggregation, or clustering) plays numerous roles and underlies the formation of a wide variety of biological complexes. For example, the self-assembly of proteins into highly ordered structures is both central to normal biology (e.g., the dynamics of cytoskeleton, such as microtubules) and a dominant feature in disease (e.g., the formation of β-amyloid in Alzheimer’s disease). The realization that enzymatic reactions govern most of the self-assembly processes has fascinated and inspired researchers to exploit Nature’s principles for studying and developing the small molecular self-assembly process by enzymatic transformation in the past decade. As shown in Figure 3A, in essence, enzymes initiate self-assembly by simply converting a non-self-assembling precursor into a self-assembling molecule via bond cleavage or formation. Such self-assembly of small molecules usually results in the formation of supramolecular nanoscale assemblies (e.g., nanofibers or nanoparticles) in water, and the nanoscale assemblies, above a certain threshold concentration, entangle to form a network and cause hydrogelation in most cases. Thus, the self-assembling molecule usually acts as a hydrogelator.31,32 Based on the above principles and using a simple Fmoc-phosphotyrosine (1), we reported the first example of EISA of small molecules.33 Specifically, we used alkaline phosphatase (ALP), a component of the canonical kinase/phosphatase switch,34 that is readily available and exhibits high catalytic efficiency, as the enzyme to instruct molecular self-assembly. As shown in Figure 3B, enzymatic dephosphorylation by ALP converts the precursor (1) into its hydrogelator (2), which, compared with 1, is more hydrophobic and self-assembles to form nanofibers/hydrogel.33 Yang et al. further explored this process by studying the EISA of methylated form of 1 on its C-terminal and demonstrated that EISA offers a sole mechanism to result in the corresponding hydrogel made of methylated 2.35
Figure 3.
(A) Schematic illustration of EISA of small molecules in water that usually results in supramolecular nanofibers/hydrogels. (B) Some representative small molecules used for EISA.
The strategy that converts precursors to hydrogelators by enzymatic transformation to induce self-assembly for the formation of supramolecular nanofibers is not limited to phosphatases. For example, we reported the use of matrix metalloprotease-9 (MMP-9) to instruct the self-assembly of hydrogelators, which form nanofibers and result in a hydrogel. Figure 3B illustrates the rational design of the short peptide (FFFFCGLDD (3)), a substrate of MMP-9. The pentapeptide, CGLDD, provides an enzyme cleavage site of MMP-9. The removal of the hydrophilic LDD (5) from 3 generates a more hydrophobic amphiphile FFFFCG (4) with balanced hydrophobic and hydrophilic interactions, which results in a hydrogelator to self-assemble into supramolecular nanofibers and afford a hydrogel.36
Enzymatic transformation to induce self-assembly for the formation of supramolecular nanofibers is also applicable to certain d-peptides. d-Peptides, as the enantiomers of the naturally occurring l-peptides, usually resist endogenous proteases and are presumably insensitive to most enzymatic transformations. We demonstrated that the chirality of the precursors derived from a tetrapeptide hardly affects the ALP-instructed self-assembly resulting from the removal of phosphate from a tyrosine phosphate residue.37 This work, as a systematic study of supramolecular hydrogelation by enzymatic dephosphorylation of ultrashort d-peptides, establishes a useful approach to generate supramolecular hydrogels that have both biostability and desired functions.38−40
Unlike phosphatases or MMPs, which break/hydrolyze a phosphoester bond or a peptide bond, respectively, to instruct self-assembly, thermolysin controls the self-assembly by catalyzing the formation of a covalent bond between two substrates, as first demonstrated by Ulijn et al.41 They reported the use of thermolysin for reverse hydrolysis to produce amphiphilic peptide hydrogelators (6) that self-assemble to form nanofibers (Figure 3). This strategy has become a powerful approach to create a dynamic library of small peptide from screening hydrogelators.
Because EISA can take place at physiological conditions, its application in biology and biomedicine is not surprising. Over the past decade, an increased number of enzymes, which catalyze bond cleavage (e.g., β-lactamase,42 esterase,41,43−45 α-chymotrypsin,46 thrombin,47 or chymotrypsin47), bond formation (e.g., lipase,48 microbial transglutaminase (MTGase),49 thermolysin41,43−45), or substrate oxidation (e.g., glucose oxidase,50 peroxidase,51−53 and tyrosinase54), have been used to instruct the self-assembly of small molecules for a wide range of applications (e.g., enzyme inhibitor screening55 and biomineralization56). These developments have largely benefited from gelation acting as a simple assay to report molecular self-assembly in a solvent.57,58 This unique feature of supramolecular gelation and the above investigations have laid a solid molecular foundation for the exploration of EISA to inhibit cancer cells.
EISA for Targeting Cancer Cells
Actually, besides being an intrinsic feature of apoptosis, the concept of enzyme transformation has already found applications in clinical medicine for converting prodrugs (e.g., cyclophosphamide, fludarabine phosphate, and irinotecan) into drugs through a normal metabolic process (e.g., hydrolysis of an ester bond).59 In the case of self-assembly,60 Svanborg et al. have made a seminal discovery61 in which the protein aggregates formed by the self-assembly of partially unfolded α-lactalbumin (HAMLET) induce apoptosis of tumor cells via multiple mechanisms,62−64 and have validated this approach in a human trial for treating skin papillomas.65 Wells et al. reported that the nanofibrils formed by self-assembly of small molecules initiate apoptosis via multiple mechanisms.66−68 In terms of the EISA, we have demonstrated EISA to inhibit tumor cells selectively in vitro and in vivo.37,38,40,69−75 These studies suggest that EISA, as a multistep process (not a single or several compounds), is an emerging and paradigm-shift approach for developing cancer therapy. In the following, we highlight some representative examples to illustrate the concept of EISA as an unprecedented process for cancer therapy.
We designed and developed 7 (a substrate of carboxylesterase) as a precursor of hydrogelator (8).69 As shown in Figure 4A, this precursor (7) hardly self-assembles extracellularly, but is able to enter cells. Once the precursor is inside cells, an endogenous esterase converts it into a hydrogelator (8) that self-assembles into nanofibers. The formation of nanofibers then induces hydrogelation, which exerts stresses on the cell and causes cell death. Interestingly, the precursor (7), being innocuous to mouse fibroblast cells (NIH3T3) (Figure 4B), kills about 80% of human cancer cells (HeLa) (Figure 4C) after a two-day incubation. One plausible explanation would be the higher expression of the esterase in HeLa cell than in NIH3T3 cell, which is supported by a fluorescence assay of esterase in the cells.69 Although other differences of these two cell lines might also contribute to the apparent low toxicity of 7 against NIH3T3 cells, this result, unambiguously, indicates that the use of enzymatic reaction (rather than enzyme inhibition) to generate nanoscale assembly is specific to different types of cells, and EISA of small molecules offers a fundamentally new way to control the fate of cells,76 including inhibiting cancer cells.69
Figure 4.
(A) Schematic illustration of intracellular formation of nanofibers that leads to hydrogelation and cell death. (B, C) Cell viability (measured by MTT assay) of (B) NIH3T3 cells (a normal cell line) and (C) HeLa cells (a cancer cell line) treated with 7 at concentrations of 0.08, 0.04, and 0.02 wt % (Adapted from ref (69), Copyright 2007 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim).
Maruyama et al. recently reported an innovative approach that uses extracellular enzymes to instruct the intracellular self-assembly of a peptide lipid, which also is a hydrogelator, to initiate cancer cell death.77 Specifically, the peptide lipid precursor (9) undergoes enzymatic transformation catalyzed by matrix metalloproteinase-7 (MMP-7) to produce a hydrogelator (10), which is taken up by cancer cells. Once inside the cells, the hydrogelator (10) self-assembles to form nanofibers that critically impairs cellular function and induces death of the cancer cells (Figure 5A). Using HeLa cells as a model for human cancer cells and human dermal microvascular endothelial cells (MvE cells) as a model for normal human cells, Maruyama et al. evaluated the cell inhibitory effect of both the precursor (9) and the hydrogelator (10) with live/dead assay. They found that 9 kills most HeLa cells while remaining innocuous to MvE cells, but 10 kills both HeLa and MvE cells (Figure 5B). In addition, 9 also inhibits other cancer cells, such as A431, SKBR3, and MCF-7 cells. These results indicate that the cytotoxicity of 9 is a result of the hydrolysis catalyzed by MMP-7, as confirmed by the high correlation between the viability of various cell lines in the presence of 9 and the amount of their MMP-7 secretion (Figure 5C and D). Using the lysate of dead cancer cells, the authors confirmed the nanofibers of 10. Based the observation that the same lysate forms a hydrogel and exhibits thermoreversible gel–sol transition, the authors also validate the intracellular hydrogelation. This work by Maruyama further validates the concept of EISA for selectively killing cancer cells.
Figure 5.
(A) Cancer cell death induced by self-assembly of an enzyme-responsive hydrogelator and molecular structures of ER-C16 (9), G-C16 (10), and the peptide fragment. (B) Live/dead assays of HeLa cells and MvE cells after incubation for 18 h with 9 (0.02 wt %) and 10 (0.02 wt %). (C) Viability assays of cancer cells and normal human cells after incubation with 9 (0.025 wt %). (D) MMP-7 concentration in the culture media after culturing the cells. PE represents primary human pancreatic epithelial cells. (E) TEM observation of the lysate of the dead HeLa cells. Inset is gelation test (inverted test tube) of the lysate of HeLa cells that were killed by 9. Scale bars represent 100 nm. (Adapted from ref (77), Copyright 2015 American Chemical Society.)
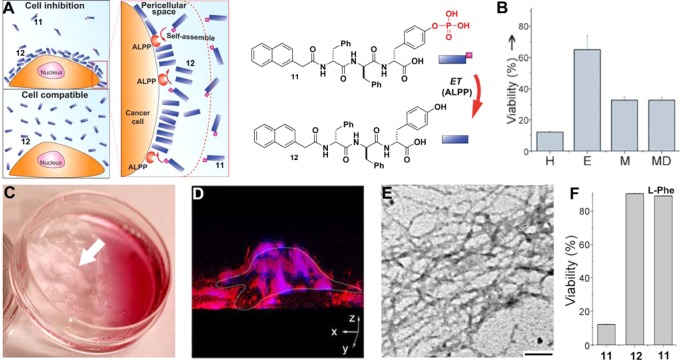
While the concept of intracellular supramolecular nanofibers formed by EISA is receiving increased exploration,37,78−80 we unexpectedly observed that EISA leads to selective inhibition of cancer cells by forming the pericellular nanofibers.38 Briefly, ectophosphatase (i.e. placental alkaline phosphatase (ALPP)) overexpressed on the cell surface of certain cancers (e.g., cervical, ovarian, stomach, endometrial, and testis81) catalytically dephosphorylates a derivative of ultrashort d-peptide (11) to form a hydrogelator (12); the accumulation of the hydrogelators results in a network of nanofibers as the matrices of a hydrogel in pericellular space, which block cellular uptake and mass exchanges, and induce cell apoptosis (Figure 6A).38 Most importantly, this d-peptide precursor (11) selectively kills cancer cells (e.g., HeLa, MES-SA, MES-SA/Dx5) due to the overexpression of ectophosphatases by the cancer cells (Figure 6B). Due to the overexpression of the ectophosphatases, the formation of nanofibers/hydrogels in pericellular space of cancer cells (i.e., HeLa cells; Figure 6C) occurs fast and locally (e.g., 4 h). Using Congo red, a dye for nanofibrils,82 together with TEM, we confirmed that the nanofibers of 12 act as the nanonets on the cell surface, which even prevent DAPI (a small nucleus dye) from entering cell (Figure 6D,E). The observation that incubating HeLa cells with 11 or 12 at the same concentration (200 μg/mL) gives apparently counterintuitive results—11 significantly inhibits the HeLa cells, while 12 remains innocuous (Figure 6F)—indicates that it is EISA of 12 that results in cell inhibition. Moreover, the addition of l-phenylalanine (l-Phe), an uncompetitive inhibitor of ALPP83 stops the dephosphorylation and abrogates the inhibitory activity of 11 (Figure 6E),40 confirming that ALPP, as the ectophosphatase, dephosphorylates 11 to result in the self-assembly of 12 on cell surface to inhibit the HeLa cells (Figure 6A). Moreover, the unassembled 12 (even at the same concentration as 11) hardly inhibit the cells, confirming that 12, as a monomer, is innocuous to cells.38 This work demonstrates an unexpected yet fundamentally new mechanism to selectively inhibit cancer cells via ectoenzyme-instructed self-assembly of small molecules.
Figure 6.
(A) ALPP-instructed formation of nanofibers of 12 on the cell surface inhibits the cancer cells, but 12, as the soluble monomer, is innocuous to cells. (B) Cell viabilities of HeLa (H), Ect1/E6E7 (E), MES-SA (M), and MES-SA/Dx5 (MD) cells treated 217 μg/mL of 11 for 48 h. (C) Optical image and (D) 3D stacked Z-scan fluorescent images of Congo red stained nanofibers/hydrogels on HeLa cells incubated with 11 (400 μg/mL). (E) TEM images of the nanofibers of 12 on the cells (scale bar = 100 nm). (F) Cell viabilities of HeLa treated by 11 or 12 at 200 μg/mL, or 11 (217 μg/mL) plus l-Phe (54 μg/mL) for 48 h (Adapted from ref (38), Copyright 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim; and ref (40), Copyright 2014 American Chemical Society).
In another work, we examined the cellular response to the EISA using the derivatives of 11, which are derived from systematically replacing d-amino acid in 11 by l-amino acid.40 Importantly, we confirmed that ALPP, as the ectoenzyme, converts the precursors to the hydrogelators to inhibit cancer cells. Moreover, these precursors exhibit completely different cytotoxicity from those of the hydrogelators, which indicates that EISA is a general process for different substrates of ALPP. This generality is further validated by the following two cases.
Pires and Ulijn et al. recently presented an aromatic carbohydrate amphiphile (N-(fluorenylmethoxycarbonyl)-glucosamine-6-phosphate (13)), as an alternative substrate of phosphatase for EISA.84 They demonstrated that a simple carbohydrate phosphate derivative (13) can be converted into the self-assembling hydrogelator 14 in situ by membrane-bound ALP, which are highly expressed on the osteosarcoma cells (SaOs2 cells). Upon conversion, 14 forms nanofibers/hydrogel surrounding the cells, which induces cytotoxicity. They also proved that this process is cell specific, as another type of cells (e.g., ATDC5) with lower level membrane-bound ALP is not affected by the addition of 13 (Figure 7A). As shown in Figure 7B, while the presence of 13 hardly affects the metabolic activity of ATDC5 cell, it drastically decreases the metabolic activity of SaOs2 cells. By quantifying both membrane-bound and extracellular ALP in SaOs2 and ATDC5 cells cultures, they found that membrane bound ALP has 15–20 times higher value and extracellular ALP has 1.5–2 times higher value for SaOs2 compared to those of ATCD5, indicating that membrane bound ALP is mainly responsible for the cytotoxicity of 13 (Figure 7C). In addition, they also confirmed that the conversion of 13 to 14 by SaOs2 cells results in pericellular nanonets (Figure 7E), while these nanonets are absent without the addition of 13 (Figure 7D). The successful use of carbohydrate derivatives on osteosarcoma cells, undoubtedly, promises new possibilities for developing new cancer therapeutics based on EISA.
Figure 7.
(A) Illustration of enzymatic transformation of 13 to 14 catalyzed by phosphatases (e.g., ALP). (B) Metabolic activity of SaOs2 and ATDC5 monolayer cultures in the presence of different concentrations of 13 for 7 h (control 0 mM, 0.5 mM, and 1 mM), without (−I) and with (+I) phosphatase inhibitors. (C) Activity of the membrane bound and extracellular ALP in the SaOs2 and ATDC5 cell cultures as a function of the concentration of 13. Quantification at 7 h of cell culture. (D, E) SEM images of SaOs2 cells, cultured during 7 h, in the (D) absence and (E) presence of 13 (0.5 mM). (Adapted from ref (84), Copyright 2014 American Chemical Society.)
We also successfully applied the concept of EISA on other self-assembling entities, such as nanoparticles.73 By decorating commercially available iron oxide nanoparticles with a simple amino acid, d-tyrosine phosphate, we obtained d-tyrosine phosphate modified magnetic nanoparticles (15). The overexpressed ALPP on cancer cells converts 15 to 16, causing 16 to assemble and adhere on the cancer cells. This EISA process enables magnetic separation of cancer cells from mixed population of cells (i.e., cocultured cancer cell (HeLa-GFP) and stromal cells (HS-5)) (Figure 8A). Figure 8B confirms the selectivity of 15 toward cancer cells, because after the cells (i.e., GFP-HeLa and HS-5, respectively) were incubated with 15 and subjected to magnetic sorting, the extraction only contains HeLa-GFP cells. Moreover, we found that enzymatic transformation is essential since the nanoparticles of 16 alone fail to sort the cancer cell from the coculture. Figure 8C shows cell capture efficiency of 15 toward GFP-HeLa, HS-5, and coculture of these two cells. Based on the proliferation rate of GFP-HeLa and HS-5 cells, we estimated that this method separates over 90% of cancer cells from the coculture. In addition, using a vibrating sample magnetometer (VSM) to quantify the amount of 15 remaining on the cells, we found that more than 63% of nanoparticles adhere to the HeLa-GFP in the culture of homogeneous GFP-HeLa culture or the coculture of GFP-HeLa and HS-5. Although there are few control iron oxide nanoparticles remaining on the surface of either cell (Figure 8D), all these results indicate that enzymatic transformation of 15 by overexpressed ALPP at the surface of cancer cells, not 16 itself, confers the magnetic separation. Besides selectively capturing cancer cells in the coculture, 15 also selectively inhibits the proliferation of HeLa-GFP cells (Figure 8E) with the IC50 of 12 μg/mL and IC90 of ∼40 μg/mL in cell viability assay. This work suggests that EISA of magnetic nanoparticles can selectively sort and inhibit cancer cells without involving specific ligand–receptor interactions or the use of antibodies.73 It is noteworthy that the IC50 of magnetic nanoparticles is already an order of magnitude lower than those of the small molecule precursors (11 and 13). This observation suggests that the introduction of multivalency to the self-assembling small molecules may enhance the potency of EISA, which remains to be explored.
Figure 8.
(A) EISA of magnetic nanoparticles for selectively sorting cancer cells. (B) Overlaid bright field and fluorescent microscopy images of the HeLa-GFP cells (top) and HS-5 cells (bottom) magnetically captured by incubating the cells with 15 (left) and 16 (right). The scale bar is 100 μm. (C) Relative amounts of cells (%) in the extraction or supernatant of all the cells collected after the treatment by 40 μg/mL 15 and the magnetic capture. (D) Relative amounts of nanoparticles remaining on the cells. (E) Relative cell viability of coculture of HeLa-GFP and HS-5 cells, HeLa-GFP cells, and HS-5 cells incubated with 15 at the concentrations of 4 and 40 μg/mL. The initial number of cells is 1.0 × 104/well (Adapted from ref (73), Copyright 2014 American Chemical Society).
Besides the representative examples above, there are increased reports on the use of enzymatic transformation and/or self-assembly to inhibit cancer cells or image cancer cells. For example, the self-assembly of a small diphenylalanine derivative accumulates selectively in cancer cells to form intracellular nanofibers that promiscuously interact with tubulins, actins, and vimentins to impede cytoskeleton dynamics.72 These promiscuous interactions are able to inhibit the growth of glioblastoma cells, but not neuronal cells.72,75 Recently, Zheng et al. reported the self-assembly of a porphyrin derivative to form liposomes and exhibit high efficiency for photodynamic therapy.85 Moreover, EISA to form the nanofibers of peptide derivatives, especially biostable d-peptides, prevents the diffusion and sometimes the degradation of the fluorescent probes, thus achieving high spatiotemporal resolution.37,78−80,86−90 In addition to multiple choices of enzymes, another unique advantage of EISA is that this methodology is not limited to peptides and their derivatives. The hybrid of different molecular building blocks (e.g., amino acids, nucleobases, carbohydrates, and many other motifs) can also result in nanofibers/hydrogel via enzymatic transformation, providing that there are sufficient intermolecular interactions.74,91−93 In addition to in vitro inhibition, our preliminary results indicate that the EISA of 11 also inhibit xenograft tumor of drug-resistant cell lines in vivo, further confirming EISA as a promising approach to potential cancer therapy.
The ALPP-instructed self-assembly to inhibit cancer cells is particularly revealing. ALPP has been identified as a cancer biomarker by Fishman over 50 years ago,94 but the monitoring of ALPP in human serum for detecting cancer has been unable to achieve high accuracy because ALPP is an ectoenzyme. Recent proteomic study reveals that the overexpression of ALPP is a generic difference between cancer and normal tissues.81,94 But the development of the inhibitors of ALPP and other phosphatases has been largely unsuccessful due to the poor selectivity of the inhibitors and the physiological importance of phosphatases. Thus, phosphatases have earned the reputation of “undruggable” enzymes.95 Using the combination of enzymatic transformation and self-assembly, we and others demonstrated that the overexpression of ALPP (or membrane-bound ALP) can be targeted via EISA.38,84 The principle established in these works may find applications to target other “undruggable” targets or “untargetable” features of cancer cells.
Future Perspective
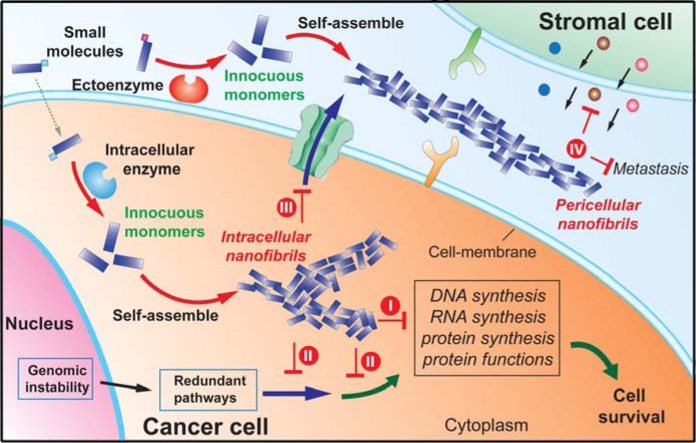
The merits of EISA as mentioned in this Review promise it to be a paradigm-shift approach for potential cancer therapy. As the case of nanofibers shown in Figure 9, EISA to generate pericellular and intracellular supramolecular nanofibers (or aggregates) can effectively modulate the pericellular and intracellular microenvironments, respectively. The intracellular nanofibers have large sizes to disfavor efflux, to interrupt protein interaction networks, and to block multiple cellular pathways,72 thus preventing cell survival via efflux pumps and evolutionary redundancy. The pericellular nanofibers entrap and isolate cancer cells to disrupt the microenvironment of cancer cells globally, and thus should directly block intercellular communications between cancer cells and the stromal cells, inhibit cancer cells, and prevent metastasis. Differing from the conventional approaches of enzyme inhibition, the use of enzyme catalysis to convert innocuous small molecules to cancer-inhibiting supramolecular nanofibers is unlikely to cause selective pressures for mutation, thus avoiding acquired drug resistance. Using the strategy of enzymatic transformation, the self-assembly/aggregation only occurs and elicits its desired functions/effects at a specific location where the enzyme of interest is overexpressed. This feature helps reduce side effects and improves selectivity—one of the reasons that EISA is superior to the direct use of the assemblies or aggregates of proteins61 or small molecules.66 Moreover, because of their supramolecular nature (i.e., formed by noncovalent interaction), the nanofibers can easily dissociate to innocuous monomers again after killing the cancer cells. This unique transitory feature is fundamentally different from conventional chemotherapy agents (e.g., cisplatin), thus greatly reducing or eliminating the systemic toxic effects in comparison to conventional cancer therapy.
Figure 9.
Plausible actions of the EISA to inhibit cancer cells: (I) interacting with multiple proteins to interrupt multiple proliferation processes and (II) interfering with redundant pathways, (III) eluding efflux pump, and (IV) blocking cellular mass exchange, prosurvival signals, and metastasis.
EISA holds great promise, especially in the context of engineering cell death signals for selectively killing cancer cells via apoptosis.96 Table 1 compares some inherent features in apoptosis and EISA that are being explored for potential cancer therapy. One fundamental conceptual advance in the study of apoptosis,97 especially in the tumor necrosis factor (TNF) superfamily of ligands and receptors, is that, even with the same ligand, paracrine and juxtacrine signaling may result paradoxical phenotypes. For example, transmembrane CD95L (as a juxtacrine) is proinflammatory, but soluble CD95L (as a paracrine) inhibits inflammation.98 In fact, the interaction between the cells and the self-assembled nanofibers resembles more juxtacrine than paracrine signaling; thus, the use of ectoenzymes99 of cancer cells to generate pericellular nanofibers (or aggregates) with spatiotemporal control offers a unique way to selective inhibit cancer cells. In that sense, supramolecular nanofibers of small molecules are a new class of signaling entities, and we propose to term them “pseudocrines” to reflect that they differ fundamentally from the prodrugs/drugs.
Table 1. Comparison of Some Key Components and Steps of Apoptosis and EISA.
| apoptosis |
EISA |
||||||
|---|---|---|---|---|---|---|---|
| locations | intrinsic | extrinsic | intracellular | pericellular | |||
| enzymes | Apaf-124 | ADAM1028 | MMP1429 | ADAM1730 | CES169 | MMP-777 | ALPP38,40,73,84 |
| substrates | dATP | mCD95L | mTRAIL | proTNF | 7 | 9 | 11, 13, 15 |
| products | dADP | CD95L | TRAIL | TNF | 8 | 10 | 12, 14, 16 |
| assemblies | apoptosomes | oligomers of death receptors | nanofibers of 8 | nanofibers of 10 | nanofibers of 12 or 14; aggregates of 16 | ||
Clearly, the use of EISA to generate pseudocrines for modulating cellular growth is just at its very beginning. There are still many challenges to develop EISA of small molecules for cancer therapy, but they will likely be met in the near future because of the rapid advancements in many other fields of science. For example, there is limited knowledge of the enzymes overexpressed on or in cancer cells. Encourgingly, the information accumulated in proteomics research is filling this gap.81 There are few methods and techniques readily available for studying the mechanisms of the EISA for selectively inhibiting cancer cells, though they significantly depart from the conventional chemotherapy and immunotherapy. However, the fast development of biomedical research agents is offering more reliable and convenient assays for the mechanistic elucidation. Although currently it is difficult to interrogate the atomistic structures of the polymorphic supramolecular nanofibers (or aggregates) formed by the self-assembling small molecules, the advancement of cryo-TEM100 and X-ray crystallography101 likely will come to the rescue. Despite the protocols for identifying the protein targets of the supramolecular nanofibers102,103 still being imperfect, the improvement of top-down mass spectrometry104 will help address this issue. The concentration required to inhibit cancer cells is still high, but the optimal balance among EISA, ligand–receptor interactions, and multivalency likely will provide a solution to this problem.
Therefore, EISA holds great potential, and the further exploration of this multiple step process, by addressing the above challenges, will eventually open up new directions for developing anticancer therapy to address the immense complexity of cancer. Moreover, considering that the self-assembly (or aggregation) of small molecules is such a prevalent phenomenon and more important66,105−110 than one thought, we expect that the development of EISA will go beyond cancer therapy, and find applications in fields of molecular imaging,86 antibacterial medicine,42,76 immunomodulation,111−115 wound healing,116,117 neurodegenerative diseases,118 tissue regeneration,119,120 and signaling transduction.121 Most importantly, the concept of EISA departs from the dogma of “lock and key”. The paradigm shift from a pair of molecules (i.e., inhibitor/enzyme or ligand/receptor) to an integrated multiple step process (i.e., enzymatic reaction, molecular aggregation, and promiscuous interactions) promises solutions to many unsolved problems in biological and medical science.
Acknowledgments
This work was partially supported by NIH (CA142746). J.Z. is a Howard Hughes Medical Institute (HHMI) International Research Fellow.
The authors declare no competing financial interest.
References
- American Cancer Society (2014) American Cancer Society. Cancer Facts & Figures 2014, American Cancer Society, Atlanta. [Google Scholar]
- Hanahan D.; Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144 (5), 646–674. [DOI] [PubMed] [Google Scholar]
- Doroshow J. H. (2013) Overcoming resistance to targeted anticancer drugs. N. Engl. J. Med. 369 (19), 1852–1853. [DOI] [PubMed] [Google Scholar]
- Holohan C.; Van Schaeybroeck S.; Longley D. B.; Johnston P. G. (2013) Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13 (10), 714–726. [DOI] [PubMed] [Google Scholar]
- Gupta G. P.; Massague J. (2006) Cancer metastasis: Building a framework. Cell 127 (4), 679–695. [DOI] [PubMed] [Google Scholar]
- Schreiber R. D.; Old L. J.; Smyth M. J. (2011) Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 331 (6024), 1565–1570. [DOI] [PubMed] [Google Scholar]
- Monteiro J. (2015) Cancer immunotherapy scores again. Cell 160 (1–2), 7–9. [Google Scholar]
- Aller S. G.; Yu J.; Ward A.; Weng Y.; Chittaboina S.; Zhuo R. P.; Harrell P. M.; Trinh Y. T.; Zhang Q. H.; Urbatsch I. L.; Chang G. (2009) Structure of p-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323 (5922), 1718–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna R.; Mayer L. D. (2000) Multidrug resistance (MDR) in cancer - Mechanisms, reversal using modulators of MDR and the role of MDR modulators in influencing the pharmacokinetics of anticancer drugs. Eur. J. Pharm. Sci. 11 (4), 265–283. [DOI] [PubMed] [Google Scholar]
- Wu A. M.; Senter P. D. (2005) Arming antibodies: prospects and challenges for immunoconjugates. Nat. Biotechnol. 23 (9), 1137–1146. [DOI] [PubMed] [Google Scholar]
- Armstrong D. K.; Bundy B.; Wenzel L.; Huang H. Q.; Baergen R.; Lele S.; Copeland L. J.; Walker J. L.; Burger R. A. (2006) Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N. Engl. J. Med. 354 (1), 34–43. [DOI] [PubMed] [Google Scholar]
- Yu M.; Ocana A.; Tannock I. F. (2013) Reversal of ATP-binding cassette drug transporter activity to modulate chemoresistance: why has it failed to provide clinical benefit?. Cancer Metastasis Rev. 32 (1–2), 211–227. [DOI] [PubMed] [Google Scholar]
- Rebucci M.; Michiels C. (2013) Molecular aspects of cancer cell resistance to chemotherapy. Biochem. Pharmacol. 85 (9), 1219–1226. [DOI] [PubMed] [Google Scholar]
- Brunton L. L., Parker K. L., Murri N., Buxton I., and Blumenth D. K. (2005) Goodman & Gilman’s The Pharmacological Basis of Therapeutics, 11th ed., McGraw-Hill, New York. [Google Scholar]
- Rodland K. D.; Bollinger N.; Ippolito D.; Opresko L. K.; Coffey R. J.; Zangar R.; Wiley H. S. (2008) Multiple Mechanisms Are Responsible for Transactivation of the Epidermal Growth Factor Receptor in Mammary Epithelial Cells. J. Biol. Chem. 283 (46), 31477–31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzetti S.; Ferlini C.; Concolino P.; Filippetti F.; Raspaglio G.; Prislei S.; Gallo D.; Martinelli E.; Ranelletti F. O.; Ferrandina G.; Scambia G. (2005) Class III beta-tubulin overexpression is a prominent mechanism of paclitaxel resistance in ovarian cancer patients. Clin. Cancer Res. 11 (1), 298–305. [PubMed] [Google Scholar]
- Samimi G.; Safaei R.; Katano K.; Holzer A. K.; Rochdi M.; Tomioka M.; Goodman M.; Howell S. B. (2004) Increased expression of the copper efflux transporter ATP7A mediates resistance to cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells. Clin. Cancer Res. 10 (14), 4661–4669. [DOI] [PubMed] [Google Scholar]
- Pyragius C. E.; Fuller M.; Ricciardelli C.; Oehler M. K. (2013) Aberrant lipid metabolism: an emerging diagnostic and therapeutic target in ovarian cancer. Int. J. Mol. Sci. 14 (4), 7742–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. M. (2002) Mechanisms of cancer drug resistance. Annu. Rev. Med. 53 (1), 615–627. [DOI] [PubMed] [Google Scholar]
- McMillin D. W.; Negri J. M.; Mitsiades C. S. (2013) The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat. Rev. Drug Discovery 12 (3), 217–228. [DOI] [PubMed] [Google Scholar]
- Liu X.; Chan D.; Ngan H. (2012) Mechanisms of chemoresistance in human ovarian cancer at a glance. OB/GYN 2 (3), 100–104. [Google Scholar]
- Gerlinger M.; Rowan A. J.; Horswell S.; Larkin J.; Endesfelder D.; Gronroos E.; Martinez P.; Matthews N.; Stewart A.; Tarpey P.; Varela I.; Phillimore B.; Begum S.; McDonald N. Q.; Butler A.; Jones D.; Raine K.; Latimer C.; Santos C. R.; Nohadani M.; Eklund A. C.; Spencer-Dene B.; Clark G.; Pickering L.; Stamp G.; Gore M.; Szallasi Z.; Downward J.; Futreal P. A.; Swanton C. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366 (10), 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S. A.; Yang J. C.; Sherry R. M.; Kammula U. S.; Hughes M. S.; Phan G. Q.; Citrin D. E.; Restifo N. P.; Robbins P. F.; Wunderlich J. R. (2011) Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin. Cancer Res. 17 (13), 4550–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner M. O. (2000) The biochemistry of apoptosis. Nature 407 (6805), 770–776. [DOI] [PubMed] [Google Scholar]
- Shi Y. (2006) Mechanical aspects of apoptosome assembly. Curr. Opin. Cell Biol. 18 (6), 677–684. [DOI] [PubMed] [Google Scholar]
- Baud V.; Karin M. (2001) Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11 (9), 372–377. [DOI] [PubMed] [Google Scholar]
- Wajant H.; Pfizenmaier K.; Scheurich P. (2003) Tumor necrosis factor signaling. Cell Death Differ. 10 (1), 45–65. [DOI] [PubMed] [Google Scholar]
- Schulte M.; Reiss K.; Lettau M.; Maretzky T.; Ludwig A.; Hartmann D.; de Strooper B.; Janssen O.; Saftig P. (2007) ADAM10 regulates FasL cell surface expression and modulates FasL-induced cytotoxicity and activation-induced cell death. Cell Death Differ. 14 (5), 1040–1049. [DOI] [PubMed] [Google Scholar]
- Hanada K.-i.; Wang Q. J.; Inozume T.; Yang J. C. (2011) Molecular identification of an MHC-independent ligand recognized by a human α/β T-cell receptor. Blood 117 (18), 4816–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black R. A.; Rauch C. T.; Kozlosky C. J.; Peschon J. J.; Slack J. L.; Wolfson M. F.; Castner B. J.; Stocking K. L.; Reddy P.; Srinivasan S.; Nelson N.; Boiani N.; Schooley K. A.; Gerhart M.; Davis R.; Fitzner J. N.; Johnson R. S.; Paxton R. J.; March C. J.; Cerretti D. P. (1997) A metalloproteinase disintegrin that releases tumor-necrosis factor-α from cells. Nature 385 (6618), 729–733. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Liang G.; Xu B. (2008) Enzymatic hydrogelation of small molecules. Acc. Chem. Res. 41 (2), 315–326. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Yang Z.; Kuang Y.; Ma M.-L.; Li J.; Zhao F.; Xu B. (2010) Enzyme-instructed self-assembly of peptide derivatives to form nanofibers and hydrogels. Biopolymers 94 (1), 19–31. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Gu H.; Fu D.; Gao P.; Lam J. K.; Xu B. (2004) Enzymatic formation of supramolecular hydrogels. Adv. Mater. 16 (16), 1440–1444. [Google Scholar]
- Yang Z. M.; Liang G. L.; Wang L.; Xu B. (2006) Using a kinase/phosphatase switch to regulate a supramolecular hydrogel and forming the supramoleclar hydrogel in vivo. J. Am. Chem. Soc. 128 (9), 3038–3043. [DOI] [PubMed] [Google Scholar]
- Gao J.; Wang H.; Wang L.; Wang J.; Kong D.; Yang Z. (2009) Enzyme promotes the hydrogelation from a hydrophobic small molecule. J. Am. Chem. Soc. 131 (32), 11286–11287. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Ma M.; Xu B. (2009) Using matrix metalloprotease-9 (MMP-9) to trigger supramolecular hydrogelation. Soft Matter 5 (13), 2546–2548. [Google Scholar]
- Li J. Y.; Gao Y.; Kuang Y.; Shi J. F.; Du X. W.; Zhou J.; Wang H. M.; Yang Z. M.; Xu B. (2013) Dephosphorylation of D-peptide derivatives to form biofunctional, supramolecular nanofibers/hydrogels and their potential applications for intracellular imaging and intratumoral chemotherapy. J. Am. Chem. Soc. 135 (26), 9907–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y.; Shi J.; Li J.; Yuan D.; Alberti K. A.; Xu Q.; Xu B. (2014) Pericellular hydrogel/nanonets inhibit cancer cells. Angew. Chem., Int. Ed. 53 (31), 8104–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Kuang Y.; Gao Y.; Du X.; Shi J.; Xu B. (2013) D-amino acids boost the selectivity and confer supramolecular hydrogels of a nonsteroidal anti-inflammatory drug (NSAID). J. Am. Chem. Soc. 135 (2), 542–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Du X.; Yuan D.; Zhou J.; Zhou N.; Huang Y.; Xu B. (2014) D-Amino acids modulate the cellular response of enzymatic-instructed supramolecular nanofibers of small peptides. Biomacromolecules 15 (10), 3559–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano S.; Williams R. J.; Jayawarna V.; Ulijn R. V. (2006) Enzyme-triggered self-assembly of peptide hydrogels via reversed hydrolysis. J. Am. Chem. Soc. 128 (4), 1070–1071. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Ho P.-L.; Liang G.; Chow K. H.; Wang Q.; Cao Y.; Guo Z.; Xu B. (2007) Using beta-lactamase to trigger supramolecular hydrogelation. J. Am. Chem. Soc. 129 (2), 266–267. [DOI] [PubMed] [Google Scholar]
- Guilbaud J.-B.; Vey E.; Boothroyd S.; Smith A. M.; Ulijn R. V.; Saiani A.; Miller A. F. (2010) Enzymatic catalyzed synthesis and triggered gelation of ionic peptides. Langmuir 26 (13), 11297–11303. [DOI] [PubMed] [Google Scholar]
- Das A. K.; Collins R.; Ulijn R. V. (2008) Exploiting enzymatic (reversed) hydrolysis in directed self-assembly of peptide nanostructures. Small 4 (2), 279–287. [DOI] [PubMed] [Google Scholar]
- Williams R. J.; Gardiner J.; Sorensen A. B.; Marchesan S.; Mulder R. J.; McLean K. M.; Hartley P. G. (2013) Monitoring the early stage self-assembly of enzyme-assisted peptide hydrogels. Aust. J. Chem. 66 (5), 572–578. [Google Scholar]
- Qin X.; Xie W.; Tian S.; Cai J.; Yuan H.; Yu Z.; Butterfoss G. L.; Khuong A. C.; Gross R. A. (2013) Enzyme-triggered hydrogelation via self-assembly of alternating peptides. Chem. Commun. 49 (42), 4839–4841. [DOI] [PubMed] [Google Scholar]
- Bremmer S. C.; Chen J.; McNeil A. J.; Soellner M. B. (2012) A general method for detecting protease activity via gelation and its application to artificial clotting. Chem. Commun. 48 (44), 5482–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronopoulou L.; Lorenzoni S.; Masci G.; Dentini M.; Togna A. R.; Togna G.; Bordi F.; Palocci C. (2010) Lipase-supported synthesis of peptidic hydrogels. Soft Matter 6 (11), 2525–2532. [Google Scholar]
- Song F.; Zhang L.-M. (2008) Enzyme-catalyzed formation and structure characteristics of a protein- based hydrogel. J. Mater. Chem. B 112 (44), 13749–13755. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Javvaji V.; Raghavan S. R.; Bentley W. E.; Payne G. F. (2012) Glucose oxidase-mediated gelation: a simple test to detect glucose in food products. J. Agric. Food Chem. 60 (36), 8963–8967. [DOI] [PubMed] [Google Scholar]
- Sakai S.; Komatani K.; Taya M. (2012) Glucose-triggered co-enzymatic hydrogelation of aqueous polymer solutions. RSC Adv. 2 (4), 1502–1507. [Google Scholar]
- Ogushi Y.; Sakai S.; Kawakami K. (2007) Synthesis of enzymatically-gellable carboxymethylcellulose for biomedical applications. J. Biosci. Bioeng. 104 (1), 30–33. [DOI] [PubMed] [Google Scholar]
- Sakai S.; Ogushi Y.; Kawakami K. (2009) Enzymatically crosslinked carboxymethylcellulose-tyramine conjugate hydrogel: cellular adhesiveness and feasibility for cell sheet technology. Acta Biomater. 5 (2), 554–559. [DOI] [PubMed] [Google Scholar]
- Choi Y. C.; Choi J. S.; Jung Y. J.; Cho Y. W. (2014) Human gelatin tissue-adhesive hydrogels prepared by enzyme-mediated biosynthesis of DOPA and Fe3+ ion crosslinking. J. Mater. Chem. B 2 (2), 201–209. [DOI] [PubMed] [Google Scholar]
- Yang Z.; Xu B. (2004) A simple visual assay based on small molecule hydrogels for detecting inhibitors of enzymes. Chem. Commun. 21, 2424–2425. [DOI] [PubMed] [Google Scholar]
- Schnepp Z. A. C.; Gonzalez-McQuire R.; Mann S. (2006) Hybrid biocomposites based on calcium phosphate mineralization of self-assembled supramolecular hydrogels. Adv. Mater. 18 (14), 1869–1872. [Google Scholar]
- Estroff L. A.; Hamilton A. D. (2004) Water gelation by small organic molecules. Chem. Rev. 104 (3), 1201–1217. [DOI] [PubMed] [Google Scholar]
- Terech P.; Weiss R. G. (1997) Low molecular mass gelators of organic liquids and the properties of their gels. Chem. Rev. 97 (8), 3133–3159. [DOI] [PubMed] [Google Scholar]
- Parker K., and Brunton L. (2008) Goodman and Gilman’s manual of pharmacology and therapeutics, McGraw-Hill Medical. [Google Scholar]
- Ulijn R. V. (2015) Molecular self-assembly: Best of both worlds, Nat. Nanotechnol. [Online early access]. [DOI] [PubMed] [Google Scholar]
- Hakansson A.; Zhivotovsky B.; Orrenius S.; Sabharwal H.; Svanborg C. (1995) Apoptosis induced by a human-milk protein. Proc. Natl. Acad. Sci. U. S. A. 92 (17), 8064–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duringer C.; Hamiche A.; Gustafsson L.; Kimura H.; Svanborg C. (2003) HAMLET interacts with histones and chromatin in tumor cell nuclei. J. Biol. Chem. 278 (43), 42131–42135. [DOI] [PubMed] [Google Scholar]
- Svanborg C.; Agerstam H.; Aronson A.; Bjerkvig R.; Duringer C.; Fischer W.; Gustafsson L.; Hallgren O.; Leijonhuvud I.; Linse S.; Mossberg A. K.; Nilsson H.; Pettersson J.; Svensson M. (2003) HAMLET kills tumor cells by an apoptosis-like mechanism - Cellular, molecular, and therapeutic aspects. Adv. Cancer Res. 88, 1–29. [DOI] [PubMed] [Google Scholar]
- Storm P.; Klausen T. K.; Trulsson M.; Ho C. S. J.; Dosnon M.; Westergren T.; Chao Y. X.; Rydstrom A.; Yang H.; Pedersen S. F.; Svanborg C. (2013) A unifying mechanism for cancer cell death through ion channel activation by HAMLET. PLoS One 8 (3), 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson L.; Leijonhufvud I.; Aronsson A.; Mossberg A.; Svanborg C. (2004) Treatment of skin papillomas with topical alpha-lactalbumin-oleic acid. New Engl. J. Med. 350 (26), 2663–2672. [DOI] [PubMed] [Google Scholar]
- Zorn J. A.; Wille H.; Wolan D. W.; Wells J. A. (2011) Self-assembling small molecules form nanofibrils that bind procaspase-3 to promote activation. J. Am. Chem. Soc. 133 (49), 19630–19633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien O.; Kampmann M.; Bassik M. C.; Zorn J. A.; Venditto V. J.; Shimbo K.; Agard N. J.; Shimada K.; Rheingold A. L.; Stockwell B. R.; Weissman J. S.; Wells J. A. (2014) Unraveling the mechanism of cell death induced by chemical fibrils. Nat. Chem. Biol. 10 (11), 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn J. A.; Wolan D. W.; Agard N. J.; Wells J. A. (2012) Fibrils colocalize caspase-3 with procaspase-3 to foster maturation. J. Biol. Chem. 287 (40), 33781–33795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. M.; Xu K. M.; Guo Z. F.; Guo Z. H.; Xu B. (2007) Intracellular enzymatic formation of nanofibers results in hydrogelation and regulated cell death. Adv. Mater. 17, 3152–3156. [Google Scholar]
- Gao Y.; Kuang Y.; Guo Z.-F.; Guo Z.; Krauss I. J.; Xu B. (2009) Enzyme-instructed molecular self-assembly confers nanofibers and a supramolecular hydrogel of taxol derivative. J. Am. Chem. Soc. 131 (38), 13576–13577. [DOI] [PubMed] [Google Scholar]
- Kuang Y.; Du X.; Zhou J.; Xu B. (2014) Supramolecular nanofibrils inhibit cancer progression in vitro and in vivo. Adv. Healthc. Mater. 3, 1217–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y.; Long M. J.; Zhou J.; Shi J.; Gao Y.; Xu C.; Hedstrom L.; Xu B. (2014) Prion-like nanofibrils of small molecules (prism) selectively inhibit cancer cells by impeding cytoskeleton dynamics. J. Biol. Chem. 289, 29208–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X.; Zhou J.; Wu L.; Sun S.; Xu B. (2014) Enzymatic transformation of phosphate decorated magnetic nanoparticles for selectively sorting and inhibiting cancer cells. Bioconjugate Chem. 25 (12), 2129–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D.; Zhou R.; Shi J.; Du X.; Li X.; Xu B. (2014) Enzyme-instructed self-assembly of hydrogelators consisting of nucleobases, amino acids, and saccharide. RSC Adv. 4 (50), 26487–26490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y.; Xu B. (2013) Disruption of the dynamics of microtubules and selective inhibition of glioblastoma cells by nanofibers of small hydrophobic molecules. Angew. Chem., Int. Ed. 52 (27), 6944–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Liang G.; Guo Z.; Guo Z.; Xu B. (2007) Intracellular hydrogelation of small molecules inhibits bacterial growth. Angew. Chem., Int. Ed. 46 (43), 8216–8219. [DOI] [PubMed] [Google Scholar]
- Tanaka A.; Fukuoka Y.; Morimoto Y.; Honjo T.; Koda D.; Goto M.; Maruyama T. (2015) Cancer cell death induced by the intracellular self-assembly of an enzyme-responsive supramolecular gelator. J. Am. Chem. Soc. 137 (2), 770–775. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Shi J.; Yuan D.; Xu B. (2012) Imaging enzyme-triggered self-assembly of small molecules inside live cells. Nat. Commun. 3, 2040/1–2040/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Kuang Y.; Du X.; Zhou J.; Chandran P.; Horkay F.; Xu B. (2013) Imaging self-assembly dependent spatial distribution of small molecules in a cellular environment. Langmuir 29 (49), 15191–15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Berciu C.; Kuang Y.; Shi J.; Nicastro D.; Xu B. (2013) Probing nanoscale self-assembly of nonfluorescent small molecules inside live mammalian cells. ACS Nano 7 (10), 9055–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M.; Fagerberg L.; Hallstroem B. M.; Lindskog C.; Oksvold P.; Mardinoglu A.; Sivertsson A.; Kampf C.; Sjoestedt E.; Asplund A.; Olsson I.; Edlund K.; Lundberg E.; Navani S.; Szigyarto C. A.-K.; Odeberg J.; Djureinovic D.; Takanen J. O.; Hober S.; Alm T.; Edqvist P.-H.; Berling H.; Tegel H.; Mulder J.; Rockberg J.; Nilsson P.; Schwenk J. M.; Hamsten M.; von Feilitzen K.; Forsberg M.; Persson L.; Johansson F.; Zwahlen M.; von Heijne G.; Nielsen J.; Ponten F. (2015) Tissue-based map of the human proteome. Science 347 (6220), 1260419–1–1260419–9. [DOI] [PubMed] [Google Scholar]
- Yang Z. M.; Liang G. L.; Xu B. (2007) Enzymatic control of the self-assembly of small molecules: a new way to generate supramolecular hydrogels. Soft Matter 3 (5), 515–520. [DOI] [PubMed] [Google Scholar]
- Fernley H. N.; Walker P. G. (1970) Inhibition of alkaline phosphatase by L-phenylalanine. Biochem. J. 116 (3), 543–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires R. A.; Abul-Haija Y. M.; Costa D. S.; Novoa-Carballal R.; Reis R. L.; Ulijn R. V.; Pashkuleva I. (2015) Controlling cancer cell fate using localized biocatalytic self-assembly of an aromatic carbohydrate amphiphile. J. Am. Chem. Soc. 137 (2), 576–579. [DOI] [PubMed] [Google Scholar]
- Lovell J. F.; Jin C. S.; Huynh E.; Jin H.; Kim C.; Rubinstein J. L.; Chan W. C. W.; Cao W.; Wang L. V.; Zheng G. (2011) Porphysome nanovesicles generated by porphyrin bilayers for use as multimodal biophotonic contrast agents. Nat. Mater. 10 (4), 324–332. [DOI] [PubMed] [Google Scholar]
- Ye D.; Shuhendler A. J.; Cui L.; Tong L.; Tee S. S.; Tikhomirov G.; Felsher D. W.; Rao J. (2014) Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo. Nat. Chem. 6 (6), 519–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D.; Shuhendler A. J.; Pandit P.; Brewer K. D.; Tee S. S.; Cui L.; Tikhomirov G.; Rutt B.; Rao J. (2014) Caspase-responsive smart gadolinium-based contrast agent for magnetic resonance imaging of drug-induced apoptosis. Chem. Sci. 5 (10), 3845–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A.; Mei B.; Wang W.; Hu W.; Li F.; Zhou J.; Yang Q.; Cui H.; Wu M.; Liang G. (2013) FITC-quencher based caspase 3-activatable nanoprobes for effectively sensing caspase 3 in vitro and in cells. Nanoscale 5 (19), 8963–8967. [DOI] [PubMed] [Google Scholar]
- Shen B.; Jeon J.; Palner M.; Ye D.; Shuhendler A.; Chin F. T.; Rao J. (2013) Positron emission tomography imaging of drug-induced tumor apoptosis with a caspase-triggered nanoaggregation probe. Angew. Chem., Int. Ed. 52 (40), 10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.; Wang X.; Wang D.; Liu F.; Mei B.; Tang A.; Jiang J.; Liang G. (2013) Multifunctional fluorescent probe for sequential detections of glutathione and caspase-3 in vitro and in cells. Anal. Chem. 85 (13), 6203–6207. [DOI] [PubMed] [Google Scholar]
- Li X.; Kuang Y.; Lin H.-C.; Gao Y.; Shi J.; Xu B. (2011) Supramolecular nanofibers and hydrogels of nucleopeptides. Angew. Chem., Int. Ed. 50 (40), 9365–9369. S9365/1-S9365/18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G.; Xu K.; Li L.; Wang L.; Kuang Y.; Yang Z.; Xu B. (2007) Using Congo red to report intracellular hydrogelation resulted from self-assembly of small molecules. Chem. Commun. 40, 4096–4098. [DOI] [PubMed] [Google Scholar]
- Du X.; Li J.; Gao Y.; Kuang Y.; Xu B. (2012) Catalytic dephosphorylation of adenosine monophosphate (AMP) to form supramolecular nanofibers/hydrogels. Chem. Commun. 48 (15), 2098–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman W. H.; Inglis N. R.; Green S.; Anstiss C. L.; Gosh N. K.; Reif A. E.; Rustigian R.; Krant M. J.; Stolbach L. L. (1968) Immunology and biochemistry of Regan isoenzyme of alkaline phosphatase in human cancer. Nature 219 (5155), 697–9. [DOI] [PubMed] [Google Scholar]
- Blaskovich M. A. T. (2009) Drug discovery and protein tyrosine phosphatases. Curr. Med. Chem. 16 (17), 2095–2176. [DOI] [PubMed] [Google Scholar]
- Wajant H.; Gerspach J.; Pfizenmaier K. (2013) Engineering death receptor ligands for cancer therapy. Cancer Lett. 332 (2), 163–174. [DOI] [PubMed] [Google Scholar]
- Nair P.; Lu M.; Petersen S.; Ashkenazi A. (2014) Apoptosis initiation through the cell-extrinsic pathway. Methods Enzymol. 544, 99–128. [DOI] [PubMed] [Google Scholar]
- Hohlbaum A. M.; Moe S.; Marshak-Rothstein A. (2000) Opposing effects of transmembrane and soluble Fas ligand expression on inflammation and tumor cell survival. J. Exp. Med. 191 (7), 1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalak G.; Ehrlich Y. H.; Olsen B. R. (2014) Ecto-protein kinases and phosphatases: an emerging field for translational medicine. J. Transl. Med. 12, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W. (2014) Cryo-EM enters a new era. eLife 3 (e03665), e03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsko G. A. (2014) 100 years of X-ray crystallography. Chem. Eng. News 92 (32), 42–43. [Google Scholar]
- Kuang Y.; Yuan D.; Zhang Y.; Kao A.; Du X.; Xu B. (2013) Interactions between cellular proteins and morphologically different nanoscale aggregates of small molecules. RSC Adv. 3 (21), 7704–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Long M. J. C.; Shi J.; Hedstrom L.; Xu B. (2012) Using supramolecular hydrogels to discover the interactions between proteins and molecular nanofibers of small molecules. Chem. Commun. 48 (67), 8404–8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr A. (2008) Top-down mass spectrometry. Nat. Methods 5 (1), 24. [Google Scholar]
- McGovern S. L.; Caselli E.; Grigorieff N.; Shoichet B. K. (2002) A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 45 (8), 1712–1722. [DOI] [PubMed] [Google Scholar]
- Owen S. C.; Doak A. K.; Ganesh A. N.; Nedyalkova L.; McLaughlin C. K.; Shoichet B. K.; Shoichet M. S. (2014) Colloidal drug formulations can explain ″bell-shaped″ concentration-response curves. ACS Chem. Biol. 9 (3), 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien O.; Kampmann M.; Bassik M. C.; Zorn J. A.; Venditto V. J.; Shimbo K.; Agard N. J.; Shimada K.; Rheingold A. L.; Stockwell B. R.; Weissman J. S.; Wells J. A. (2014) Unraveling the mechanism of cell death induced by chemical fibrils. Nat. Chem. Biol. 10 (11), 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M.; Han T. N. W.; Xie S. H.; Shi K.; Du X. L.; Wu L. C.; Mirzaei H.; Goldsmith E. J.; Longgood J.; Pei J. M.; Grishin N. V.; Frantz D. E.; Schneider J. W.; Chen S.; Li L.; Sawaya M. R.; Eisenberg D.; Tycko R.; McKnight S. L. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149 (4), 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing B. G.; Yu C. W.; Chow K. H.; Ho P. L.; Fu D. G.; Xu B. (2002) Hydrophobic interaction and hydrogen bonding cooperatively confer a vancomycin hydrogel: A potential candidate for biomaterials. J. Am. Chem. Soc. 124 (50), 14846–14847. [DOI] [PubMed] [Google Scholar]
- Frenkel Y. V.; Gallicchio E.; Das K.; Levy R. M.; Arnold E. (2009) Molecular dynamics study of non-nucleoside reverse transcriptase inhibitor 4- 4- 4- (E)-2-cyanoethenyl −2,6-dimethylphenyl amino −2-pyrimidinyl a mino benzonitrile (TMC278/Rilpivirine) aggregates: correlation between amphiphilic properties of the drug and oral bioavailability. J. Med. Chem. 52 (19), 5896–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F.; Heesters B. A.; Chiu I.; Gao Y.; Shi J.; Zhou N.; Carroll M. C.; Xu B. (2014) l-Rhamnose-containing supramolecular nanofibrils as potential immunosuppressive materials. Org. Biomol. Chem. 12 (35), 6816–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F.; Li J.; Zhou N.; Sakai J.; Gao Y.; Shi J.; Goldman B.; Browdy H. M.; Luo H. R.; Xu B. (2014) De novo chemoattractants form supramolecular hydrogels for immunomodulating neutrophils in vivo. Bioconjugate Chem. 25 (12), 2116–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra J. S.; Tian Y. F.; Jung J. P.; Collier J. H. (2010) A self-assembling peptide acting as an immune adjuvant. Proc. Natl. Acad. Sci. U. S. A. 107 (2), 622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesson C. B.; Huelsmann E. J.; Lacek A. T.; Kohlhapp F. J.; Webb M. F.; Nabatiyan A.; Zloza A.; Rudra J. S. (2014) Antigenic peptide nanofibers elicit adjuvant-free CD8+ T cell responses. Vaccine 32 (10), 1174–1180. [DOI] [PubMed] [Google Scholar]
- Rudra J. S.; Mishra S.; Chong A. S.; Mitchell R. A.; Nardin E. H.; Nussenzweig V.; Collier J. H. (2012) Self-assembled peptide nanofibers raising durable antibody responses against a malaria epitope. Biomaterials 33 (27), 6476–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.-Y. C.; McKeon R.; Goussev S.; Werb Z.; Lee J.-U.; Trivedi A.; Noble-Haeusslein L. J. (2006) Matrix metalloproteinase-2 facilitates wound healing events that promote functional recovery after spinal cord injury. J. Neurosci. 26 (39), 9841–9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-McCaw A.; Ewald A. J.; Werb Z. (2007) Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 8 (3), 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U. S. A 95 (23), 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T.; Morrison S. J.; Clarke M. F.; Weissman I. L. (2001) Stem cells, cancer, and cancer stem cells. Nature 414 (6859), 105–111. [DOI] [PubMed] [Google Scholar]
- Langer R.; Vacanti J. P. (1993) Tissue engineering. Science 260 (5110), 920–926. [DOI] [PubMed] [Google Scholar]
- Wu H. (2013) Higher-order assemblies in a new paradigm of signal transduction. Cell 153 (2), 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]