Abstract
Development of a prophylactic vaccine against hepatitis C virus (HCV) has been hampered by the extraordinary viral diversity and the poor host immune response. Scaffolding, by grafting an epitope onto a heterologous protein scaffold, offers a possible solution to epitope vaccine design. In this study, we designed and characterized epitope vaccine antigens for the antigenic sites of HCV envelope glycoproteins E1 (residues 314–324) and E2 (residues 412–423), for which neutralizing antibody-bound structures are available. We first combined six structural alignment algorithms in a “scaffolding meta-server” to search for diverse scaffolds that can structurally accommodate the HCV epitopes. For each antigenic site, ten scaffolds were selected for computational design, and the resulting epitope scaffolds were analyzed using structure-scoring functions and molecular dynamics simulation. We experimentally confirmed that three E1 and five E2 epitope scaffolds bound to their respective neutralizing antibodies, but with different kinetics. We then investigated a “multivalent scaffolding” approach by displaying 24 copies of an epitope scaffold on a self-assembling nanoparticle, which markedly increased the avidity of antibody binding. Our study thus demonstrates the utility of a multi-scale scaffolding strategy in epitope vaccine design and provides promising HCV immunogens for further assessment in vivo.
Hepatitis C is a global health burden with approximately 180 million chronic carriers of hepatitis C virus (HCV) worldwide1,2,3,4. Although small-molecule drugs are available for treatment5, a prophylactic vaccine will offer a cost-effective means to prevent HCV transmission and to eventually eradicate the virus6,7,8. However, due to the poor fidelity of the viral RNA polymerase9 and the high replication rate (1012 virions/day), HCV exhibits extraordinary genetic diversity with over 30% sequence difference between genotypes10, posing a significant challenge to the development of a broadly effective vaccine.
HCV entry is mediated by the interactions between viral envelope glycoproteins and cell surface receptors such as CD81, scavenger receptor class B type I (SR-BI), and other host entry factors11,12. While it is known that glycoproteins E1 and E2 form heterodimers on the surface of a virion, how they mediate viral membrane fusion and cell entry remain elusive13,14,15,16. Neutralizing antibodies (nAbs) induced during natural infection have been mapped onto both E1 and E2. Early studies in infected humans and chimpanzees showed that the highly variable region 1 (HVR1) located at the N-terminus of E2 (residues 384–410) is a major target for the antibody response17,18,19,20,21,22. Recently, several E2-directed broadly neutralizing antibodies (bnAbs) have been identified23,24,25,26,27. Structures of the E1 antigenic site (residues 314–324) in complex with antibody IGH52628,29 and the E2 antigenic site (residues 412–423) bound to murine and human bnAbs26,30,31,32,33,34 have revealed critical details of HCV epitope recognition by the humoral immune system. More recently, Kong et al. reported a 2.65 Å crystal structure of the E2 core domain (E2c) in complex with a human bnAb AR3C35, which showed a stark difference from the class II fusion protein model found in other Flaviviridae viruses36. This finding was confirmed by another E2c structure determined by Khan et al.13 It is foreseeable that this ever-accumulating structural information will enable an in-depth understanding of HCV biology as well as structure-based design of epitope vaccines.
Conventional approaches have failed to produce broadly effective vaccines for highly variable viral pathogens such as HCV and human immunodeficiency virus type-1 (HIV-1), which have evolved various mechanisms such as glycan shielding and immune decoys to evade host immunity37,38. Epitope grafting, or scaffolding, has been proposed as a possible solution to epitope vaccine design by transferring a conserved neutralizing epitope onto heterologous protein scaffolds39. For HIV-1, scaffold antigens have been designed for two linear epitopes in the membrane proximal external region (MPER) that are recognized by bnAbs 4E1040 and 2F541, respectively. A large-scale in-silico design, coupled with high-throughput antigenic screening, was recently reported for three major HIV-1 epitopes, with up to 50% of the designs able to bind their respective bnAbs42. Scaffolding was also applied to the antigenic site II of the fusion (F) protein of respiratory syncytial virus (RSV)43, and neutralizing antibodies were induced by a scaffolded epitope in macaques44. Based on these studies, a general strategy consisting of epitope identification, scaffolding, particulate presentation of the designed antigen, animal immunization, and quantitative analysis of immune response has been proposed for epitope vaccine design and evaluation45.
In this study, we sought to develop robust scaffolding and particulate display methods for epitope vaccine design and with which to design epitope vaccine immunogens for the antigenic sites of HCV glycoproteins E1 (residues 314–324) and E2 (residues 412–423). We first developed a scaffolding meta-server by combining six structural alignment algorithms for scaffold search. We demonstrated increased coverage and diversity of the meta-server for an HIV-1 epitope and obtained a large number of scaffolds for the two HCV antigenic sites studied. We grafted the E1 and E2 antigenic sites onto twenty selected scaffolds, ten for each site, and analyzed the resulting antigens using residue-based quality scores and explicit-water molecular dynamics simulation. We characterized these epitope scaffolds by mammalian expression and antibody binding assays, with antigenicity confirmed for three E1 and five E2 designs. We then explored a multivalent scaffolding approach by displaying 24 copies of an epitope scaffold on a self-assembling ferritin nanoparticle. For both antigenic sites, the nanoparticles exhibited slow off-kinetics, suggesting a strong avidity effect due to multivalent antibody binding. We expect that the antigenic epitope scaffolds and nanoparticles will provide promising candidates for animal studies, while the new scaffolding methods will find many applications in epitope vaccine design.
Results
Development of a scaffolding meta-server
Scaffold search is the first step of epitope scaffold design, preceding epitope grafting and antigen optimization45. To date, most design studies have been conducted using a single structural alignment algorithm for scaffold search, which sampled only a subspace of protein structures in an algorithm-specific manner40,41,42,43,44,46,47. As a large number of structural alignment tools can be adapted for scaffolding, and meta-servers that combine output from multiple methods are widely accepted as a practical solution to difficult structure prediction problems48, we sought to develop a scaffolding meta-server to facilitate epitope vaccine design. We first examined the coverage of two previously reported scaffolding algorithms. Ofek et al. used MAMMOTH49 to search for scaffolds for the HIV-1 epitope recognized by 2F541, whereas Zhou et al. utilized TM-align50 to identify scaffolds for three major HIV-1 bnAb epitopes42. In our benchmark test, MAMMOTH and TM-align identified 273 and 262 scaffolds for the 2F5 epitope, respectively, with a Cα root-mean-square deviation (RMSD) of 1.5 Å or lower in the epitope-matching region. Of these scaffolds, 79 are identical, suggesting that the two algorithms identified a set of different but overlapping scaffolds. We then included four more structural alignment algorithms—a C implementation of TM-align50, SPalign51, CLICK52, and FAST53, which identified 539, 331, 74, and 152 scaffolds, respectively (Fig. S1). Together, six scaffolding algorithms yielded 1,003 non-redundant scaffolds with a pairwise overlap of 22–202, indicating that a large protein structural space has been covered in the search. Our results thus illustrate the improved coverage by combining six scaffolding algorithms in a meta-server and a consensus-based approach for scaffold selection. An epitope scaffold design pipeline was devised based on the scaffolding meta-server (Fig. 1, see Methods) and applied to the two HCV antigenic sites.
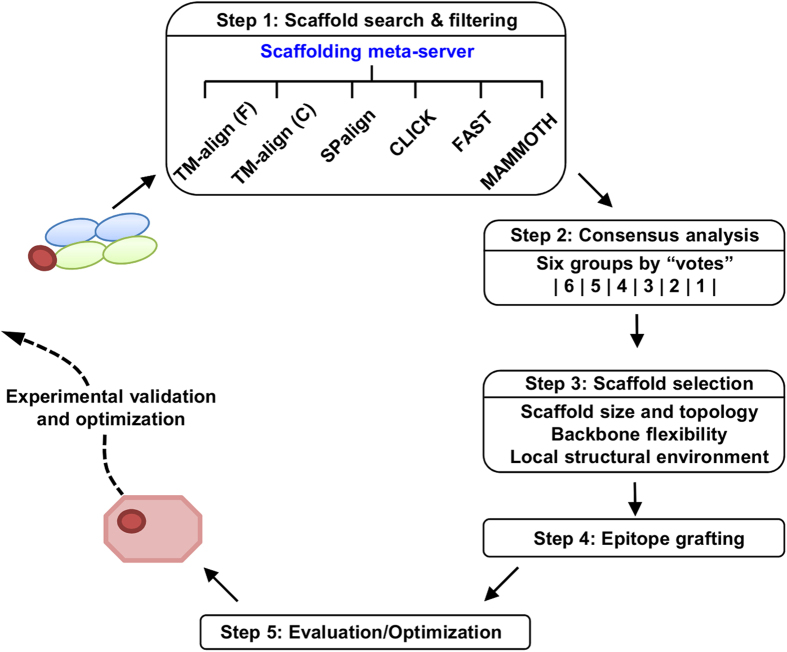
Figure 1. Schematic flow of an epitope scaffold design pipeline based on the scaffolding meta-server (step 1), a consensus-based analysis (step 2), scaffold selection (step 3), side chain-based epitope grafting (step 4), and evaluation and optimization of the designed antigens (step 5).
Epitope scaffold design for the E1 antigenic site 314–324
A 1.75 Å crystal structure was recently determined for an E1 antigenic site (314TGHRMAWDMMM324) in complex with human nAb IGH52628,29. (n.b. residue range is indicated by the superscripts hereafter). The crystal structure (PDB ID 4N0Y) revealed an 11-aa helical peptide bound to a hydrophobic groove formed by the complementarity determining regions of the heavy chain (HCDRs) and light chain (LCDRs) of IGH526 (Fig. S2). The phenyl rings of a tyrosine (Y100E) and a phenylalanine (F98) in HCDR3 interact with a histidine (H316) and the aliphatic portion of an arginine (R317) in the E1 epitope, respectively, with additional stabilizing contacts provided by the base of LCDR3 (W91 and N95) and the last residue (W50) of framework 2 (FW2). Based on the structural and functional information, we hypothesized that a scaffold that can present this E1 helical epitope in the IGH526-bound conformation may elicit E1-specific nAbs, and furthermore, a β-sheet-rich scaffold environment surrounding the grafted E1 helical epitope may enhance the specificity of elicited antibody response as antibodies often employ different types of interaction to recognize epitopes of different secondary structures45.
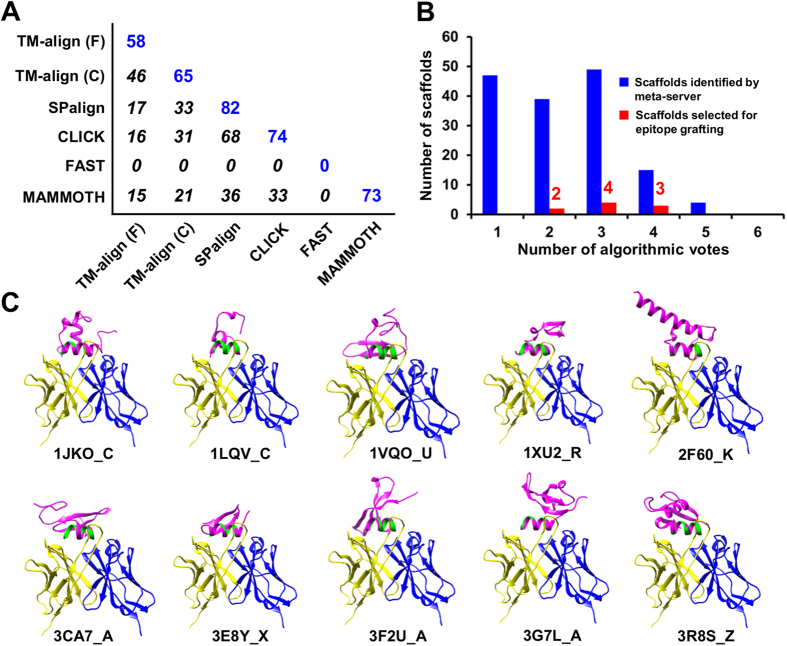
Collectively, the meta-server identified 704 non-redundant scaffolds, using input parameters consisting of a protein size of 30–60 aa, an epitope matching length of 10–11 aa, and a Cα RMSD cutoff of 2.0 Å. After removing scaffolds with a clash score over 5.0 with the docked IGH526, 154 candidates remained. We then calculated the number of scaffolds contributed by each algorithm and the overlap between any two algorithms (Fig. 2A). Similar to the 2F5 case, we observed an improved coverage of scaffold search. Among the six tested algorithms, SPalign appeared to be the best performer by providing 82 scaffolds, while the two versions of TM-align yielded 77 non-redundant scaffolds, accounting for 50% of the group. Surprisingly, FAST did not find any scaffolds, indicating that this algorithm may be less advantageous for short helices than for full-length proteins with distinctive structural features. We next ranked the scaffolds according to the count of algorithms (“votes”) by which they were identified (Figs 1, 2B). As expected, fewer scaffolds were found as the count increases, with 49 by three, 15 by four, 4 by five, and none by all six algorithms. After manual inspection, a total of 9 scaffolds were selected from the voting groups with 2, 3 and 4 counts (Fig. 2B). Criteria such as scaffold size, topology, flexibility, and structural environment of the epitope were major considerations in the selection45. We also included a cysteine knot, 3CA7_A, which has been used to scaffold the HIV-1 V3 epitope42 (n.b. a scaffold is denoted by the PDB ID followed by chain ID hereafter, which specifies a unique entry in the non-redundant protein database compiled by PISCES54, see Methods). Together, ten scaffolds were advanced to the design stage (Table 1, Table S1 and Fig. 2C), with 8–11 residues matched to the epitope by a Cα RMSD of 1.6 Å or lower and an epitope exposure ratio of 0.55 or greater in the scaffold context, except for 1JKO_C.
Figure 2. Computational design of epitope scaffolds for HCV E1 antigenic site 314–324.
(A) Coverage matrix of scaffolds identified by six structural alignment algorithms used in the scaffolding meta-server. The number of scaffolds identified by each individual algorithm is shown in blue and the overlap between two algorithms is shown in italic. (B) Consensus analysis of the scaffolds identified by the scaffolding meta-server after filtering (blue) and manually selected for epitope grafting (red). The number of scaffolds is plotted as a function of the number of algorithms (votes) by which a scaffold is identified. A “cysteine knot” (PDB ID: 3CA7) is included to render a total of 10 scaffolds for epitope grafting, energy evaluation and explicit-water MD simulation (Figs S4–S6). (C) Ten protein scaffolds identified by the scaffolding meta-server and manual selection are superimposed onto the E1 epitope in complex with the human neutralizing antibody IGH526. All protein structures are shown as a ribbon model with the scaffold colored in magenta, epitope in green.
Table 1. Protein scaffolds identified for HCV glycoprotein E1 antigenic site 314–324a.
| # | PDB ID | Chain | Selection | Nres | Nali | Cα RMSD (Å) | TM-score | SA ratio | Clash | Fitting | Votes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1JKO | C | Meta-server | 46 | 10 | 0.41 | 0.32 | 0.39 | 0.00 | TM-align (F) | 2 |
| 2 | 1LQV | C | Meta-server | 33 | 11 | 0.57 | 0.33 | 0.62 | 1.53 | TM-align (F) | 4 |
| 3 | 1VQO | U | Meta-server | 53 | 10 | 1.59 | N/Ac | 0.57 | 0.00 | MAMMOTH | 3 |
| 4 | 1XU2 | R | Meta-server | 36 | 11 | 1.24 | 0.26 | 0.82 | 1.11 | TM-align (F) | 3 |
| 5 | 2F60 | K | Meta-server | 60 | 11 | 0.28 | 0.31 | 0.56 | 1.05 | TM-align (F) | 4 |
| 6 | 3CA7b | A | Cysteine knot | 50 | 8 | 0.25 | 0.26 | 0.55 | 2.69 | TM-align (F) | − |
| 7 | 3E8Y | X | Meta-server | 30 | 10 | 0.24 | 0.42 | 0.63 | 4.67 | TM-align (F) | 2 |
| 8 | 3F2U | A | Meta-server | 51 | 11 | 1.23 | 0.31 | 0.70 | 0.00 | TM-align (F) | 3 |
| 9 | 3G7L | A | Meta-server | 55 | 10 | 0.16 | 0.30 | 0.67 | 1.99 | TM-align (F) | 3 |
| 10 | 3R8S | Z | Meta-server | 58 | 11 | 0.49 | 0.30 | 0.57 | 0.00 | TM-align (F) | 4 |
aListed items include scaffold index, PDB identifier, selection method, chain name, number of residues in the scaffold, number of scaffold residues aligned to the epitope, Cα RMSD of aligned residues, solvent accessibility ratio of the epitope-matching region in the scaffold context versus the epitope alone, scaffold-antibody clash score after docking the scaffold into the epitope-antibody complex, fitting algorithm used in the scaffold docking, and the number of algorithmic votes each scaffold received.
bSelected from the “cysteine knot” structural superfamily or a set of scaffolds identified for the HIV-1 epitope at the V3 base recognized by a broadly neutralizing antibody PGT128 (Ref. 41).
cTM-score was only calculated for the scaffolds identified and fitted by TM-align and SPalign, which provide TM-score as part of the output.
A key requirement for epitope grafting is to maintain epitope-antibody interactions and to minimize unfavorable interactions between the graft and the scaffold environment. As indicated by the sequence alignment of epitope-matching regions (Fig. S3), on average 7–8 critical residues in the E1 antigenic site were grafted onto a scaffold by superimposing the backbone atoms (N, Cα and C). Up to four mutations were introduced to avoid the scaffold side chains occluding antibody access or affecting protein stability due to exposed hydrophobic patches or buried charges (Fig. 2C). The flexible N- or C-terminus that may interfere with IGH526 binding was also truncated. The designed epitope scaffolds, indicated by the postfix “_ES” hereafter, were then evaluated in the presence of IGH526 using two residue-based local quality functions, a statistical potential DFIRE55, and a tabulated soft-core van der Waals (vdW) function (Fig. S4). Both functions have been used to detect local structural errors in homology models56. For these antigens, the local DFIRE score ranges from −1.41 to 4.20 with an average score of 0.75 and, in most cases, is below the 2.0 cutoff except for two regions (residues 5–7 and 22–24) in 3CA7_A_ES. The local vdW score, however, reported potential clashes between the grafted epitope residues and IGH526 or, in some cases, the scaffold (Fig. S4). These clashes could be readily removed by side-chain remodeling and energy minimization (see Methods), suggesting they were likely caused by rigid-body grafting rather than structural errors. MD simulations were performed to study the dynamics of epitope scaffolds in solution. The average Cα RMSD showed different levels of conformational differences relative to the initial model, ranging from 0.83 to 3.72 Å (Fig. S5). This global structural mobility was confirmed by the Cα RMS fluctuation (Cα RMSF) calculated from the conformational ensemble generated for each designed antigen (Fig. S6). The grafted epitope appeared to remain stable relative to the scaffold, showing an average Cα RMSF of 0.90 Å, compared to 1.81 Å for the whole protein.
Experimental validation and case analysis of the E1 epitope scaffolds
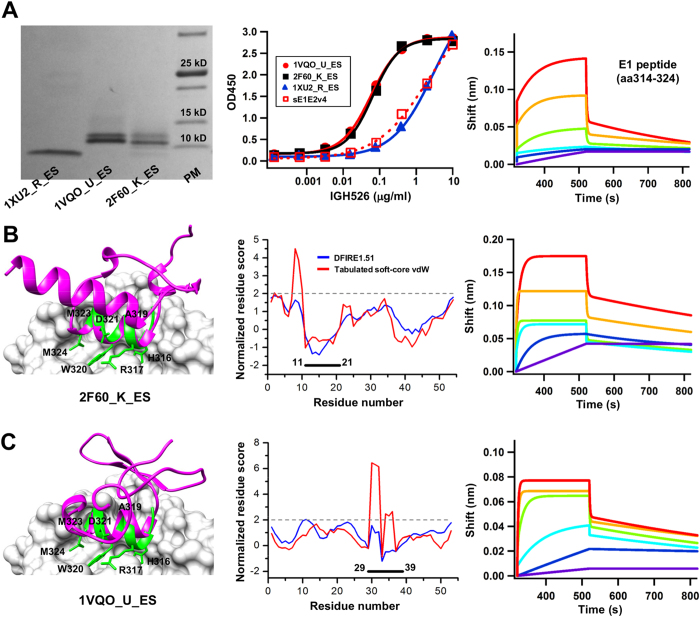
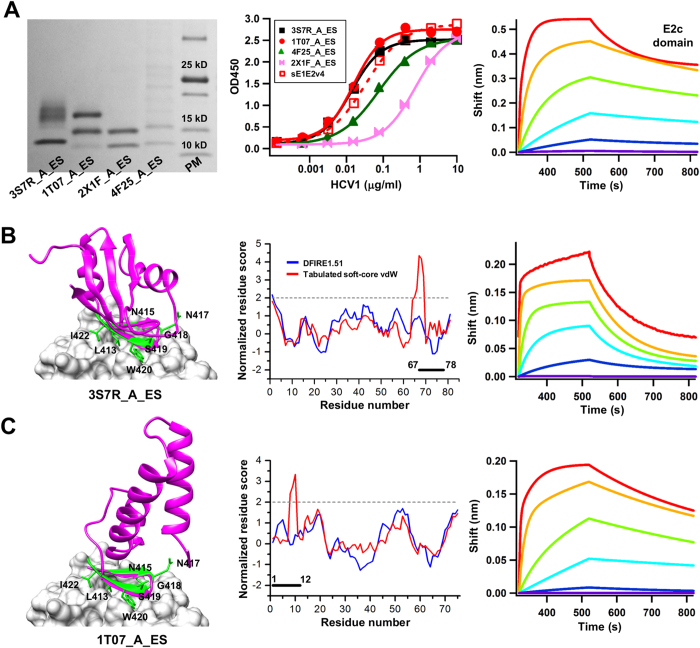
His-tagged epitope scaffolds were expressed transiently in HEK293F cells, purified with size exclusion chromatography (SEC), and analyzed by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Three epitope scaffolds, 1VQO_U_ES, 1XU2_R_ES and 2F60_K_ES, showed expression on SDS-PAGE (Fig. 3A, left). The antigenic properties were assessed by enzyme-linked immunosorbent assay (ELISA), which showed more pronounced IGH526 binding for 2F60_K_ES and 1VQO_U_ES than for 1XU2_R_ES (Fig. 3A, middle), using a soluble E1E257 as a control. Overall, the results indicate that the E1 antigenic site presented by these protein scaffolds can be recognized by antibody IGH526, for which the E1 peptide displayed slow-on/fast-off binding kinetics on Octet (Fig. 3A, right).
Figure 3. Experimental validation and case analysis of the E1 epitope scaffolds.
(A) SDS PAGE analysis of three expressed epitope scaffolds containing the E1 antigenic site (314–324) under reducing conditions, followed by Coomassie blue staining (left), ELISA analysis of antigen binding to the neutralizing antibody IGH526 (middle), and Octet measurement of the E1 epitope-containing peptide (right). (B,C) Structural modeling (left), local quality assessment (middle), and Octet kinetics measurement (right) of the E1 epitope scaffolds designed based on (B) 2F60_K, and (C) 1VQO_U. The epitope scaffold model (magenta) is superimposed onto the epitope (green) in complex with IGH526 (surface). The side chains of the epitope residues grafted onto the scaffolds are shown in stick model and labeled. The normalized residue-based scores calculated from DFIRE (blue) and a tabulated soft-core vdW energy function (red) are plotted as a function of the residue number. The sensorgrams from the Octet RED96 system show the binding of E1 peptide and epitope scaffolds to IGH526 using an antigen titration series of six starting at a maximum concentration of 100 μg/ml and 50 μg/ml with two- and four-fold dilutions, respectively. The antibody concentration was fixed at 5 μg/ml.
2F60_K is the binding domain (E3BD) of human dihydrolipoamide dehydrogenase (E3) binding protein (E3BP)58. Crystal structures revealed a rigid-body association of E3BD and E3, with a Cα RMSD of 0.7 Å between the bound and unbound forms of E3BD. Given a Cα RMSD of 0.28 Å between the N-terminal region (11–21) and the E1 antigenic site (Table 1), 2F60_K appeared to be an ideal scaffold to display this epitope (Fig. 3B, left). We transplanted E1 residues 316HR317, 319AWD321, and 323MM324 onto 2F60_K, and shortened the C-terminus (55–60) to reduce non-specific antibody response. We mutated the scaffold residues 8RF9 to alanine to avoid clashes with IGH526, and added a K40A mutation to reduce clashes with the grafted E1 R317 (Fig. 3B, left). The double mutation led to a more favorite DFIRE score, although the vdW score was still two-fold above the threshold (Fig. 3B, middle). In MD simulations, the grafted epitope showed an average Cα RMSF of 2.0 Å, compared to 2.7 Å for the whole protein. Using Octet, we quantified the IGH526 binding kinetics of 2F60_K_ES (Fig. 3B, right), which showed a faster on-rate than that of the E1 peptide (Fig. 3A, right).
1VQO_U is a 50S ribosomal protein (L24E) found in the large ribosomal subunit59 (Fig. 3C, left). We grafted E1 residues 316HR317, 319AWD321, and 323MM324 onto 1VQO_U, and introduced a double mutation T16G/F18A to minimize scaffold-antibody clashes. We also mutated the scaffold R41 to alanine to minimize clashes with the grafted E1 W320 and IGH526. We observed a similar pattern of local quality scores, with DFIRE indicating near-native contacts while vdW function suggesting potential clashes for residues 30–32, two of which (316HR317) were grafted from the E1 epitope (Fig. 3C, middle). MD simulations suggested that the grafted epitope would remain stable in the scaffold context, with an average Cα RMSF of 2.0 Å, compared to 2.3 Å for the whole protein. When tested for IGH526 binding using the Octet, 1VQO_U_ES showed a faster on-rate than 2F60_K_ES and the E1 peptide (Fig. 3C, right).
Taken together, the computational design coupled with experimental screening resulted in two antibody-reactive E1 epitope scaffolds with modest affinity and similar kinetics. Most scaffolds are components of large molecular complexes, explaining the difficulty in expressing these scaffold antigens as monomers (Fig. 3A, left). We also noted that the protein size cutoff (≤60 aa) might have excluded some larger scaffolds that are suitable for presentation of this epitope. Nevertheless, our results demonstrated that the E1 antigenic site can be displayed on heterologous protein scaffolds in a nAb-bound conformation.
Epitope scaffold design for the E2 antigenic site 412–423
The E2 glycoprotein is a major target for neutralizing antibodies and is responsible for facilitating cell entry by interacting with its receptor CD8160. Several cross-genotype neutralizing antibodies recognizing E2 epitopes have been identified23,24,25. Crystal structures of an E2 antigenic site (412QLINTNGSWHIN423) in complex with murine antibody AP3326,30,33 and human antibody HCV131 revealed a 12-aa epitope in the E2 N-terminus, which adopted a β-hairpin conformation with hydrophilic and hydrophobic faces on the opposing sides. Both antibodies recognize the CD81-receptor binding site (CD81bs) and neutralize HCV by blocking CD81 association. Remarkably, two independent antibody maturation pathways have arrived at a similar solution to engage the hydrophobic face of this epitope30,31. We hypothesized that a protein scaffold with an exposed β-hairpin resembling the structure of E2 antigenic site can be used to present this epitope in AP33- or HCV1-bound conformation and elicit epitope-specific neutralizing antibodies.
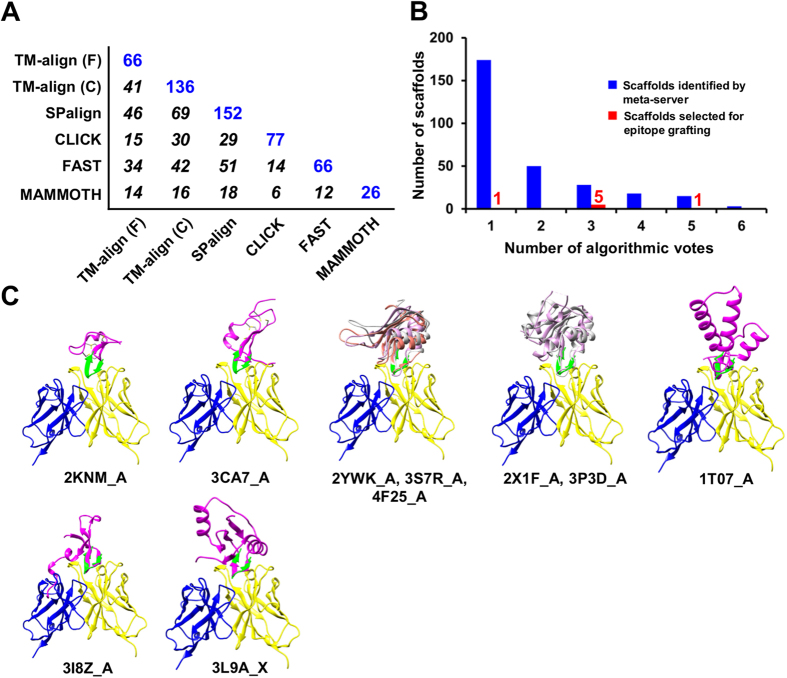
A total of 2,564 non-redundant scaffolds were identified by the scaffolding meta-server, using coordinates of the E2 epitope as a template and search criteria comprising a protein size of 40–100 aa, an epitope-match length of 10–12 aa, and a Cα RMSD of 2.0 Å or less. Nearly 89% of the scaffolds were removed in the clash filtering step. The coverage matrix of scaffold search was determined for the remaining 288 scaffolds (Fig. 4A). Among these scaffolds, SPalign contributed 152, while the two TM-align algorithms provided a combined set of 161, accounting for 56% of the scaffolds. The consensus analysis revealed a different pattern of distribution, with majority of the scaffolds identified by a single algorithm (Fig. 4B). Based on similar criteria45, we manually selected 5 and 1 scaffolds from the voting groups with 3 and 5 counts, respectively, and a top-ranking candidate from the voting group with one count (Figs 1, 4B). We also included 2KNM_A and 3CA7_A to investigate the utility of “cysteine knots” as scaffolds to present the E2 antigenic site, and 3I8Z_A from a previous design study based on the structural similarity between the E2 epitope and the HIV-1 V3 epitope42 (Table 2, Table S2, and Fig. 4C). Further analysis revealed three scaffold groups: two structural homolog groups consisting of 2YWK_A, 3S7R_A and 4F25_A, and of 2X1F_A and 3P3D_A, respectively, and a cysteine knot group of 2KNM_A and 3CA7_A (Fig. 4C).
Figure 4. Computational design of epitope scaffolds for HCV E2 antigenic site 412–423.
(A) Coverage of scaffolds identified by six structural alignment algorithms used in the scaffolding meta-server, with the number of scaffolds identified by each individual algorithm shown in blue and the overlap between two algorithms in italic. (B) Consensus analysis of the scaffolds identified by the scaffolding meta-server after filtering (blue) and manually selected for epitope grafting (red). The number of scaffolds is plotted as a function of the number of algorithms (votes) by which a scaffold is identified. Two “cysteine knots” (PDB IDs: 2KNM and 3CA7) and another protein (PDB ID: 3I8Z) are included to render a total of 10 scaffolds for epitope grafting, energy evaluation and explicit-water MD simulation (Figs S8–S10). (C) Ten protein scaffolds identified by the scaffolding meta-server and manual selection are superimposed onto the E2 epitope in complex with the human neutralizing antibody HCV1. All protein structures are shown as a ribbon model with the scaffold colored in magenta, epitope in green, with an exception for two groups of structural homologs for which the scaffolds are colored in magenta, gray, and orange.
Table 2. Protein scaffolds identified for HCV glycoprotein E2 antigenic site 412–423a.
| # | PDB ID | Chain | Selection | Nres | Nali | Cα RMSD (Å) | TM-score | SA ratio | Clash | Fitting | Votes |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2KNMb | A | Cysteine knot | 30 | 2 | 0.02 | N/Ac | 0.09 | 1.65 | Manual fitting | − |
| 2 | 3CA7b | A | Cysteine knot | 50 | 6 | 0.61 | N/Ac | 0.31 | 11.58 | Manual fitting | − |
| 3 | 2YWK | A | Meta-server | 84 | 12 | 1.26 | 0.19 | 0.57 | 0.00 | TM-align (C) | 3 |
| 4 | 3S7R | A | Meta-server | 81 | 12 | 1.48 | 0.20 | 0.56 | 0.00 | TM-align (F) | 3 |
| 5 | 4F25 | A | Meta-server | 81 | 10 | 1.10 | N/Ac | 0.58 | 0.00 | FAST | 3 |
| 6 | 2X1F | A | Meta-server | 94 | 12 | 1.43 | 0.18 | 0.72 | 0.00 | TM-align (C) | 3 |
| 7 | 3P3D | A | Meta-server | 87 | 12 | 1.11 | 0.20 | 0.60 | 0.00 | TM-align (C) | 3 |
| 8 | 1T07 | A | Meta-server | 81 | 12 | 1.77 | 0.18 | 0.51 | 0.00 | TM-align (C) | 1 |
| 9 | 3I8Zb | A | PGT128 scaffold | 50 | 12 | 0.70 | 0.32 | 0.71 | 36.78 | TM-align (F) | − |
| 10 | 3L9A | X | Meta-server | 81 | 12 | 1.18 | 0.22 | 0.64 | 1.60 | TM-align (F) | 5 |
a-cSee Table 1 for detailed explanation of listed items
Two grafting strategies were explored to design E2 epitope scaffolds: to graft the entire hairpin (413–422) onto a scaffold, as exemplified for the cysteine knot group, or to graft the hydrophobic face only, as for the other eight scaffolds (Fig. 4C). Of the 6–7 grafted E2 residues, a conserved N-linked glycosylation site was located in the turn of the E2 hairpin, 417NGS419 (Fig. S7). Other potential N-linked glycosylation sites in the scaffolds, such as N38 in 4F25_A_ES and N58 in 3L9A_X_ES, were mutated to glutamine. Up to three mutations were introduced to the non-graft regions to remove steric clashes and unfavorable interactions and to improve the protein stability. As indicated by the overall favorite DIFRE scores (Fig. S8), most E2 epitope-HCV1 interactions appeared to be preserved, although minor clashes due to rigid-body grafting were detected by the vdW function. In MD simulations, the E2 epitope scaffolds have undergone a more visible global conformational shift compared to their E1 counterparts, with the average Cα RMSD ranging from 1.5 to 3.9 Å (Fig. S9). Despite the global shift, the epitope scaffolds showed less internal variation, with a Cα RMSF of 0.6 Å for the graft and a Cα RMSF of 1.4Å for the whole protein, respectively (Fig. S10).
Experimental validation and case analysis of the E2 epitope scaffolds
The E2 epitope scaffolds were expressed transiently in HEK293F cells, SEC purified, and assessed by reducing SDS-PAGE. Four designs derived from the scaffolds 4F25_A, 1T07_A, 3S7R_A, and 2X1F_A showed detectable expression on SDS PAGE, with multiple bands corresponding to different glycoforms of N417 (and an extra N-linked glycan in 1T07_A_ES) (Fig. 5A, left). These epitope scaffolds exhibited different HCV1 binding profiles in ELISA analysis, with 1T07_A_ES being the best performer (Fig. 5A, middle). 2KNM_A_ES, an epitope scaffold derived from a cysteine knot, bound to HCV1 at a similar level to 3S7R_A_ES in ELISA, but exhibited little expression on SDS PAGE. In summary, three scaffold groups each contributed at least one antigenic epitope scaffold, with 1T07_A_ES derived from a distinct fold family. The HCV1 binding was assessed for the E2c domain using the Octet, showing a fast-one/slow-off kinetics (Fig. 5A, right) in contrast to the kinetics observed for the E1 peptide (Fig. 3A, right).
Figure 5. Experimental validation and case analysis of the E2 epitope scaffolds.
(A) SDS PAGE analysis of five expressed epitope scaffolds containing the E2 antigenic site (412–423) under reducing conditions followed by Coomassie blue staining (left), ELISA analysis of E2 antigen binding to the human neutralizing antibody HCV1 (middle), and Octet measurement of the E2c domain (right). (B,C) Structural modeling (left), quality assessment (middle) and Octet measurements (right) of E2 epitope scaffolds designed based on (B) 3S7R_A, and (C) 1T07_A. The epitope scaffold model (magenta) is superimposed onto the epitope (green) in complex with HCV1 (surface). The side chains of the epitope residues grafted onto the scaffolds are shown in stick model and labeled. The normalized residue-based scores calculated from DFIRE (blue) and a tabulated soft-core vdW energy function (red) are plotted as a function of the residue number. The sensorgrams from the Octet RED96 system show the binding of E2c domain and E2 epitope scaffolds to HCV1 using an antigen titration series of six starting at a maximum concentration of 100 μg/ml and 25 μg/ml with four-fold dilutions, respectively. The antibody concentration was fixed at 5 μg/ml.
Scaffolds 2YWK_A, 3S7R_A and 4F25_A are a group of RNA-binding proteins61 with an intra-group Cα RMSD of ~1.7 Å. Both 3S7R_A_ES and 4F25_A_ES bound to HCV1 with a similar binding profile by ELISA (Fig. 5A, middle). For scaffold 3S7R_A, we grafted epitope residues L413, N415, N417, 419SW420, and I422 to the scaffold, and mutated Q66 and K67 to alanine and glycine, respectively, to remove potential clashes with the antibody (Fig. 5B, left). Although the vdW score indicated steric clashes between 65DAGLH69 and HCV1, we observed a rather reasonable DFIRE score for this region (Fig. 5B, middle). Note that the highest-scoring residues 67GLH69 have extended into the region that anchors the β-hairpin epitope (Fig. S7). As stated previously, the high vdW scores were likely caused by rigid-body grafting rather than actual structural errors. When tested for the HCV1 binding, 3S7R_A_ES showed a less favorable binding kinetics than the E2c domain (Fig. 5B, right). For scaffold 1T07_A, a conserved protein from Pseudomonas aeruginosa with unidentified function, we grafted epitope residues L413, N415, 417NGSW420 and I422, and truncated the flexible N-terminus by four residues (Fig. 5C, left). Although the vdW function indicated some clashes between 8SWL10 and antibody HCV1, DFIRE showed a favorite local score for this region, indicating an acceptable quality of the designed interface (Fig. 5C, middle). In MD simulations, the grafted E2 epitope centered at the turn (residues 6–8) appeared to be more mobile relative to the scaffold, with a Cα RMSF of 2–4 Å, but relatively rigid within the region, with a local Cα RMSF of 0.5 Å (Fig. S10). For 1T07_A_ES, the Octet data showed slow-on/slow-off kinetics, indicative of a more desirable antigenic profile (Fig. 5C, right).
Overall, our results indicate that the E2 antigenic site is more amenable to the scaffolding approach, as half of the designed epitope scaffolds were reactive to antibody HCV1. Our results are also reminiscent of a previous scaffolding study where similar success rates were observed, with 55% for a hairpin-like epitope and 14% for a helical epitope, respectively42. The two best performers by ELISA, 3S7R_A_ES and 1T07_A_ES, showed different kinetics as determined by Octet, indicating that such biophysical measurements are critical in a quantitative assessment and comparison of scaffold antigen designs of the same epitope.
Multivalent scaffolding
With highly symmetric, repetitive antigen display on the surface, virus-like particles (VLPs) can induce potent immune response and have been used as vaccines against cognate viruses or vaccine platforms to carry foreign antigens62,63. It has been demonstrated that a minimum of 20–25 epitopes spaced by 5–10 nm are sufficient for effective B-cell activation66. Therefore, nanoparticles with similar molecular traits to that of VLPs may be used as multivalent scaffolds to present an epitope scaffold, providing useful tools to probe the immune response to the presented epitope. For example, the 60-mer lumazine synthase particle from Aquifex aeolicus has been used to display an engineered HIV-1 gp120 outer domain to target a specific antibody germline VH gene67. The 24-mer ferritin particle from Helicobacter pylori was used to present influenza hemagglutinins (HAs) and elicited H1N1-neutralzing antibodies in ferrets68. Ferritin was also used as a carrier of scaffold antigen designs of the HIV-1 V3 epitope42, suggesting that ferritin is a robust particle platform for designing and testing multivalent antigens.
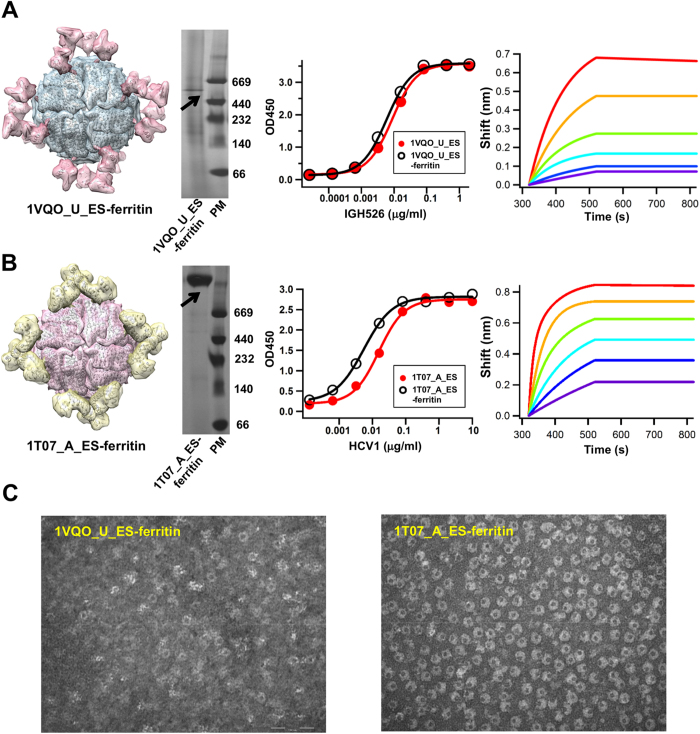
We first investigated whether ferritin particle can be used to present an E1 epitope scaffold, 1VQO_U_ES. The C-terminus of this antigen was fused to the N-terminus of a ferritin subunit (Asp5) with a 5-aa flexible linker. If such fusion protein is capable of assembling into a particle, 24 copies of the E1 epitope scaffold will be displayed on the surface (Fig. 6A, left). Indeed, blue native (BN) PAGE showed a band at the molecular weight of ~624 kD, consistent with properly formed 24-mer particles (Fig. 6A, middle left). Compared to the monomeric epitope scaffold, the nanoparticle only showed a slight shift of IGH526 binding profile by ELISA (Fig. 6A, middle right), but a significantly improved off-rate as indicated by Octet (Fig. 6A, right). The immeasurable dissociation is likely due to increased avidity (with 24 binding sites exposed on the particle surface) and consistent with a previous study42.
Figure 6. Design and characterization of nanoparticle antigens.
For E1 epitope scaffold 1VQO_U_ES (A) and E2 epitope scaffold 1T07_A_ES (B), the modeled ferritin nanoparticle displaying twenty-four copies of the antigen (left), blue native (BN) PAGE of the nanoparticle (middle left), ELISA binding of individual epitope scaffold and its particle form to the respective antibody (middle right), and Octet kinetics measurement of the nanoparticle (right) are shown. The molecular surface of the nanoparticle model is color-coded differently for ferritin and attached epitope scaffold. Coomassie blue staining was used after BN PAGE. IGH526 and HCV1 were used in ELISA and Octet experiments to test the antibody binding of E1 and E2 nanoparticle antigens, respectively. The sensorgrams show the nanoparticle-antibody binding using an antigen titration series of six starting at the maximum concentration of 10 μg/ml (calculated based on a single subunit) with two-fold dilutions. The antibody concentration was fixed at 5 μg/ml. (C) Negative-stain electron microscopy (EM) images are shown for 1VQO_U_ES-ferritin (left) and E2 epitope scaffold 1T07_A_ES-ferritin (right) at a magnification of 245K × for both particle samples.
We then extended the ferritin particle design to the E2 epitope scaffolds based on a similar fusion strategy. The chimeric protein showed comparable if not higher expression than the epitope scaffold alone, suggesting that a robust particle platform like ferritin can also be used to improve the expression of a designed antigen. Structural modeling of the 1T07_A_ES-ferritin particle (Fig. 6B, left) indicated that this E2 epitope scaffold would form propeller-shape trimers on the particle surface, with exposed E2 epitopes pointing away from the 3-fold axes. A single band was seen on the BN-PAGE gel (Fig. 6B, middle left), confirming proper assembly of antigen-carrying ferritin particles. Compared to 1VQO_U_ES, we observed a more notable shift of the HCV1 binding profile in ELISA (Fig. 6B, middle right). Remarkably, the 1T07_A_ES nanoparticle exhibited a lower than picomolar affinity on Octet, with plateaued dissociation curves under all tested concentrations, suggesting a strong avidity effect (Fig. 6B, right). A similar characterization was performed for 2KNM_A_ES-ferritin (Fig. S11), indicating that multiple E2 epitope scaffolds are suitable for particulate presentation.
We also validated the particle assembly of 1VQO_U_ES-ferritin (Fig. 6C, left) and 1T07_A_ES-ferritin (Fig. 6C, right) by negative-stain electron microscopy (EM), which showed results in agreement with their antigenic profiles. 1VQO_U_ES-ferritin particles were visible with moderate density and purity, compared to the remarkable numbers of homogeneous particles observed for 1T07_A_ES-ferritin. The “layered” structure of 1T07_A_ES-ferritin particles (Fig. 6C, right) is likely attributed to the 48 N-linked glycans displayed on the surface (2 × 24 subunits). Taken together, our results demonstrated that self-assembling nanoparticles such as ferritin can be used as a multivalent scaffold to present a neutralizing epitope in high density. Such multi-scale scaffolding strategy may prove to be useful in designing immunogens to elicit a robust antibody response to a target epitope. The HCV nanoparticles designed in this study will be further tested in vivo to examine this possibility.
Discussion
Vaccines provide cost-effective tools to improve public health and to prevent infectious diseases. Despite great success, broadly effective vaccines have not been developed for viral pathogens such as HIV-1, HCV, and influenza69. Protein engineering has been applied to the development of novel diagnostic and therapeutic agents, for which cysteine knots70,71, designed ankyrin repeat proteins (DARPins)72,73, and other unique protein folds have been used as scaffolds to display a functional site. It was only recently that the concept of scaffold was extended to epitope vaccine design by grafting an epitope of interest onto heterologous protein scaffolds. The scaffolding approach has proved to be useful for epitope vaccine design, with case studies reported for HIV-140,41,42,46,47, Flu74, and RSV43,44. For HCV, a number of cross-neutralizing epitopes have been identified and structurally characterized23,24,25,30,31,32,33,35, but not yet subjected to vaccine design by the scaffolding approach. Scaffolding-based epitope vaccine design builds upon the assumption that the available protein database contains scaffolds that can structurally accommodate an epitope of interest and a scaffolding algorithm can identify such needles from a large haystack. Since most scaffolding studies have been conducted using a single structural alignment algorithm, a large repository of other potential techniques was unexamined. In this study, we borrowed the concept of meta-server75,76 to exploit the combined strengths of six structural alignment algorithms for scaffold search. For both HCV antigenic sites studied, the scaffolding meta-server consistently provided a more complete set of scaffolds for epitope scaffold design. Biochemical and antigenic characterization then played a critical role in the screening of designed antigens (Figs 3 and 5, and Figs S12 and S13). We also attempted to bridge the gap between epitope scaffolding and the VLP vaccine concept by utilizing ferritin nanoparticle as a multivalent scaffold to present an antigenic epitope scaffold. With a diameter of ~12 nm and 24 fusion sites on the surface, ferritin particle can serve as a model system to experiment with various VLP designs without involving complex purification and polishing process typical of the VLP approaches77. Other self-assembling nanoparticles, especially those of microbial origin, may also be exploited for this purpose. To our knowledge, this study represents the first attempt to design epitope vaccines for HCV by a scaffolding approach, and a set of novel immunogen candidates is now awaiting in vivo trials.
Methods
Protein structure database
A protein structure database was compiled for scaffold search as previously described42. Briefly, the PISCES server54 was used to generate a representative, non-homologous set of Protein Databank (PDB) entries at the 90% identity level, with a resolution cutoff of 3.0 Å, and an R-factor cutoff of 1.0. Of note, these parameters were selected to increase the coverage of the resulting database. As of February, 2013, the PISCES server yielded a list of 23,576 peptide chains after screening using the above parameters, with the chemically modified residues corrected and Cα-only PDB files removed. Each entry in the PISCES database contains a PDB ID and a chain ID, which provide a unique identifier for scaffold search.
A semi-automated pipeline for epitope scaffold design
Epitope scaffold design is a critical component of the epitope vaccine strategy44. In this study, we have developed a semi-automated pipeline for epitope scaffold design (Fig. 1). Given an antibody-antigen complex with a well-defined epitope, this pipeline consists of five steps: (1) scaffold search and filtering using a scaffolding meta-server, (2) consensus analysis to determine the coverage of scaffold search and to prioritize the scaffold list, (3) scaffold selection based on a set of structural criteria, (4) epitope grafting by rigid-body fitting, and (5) energy evaluation using residue-based scoring functions and structure optimization. The output is a list of epitope scaffold models.
In step one, a scaffolding meta-server is used to identify diverse scaffolds. In this method, six structural alignment algorithms including two versions of TM-align50, SPalign51, CLICK52, FAST53, and MAMMOTH49 are combined to search the PISCES database. These algorithms are based on diverse scoring functions and search schemes: TM-align generates structural alignment by optimizing TM-score in a dynamic programming (DP) scheme; SPalign optimizes a size-independent objective function and provides both SP-score and TM-score as output; CLICK is a graph theory algorithm that takes into account structural information such as Cα coordinates, secondary structure, solvent accessibility and residue depth; FAST relies on heuristic search to approach the optimal structural alignment; MAMMOTH maximizes the overlap between two protein structures using a sequence-independent heuristic search scheme. The coordinates of the target epitope are used as input to six structural alignment algorithms for scaffold search. The output from each algorithm is processed on the fly, resulting in a list of PDB entries with detailed parameters such as protein size, number of matched residues, Cα RMSD, and epitope exposure ratio (epitope surface in the context of a scaffold versus solvent-exposed peptide). In the filtering step, scaffolds with a clash score of 5.0 or greater in the docked antibody-scaffold complex are removed. The clash score is calculated for all heavy atoms of the antibody-scaffold interface within 1.0 Å using the sum of 1/r (r: the atomic distance).
In step two, protein scaffolds identified by the meta-server are subjected to a consensus analysis to calculate the “coverage matrix”. Specifically, the number of scaffolds contributed by each algorithm and the pairwise overlap are determined (Fig. S1, Figs 2A, 4A). This matrix provides a quantitative measure of scaffold search coverage. Each scaffold is assigned to a voting group based on the number of algorithms by which the scaffold is identified. The voting count provides a basis for consensus-based scaffold selection.
In step three, the top-ranking scaffolds are manually inspected based on criteria such as protein size, topology, backbone flexibility, and structural environment of the epitope-matching region. Briefly, small, inherently flexible scaffolds with the epitope-matching region situated in a different secondary structural environment (e.g. a helical epitope in a β-rich protein) or separated from the surrounding structures with well-defined boundaries are preferred. The rationales for these selection criteria have been explained in detail45.
In step four, transplantation of epitope residues is performed by rigid-body fitting of the backbone atoms (N, Cα and C). After epitope grafting, scaffold residues with side chains within a 6.5 Å distance to the grafted epitope side chains or antibody side chains are mutated to eliminate potential steric clashes. Exposed hydrophobic residues or buried charges in the scaffold are also mutated to improve the solubility and stability of the designed antigen.
In step five, two residue-based scoring functions are used to assess the structural quality of the designed epitope scaffold antigen in the presence of the antibody after rigid-body docking. DFIRE has been used to distinguish the native structure from decoy models55, predict protein-protein interface78, and refine protein models with or without experimental constraints56,79,80,81. The tabulated soft-core vdW function has been applied to homology model refinement56. In structural quality assessment, if the normalized residue score is above 2.0, the side chains involved will be subjected to an in-house side-chain modeling program that combines an XYZ side-chain rotamer library81, DFIRE54, and a FASTER search algorithm83, to obtain the optimal conformation. A 50-step energy minimization using TINKER84 and OPLS all-atom force field85 is performed to relax the epitope scaffold structure. Side-chain modeling coupled with a short energy minimization has been found to effectively reduce the clash score.
MD simulation of epitope-scaffolds
The GROMACS 3.3.3 package86,87 was used to perform MD simulation on the designed epitope scaffolds. The GROMOS96 43a1 force field was used. The protonation state of ionizable amino acids was set for a pH of 7.0, with counterions added to neutralize the system. Each epitope scaffold was solvated in a rectangular box using the SPC water model. The minimal distance between the protein and the wall of the unit cell was set to be 10 Å. A non-bound pair-list with cutoff of 9.0 Å was used, and the pair-list was updated every five time steps. A twin-range method was used to calculate the van der Waals interactions. Interactions within the short-range cutoff (9.0 Å) were updated every step, while interactions with the long-range cutoff (14 Å) were updated every five steps together with the pair-list. The electrostatic interaction within the short-range cutoff (9.0 Å) was calculated with Columbic function while those beyond the cutoff were treated with the particle mesh Ewald method (PME)88. LINCS algorithm89 and SETTLE algorithm90 were used to constrain covalent bonds in the proteins and the geometry of the water molecules, respectively. A time step of two femtoseconds was used. The protein/ion/water system was first energy minimized and then equilibrated in a 100-picosecond MD simulation with positional restraints on the heavy atoms of the protein. The production run (5 × 5 nanoseconds) was performed at a temperature of 300 K and a constraint pressure of one bar by coupling to an external heat and an isotropic pressure bath91. Snapshots were stored every one picosecond, resulting in a total of 25,000 conformations for each epitope scaffold, which were used in the MD trajectory analysis.
Expression and purification of designed HCV antigens
The designed HCV antigens were transiently expressed in HEK293F cells (Life Technologies, CA). Briefly, 293F cells were thawed and incubated with FreeStyleTM 293 Expression Medium (Life Technologies, CA) in the Shaker incubator at 37 °C, with 120 rpm and 8% CO2. When the cells reached a density of 2.0 × 106/ml, expression medium was added to reduce cell density to 1.0 × 106/ml for transfection with polyethyleneimine (PEI) (Polysciences, Inc). 1000 μg plasmid DNA in 25 ml of Opti-MEM transfection medium (Life Technologies, CA) was mixed with 4 ml of PEI-Max (1.0 mg/ml) in 25 ml of Opti-MEM. After incubation for 25 min, the DNA-PEI-Max complex was added to 1 L 293 F cells. Culture supernatants were harvested five days after transfection, clarified by centrifugation at 3000 rpm for 10 min, and filtered using a 0.22 μm filters (Thermo Scientific). Proteins were purified from the supernatants using Ni SepharoseTM excel column (GE Healthcare) and then further purified using size exclusion chromatography (SEC) on a Superdex 200 10/300 GL column (GE Healthcare). Protein concentrations were determined using UV280 absorbance with theoretical extinction coefficients.
SDS PAGE and Blue Native (BN) PAGE
HCV antigens were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS PAGE) and BN PAGE and stained using Coomassie blue. The protein samples were mixed with loading dye and loaded onto a 4–12% Bis-Tris NuPAGE gel (Life Technologies) or a 10% Tris-Glycine Gel (Bio-Rad). The SDS PAGE gels were run for 20 min at 250 V using the SDS running buffer (Bio-Rad). BN PAGE gels were run for 1.5 h at 200 V using NativePAGETM running buffer (Life Technologies) according to the manufacturer’s instructions.
ELISA binding assays
CostarTM 96-well assay plates (Corning) were coated with the HCV antigen overnight at 4 °C. The wells were washed once with PBS + 0.05% Tween 20, and then incubated with 150 μl of blocking buffer (PBS with 5% w/v dry milk) per well for 1 hour at room temperature (RT) followed by 5 times of washing in PBS + 0.05% Tween 20. 50 μl of HCV-specific antibodies in blocking buffer were added, with a maximum concentration of 2 μg/ml and a 5-fold dilution series, and incubated for 1 hour at RT. After washing 5 times in PBS + 0.05% Tween 20, the wells were incubated with 50 ul of Peroxidase-AffiniPure Goat Anti-Human IgG antibody (Jackson ImmunoResearch Laboratories, Inc) at 1:5000 in PBS + 0.05% Tween 20 per well for 1 h at RT. After washed 5 times in PBS + 0.05% Tween 20, the wells were developed using TMB at RT for 5 min and the reaction stopped with 180 mM HCl. The readout was measured at a wavelength of 450 nm.
Octet binding assays
The kinetics of the HCV antigens binding to their respective antibodies was measured using an Octet Red96 instrument (fortéBio). All assays were performed with agitation set to 1000 rpm in fortéBIO 1 × kinetic buffer. The final volume for all the solutions was 200 μl/well. Assays were performed at 30 °C in solid black 96-well plates (Geiger Bio-One). 1 μg/ml of protein in 1 × kinetic buffer was used to load the HCV antibody IGH526 or HCV1 on the surface of anti-human Fc Capture Biosensors (AHC) for 300 s. Typical capture levels were between 0.5 and 1 nm and variability within a row of eight tips did not exceed 0.1 nm. A 60 s biosensor baseline step was applied prior to the analysis of the association of the antibody on the biosensor to the antigen in solution for 200 s. Two-fold concentration gradient of HCV antigen starting at 10 μg/ml was used in a titration series of six. The dissociation of the interaction was followed for 300 s. Correction of baseline drift was performed by subtracting the averaged shift recorded for a sensor loaded with HCV antibody but not incubated with HCV antigen, or a sensor without HCV antibody but incubated with HCV antigen. Octet data were processed by fortéBio’s data acquisition software v.8.1. Experimental data were fitted with the binding equations describing a 1:1 interaction. Local fitting of the data sets was performed to obtain the optimal results for different HCV antigens examined. The KD value was determined using the estimated response at equilibrium for each antigen concentration rather than the kon and kdis values.
Negative-stain electron microscopy (EM)
Copper grids (carbon coated, 400 mesh (Electron Microscopy Sciences, Hatfield PA)) were glow discharged and inverted on a 7 μl aliquot of sample for 3 minutes. Excess sample was removed and the grids immediately placed briefly on a droplet of double distilled water followed by 1% uranyl formate solution for 2 minutes. Excess stain was removed and the grid allowed to dry thoroughly. Grids were then examined on a Philips CM100 electron microscope (FEI, Hillsbrough OR) at 80 kv and images collected using a Megaview III ccd camera (Olympus Soft Imaging Solutions GmbH, Münster, Germany).
Additional Information
How to cite this article: He, L. et al. Approaching rational epitope vaccine design for hepatitis C virus with meta-server and multivalent scaffolding. Sci. Rep. 5, 12501; doi: 10.1038/srep12501 (2015).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants R01 AI079031, R01 AI106005, and the Scripps Center for HIV/AIDS Immunology & Immunogen Discovery (CHAVI-ID) UM1 AI-100663.
Footnotes
Author Contributions J.Z. and M.L. designed the research, with the scaffolding methods implemented by J.Z. and Y.C., antigens designed by J.Z., computational analysis performed by Y.C., construct design, ELISA and Octet binding analysis by L.H., protein production by L.H., P.A. and J.K., western blot by E.G., IGH526 structure by L.K. and I.A.W. and negative-stain EM by M.R.W. J.Z., L.H. and Y.C. wrote the paper, with all principal investigators providing comments or revisions.
References
- Lavanchy D. The global burden of hepatitis C. Liver Int. 29, 74–81, 10.1111/j.1478-3231.2008.01934.x (2009). [DOI] [PubMed] [Google Scholar]
- Ly K. N. et al. The increasing burden of mortality from viral hepatitis in the United States between 1999 and 2007. Ann. Intern. Med. 156, 271–278, 10.7326/0003-4819-156-4-201202210-00004 (2012). [DOI] [PubMed] [Google Scholar]
- Shepard C. W., Finelli L. & Alter M. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5, 558–567, 10.1016/s1473-3099(05)70216-4 (2005). [DOI] [PubMed] [Google Scholar]
- Messina J. P. et al. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61, 77–87, 10.1002/hep.27259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- deLemos A. S. & Chung R. T. Hepatitis C treatment: an incipient therapeutic revolution. Trends Mol. Med. 20, 315–321, 10.1016/j.molmed.2014.02.002 (2014). [DOI] [PubMed] [Google Scholar]
- Liang T. J. Current progress in development of hepatitis C virus vaccines. Nat. Med. 19, 869–878, 10.1038/nm.3183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M. & Abrignani S. Prospects for a vaccine against the hepatitis C virus. Nature 436, 961–966, 10.1038/nature04081 (2005). [DOI] [PubMed] [Google Scholar]
- Feinstone S. M., Hu D. J. & Major M. E. Prospects for prophylactic and therapeutic vaccines against hepatitis C virus. Clin. Infect. Dis. 55, S25–S32, 10.1093/cid/cis362 (2012). [DOI] [PubMed] [Google Scholar]
- Bowen D. G. & Walker C. M. Mutational escape from CD8(+) T cell immunity: HCV evolution, from chimpanzees to man. J. Exp. Med. 201, 1709–1714, 10.1084/jem.20050808 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. Genetic diversity and evolution of hepatitis C virus—15 years on. J. Gen. Virol. 85, 3173–3188, 10.1099/vir.0.80401-0 (2004). [DOI] [PubMed] [Google Scholar]
- Pileri P. et al. Binding of hepatitis C virus to CD81. Science 282, 938–941, 10.1126/science.282.5390.938 (1998). [DOI] [PubMed] [Google Scholar]
- Scarselli E. et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21, 5017–5025, 10.1093/emboj/cdf529 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. G. et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature 509, 381–384, 10.1038/nature13117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey T. et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 6, e1000762, 10.1371/journal.ppat.1000762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavillette D. et al. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J. Virol. 81, 8752–8765, 10.1128/jvi.02642-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel M. B., Felmlee D. J. & Baumert T. F. Hepatitis C virus entry. Curr. Top. Microbiol. Immunol. 369, 87–112, 10.1007/978-3-642-27340-7_4 (2013). [DOI] [PubMed] [Google Scholar]
- Weiner A. J. et al. Intrahepatic genetic inoculation of hepatitis C virus RNA confers cross-protective immunity. J. Virol. 75, 7142–7148, 10.1128/jvi.75.15.7142-7148.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farci P. et al. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93, 15394–15399, 10.1073/pnas.93.26.15394 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N. et al. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 189, 119–127, 10.1016/0006-291x(92)91533-v (1992). [DOI] [PubMed] [Google Scholar]
- Shimizu Y. K. et al. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology 223, 409–412, 10.1006/viro.1996.0497 (1996). [DOI] [PubMed] [Google Scholar]
- Weiner A. J. et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology 180, 842–848, 10.1016/0042-6822(91)90104-j (1991). [DOI] [PubMed] [Google Scholar]
- Farci P. et al. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288, 339–344, 10.1126/science.288.5464.339 (2000). [DOI] [PubMed] [Google Scholar]
- Broering T. J. et al. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 evelope glycoprotein of hepatitis C virus. J. Virol. 83, 12473–12482, 10.1128/jvi.01138-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck Z.-y. et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 8, e1002653, 10.1371/journal.ppat.1002653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M. et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat. Med. 14, 25–27, 10.1038/nm1698 (2008). [DOI] [PubMed] [Google Scholar]
- Owsianka A. et al. Monoclonal antibody AP33 defines a broadly neutralizing epitope on the hepatitis C virus E2 envelope glycoprotein. J. Virol. 79, 11095–11104, 10.1128/jvi.79.17.11095-11104.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson D. X. et al. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc. Natl. Acad. Sci. USA 104, 16269–16274, 10.1073/pnas.0705522104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J.-C. et al. Isolation and characterization of broadly neutralizing human monoclonal antibodies to the E1 glycoprotein of hepatitis C virus. J. Virol. 82, 966–973, 10.1128/jvi.01872-07 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Kadam R. U., Giang E., Ruwona T. B., Nieusma T., Culhane J. C. Stanfield R. L. Dawson P. E., Wilson I. A., Law M. Structure of hepatitis C virus envelope glycoprotein E1 antigenic site 314-324 in complex with antibody IGH526. J. Mol. Biol. 10.1016/j.jmb.2015.06.012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L. et al. Structure of hepatitis C virus envelope glycoprotein E2 antigenic site 412 to 423 in complex with antibody AP33. J. Virol. 86, 13085–13088, 10.1128/jvi.01939-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L. et al. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc. Natl. Acad. Sci. USA 109, 9499–9504, 10.1073/pnas.1202924109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey T. et al. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog. 9, e1003364, 10.1371/journal.ppat.1003364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter J. A. et al. Toward a hepatitis C virus vaccine: the structural basis of hepatitis C virus neutralization by AP33, a broadly neutralizing antibody. J. Virol. 86, 12923–12932, 10.1128/jvi.02052-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantua H. et al. Glycan shifting on hepatitis C virus (HCV) E2 glycoprotein is a mechanism for escape from broadly neutralizing antibodies. J. Mol. Biol. 425, 1899–1914, 10.1016/j.jmb.2013.02.025 (2013). [DOI] [PubMed] [Google Scholar]
- Kong L. et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342, 1090–1094, 10.1126/science.1243876 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvelle C., Felmlee D. J. & Baumert T. F. Unraveling hepatitis C virus structure. Cell Res. 24, 385–386, 10.1038/cr.2014.31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo C., Angus A. G. N. & Patel A. H. Hepatitis C virus evasion mechanisms from neutralizing antibodies. Viruses-Basel 3, 2280–2300, 10.3390/v3112280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle F., Duverlie G. & Dubuisson J. The hepatitis C virus glycan shield and evasion of the humoral immune response. Viruses-Basel 3, 1909–1932, 10.3390/v3101909 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton D. R. Scaffolding to build a rational vaccine design strategy. Proc. Natl. Acad. Sci. USA 107, 17859–17860, 10.1073/pnas.1012923107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia B. E. et al. Computational protein design using flexible backbone remodeling and resurfacing: case studies in structure-based antigen design. J. Mol. Biol. 405, 284–297, 10.1016/j.jmb.2010.09.061 (2011). [DOI] [PubMed] [Google Scholar]
- Ofek G. et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc. Natl. Acad. Sci. USA 107, 17880–17887, 10.1073/pnas.1004728107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T. et al. Transplanting supersites of HIV-1 vulnerability. PLoS ONE 9, e99881, 10.1371/journal.pone.0099881 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J. S. et al. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J. Mol. Biol. 409, 853–866, 10.1016/j.jmb.2011.04.044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia B. E. et al. Proof of principle for epitope-focused vaccine design. Nature 507, 201–206, 10.1038/nature12966 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L. & Zhu J. Computational tools for epitope vaccine design and evaluation. Curr. Opin. Virol. 11, 103–112, 10.1016/j.coviro.2015.03.013 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia B. E. et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 18, 1116–1126, 10.1016/j.str.2010.06.010 (2010). [DOI] [PubMed] [Google Scholar]
- Azoitei M. L. et al. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. J. Mol. Biol. 415, 175–192, 10.1016/j.jmb.2011.10.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 18, 342–348, 10.1016/i.sbi.2008.02.004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz A. R., Strauss C. E. M. & Olmea O. MAMMOTH (Matching molecular models obtained from theory): An automated method for model comparison. Protein Sci. 11, 2606–2621, 10.1110/ps.0215902 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. & Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309, 10.1093/nar/gki524 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Zhan J., Zhao H. & Zhou Y. A new size-independent score for pairwise protein structure alignment and its application to structure classification and nucleic-acid binding prediction. Proteins 80, 2080–2088, 10.1002/prot.24100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. N., Tan K. P. & Madhusudhan M. S. CLICK-topology-independent comparison of biomolecular 3D structures. Nucleic Acids Res. 39, W24–W28, 10.1093/nar/gkr393 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. H. & Weng Z. P. FAST: A novel protein structure alignment algorithm. Proteins 58, 618–627, 10.1002/prot.20331 (2005). [DOI] [PubMed] [Google Scholar]
- Wang G. L. & Dunbrack R. L. PISCES: a protein sequence culling server. Bioinformatics 19, 1589–1591, 10.1093/bioinformatics/btg224 (2003). [DOI] [PubMed] [Google Scholar]
- Zhou H. Y. & Zhou Y. Q. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 11, 2714–2726, 10.1110/ps.0217002 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Fan H., Periole X., Honig B. & Mark A. E. Refining homology models by combining replica-exchange molecular dynamics and statistical potentials. Proteins 72, 1171–1188, 10.1002/prot.22005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwona T. B., Giang E., Nieusma T. & Law M. Fine mapping of murine antibody responses to immunization with a novel soluble form of hepatitis C virus envelope glycoprotein complex. J. Virol. 88, 10459–10471, 10.1128/jvi.01584-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam C. A. et al. Structural insight into interactions between dihydrolipoamide dehydrogenase (E3) and E3 binding protein of human pyruvate dehydrogenase complex. Structure 14, 611–621, 10.1016/j.str.2006.01.001 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing T. M., Huang K. S., Kitchen D. E., Strobel S. A. & Steitz T. A. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol. Cell 20, 437–448, 10.1016/j.molcel.2005.09.006 (2005). [DOI] [PubMed] [Google Scholar]
- Bartosch B. et al. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278, 41624–41630, 10.1074/jbc.M305289200 (2003). [DOI] [PubMed] [Google Scholar]
- Safaee N. et al. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell 48, 375–386, 10.1016/j.molcel.2012.09.001 (2012). [DOI] [PubMed] [Google Scholar]
- Grgacic E. V. L. & Anderson D. A. Virus-like particles: Passport to immune recognition. Methods 40, 60–65, 10.1016/j.ymeth.2006.07.018 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig C. & Wagner R. Virus-like particles—universal molecular toolboxes. Curr. Opin. Biotechnol. 18, 537–545, 10.1016/j.copbio.2007.10.013 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings G. T. & Bachmann M. F. Coming of age of virus-like particle vaccines. Biol. Chem. 389, 521–536, 10.1515/bc.2008.064 (2008). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Limas W. A., Sekar K. & Tyo K. E. J. Virus-like particles: the future of microbial factories and cell-free systems as platforms for vaccine development. Curr. Opin. Biotechnol. 24, 1089–1093, 10.1016/j.copbio.2013.02.008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dintzis R. Z., Vogelstein B. & Dintzis H. M. Specific cellular stimulation in the primary immune response: experimental test of a quantized model. Proc. Natl. Acad. Sci. USA 79, 884–888, 10.1073/pnas.79.3.884 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J. et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716, 10.1126/science.1234150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanekiyo M. et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106, 10.1038/nature12202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. M. & Burton D. R. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr. Opin. Immunol. 22, 358–366 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craik D. J., Cemazar M. & Daly N. L. The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discovery Dev. 9, 251–260 (2006). [PubMed] [Google Scholar]
- Craik D. J., Simonsen S. & Daly N. L. The cyclotides: Novel macrocyclic peptides as scaffolds in drug design. Curr. Opin. Drug Discovery Dev. 5, 251–260 (2002). [PubMed] [Google Scholar]
- Stumpp M. T. & Amstutz P. DARPins: A true alternative to antibodies. Curr. Opin. Drug Discovery Dev. 10, 153–159 (2007). [PubMed] [Google Scholar]
- Stumpp M. T., Binz H. K. & Amstutz P. DARPins: A new generation of protein therapeutics. Drug Discovery Today 13, 695–701, 10.1016/j.drudis.2008.04.013 (2008). [DOI] [PubMed] [Google Scholar]
- Fleishman S. J. et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 332, 816–821, 10.1126/science.1202617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K., Elofsson A., Fischer D. & Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics 19, 1015–1018, 10.1093/bioinformatics/btg124 (2003). [DOI] [PubMed] [Google Scholar]
- Fischer D. 3D-SHOTGUN: A novel, cooperative, fold-recognition meta-predictor. Proteins 51, 434–441, 10.1002/prot.10357 (2003). [DOI] [PubMed] [Google Scholar]
- Lua L. H. L. et al. Bioengineering virus-like particles as vaccines. Biotechnol. Bioeng. 111, 425–440, 10.1002/bit.25159 (2014). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. Structural model of the TRPP2/PKD1 C-terminal coiled-coil complex produced by a combined computational and experimental approach. Proc. Natl. Acad. Sci. USA 108, 10133–10138, 10.1073/pnas.1017669108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. S., Fasnacht M., Zhu J., Forrest L. & Honig B. Loop modeling: Sampling, filtering, and scoring. Proteins 70, 834–843, 10.1002/prot.21612 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Xie L. & Honig B. Structural refinement of protein segments containing secondary structure elements: Local sampling, knowledge-based potentials, and clustering. Proteins 65, 463–479, 10.1002/prot.21085 (2006). [DOI] [PubMed] [Google Scholar]
- Zhu J., Cheng L., Fang Q., Zhou Z. H. & Honig B. Building and refining protein models within cryo-electron microscopy density maps based on homology modeling and multiscale structure refinement. J. Mol. Biol. 397, 835–851, 10.1016/j.jmb.2010.01.041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z. X. & Honig B. Extending the accuracy limits of prediction for side-chain conformations. J. Mol. Biol. 311, 421–430, 10.1006/jmbi.2001.4865 (2001). [DOI] [PubMed] [Google Scholar]
- Desmet J., Spriet J. & Lasters I. Fast and Accurate Side-Chain Topology and Energy Refinement (FASTER) as a new method for protein structure optimization. Proteins 48, 31–43, 10.1002/prot.10131, (2002) [DOI] [PubMed] [Google Scholar]
- Ponder J. W. & Richards F. M. An efficient Newton-like method for molecular mechanics energy minimization of large molecules. J. Comput. Chem. 8, 1016–1024, 10.1002/jcc.540080710 (1987). [DOI] [Google Scholar]
- Jorgensen W. L. & Tiradorives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 110, 1657–1666, 10.1021/ja00214a001 (1988). [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. C., Vanderspoel D. & Vandrunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56, 10.1016/0010-4655(95)00042-e (1995). [DOI] [Google Scholar]
- Lindahl E., Hess B. & van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7, 306–317, 10.1007/s008940100045 (2001). [DOI] [Google Scholar]
- Essmann U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593, 10.1063/1.470117 (1995). [DOI] [Google Scholar]
- Hess B., Bekker H., Berendsen H. J. C. & Fraaije J. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472, (1997). [DOI] [Google Scholar]
- Miyamoto S. & Kollman P. A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962, 10.1002/jcc.540130805 (1992). [DOI] [Google Scholar]
- Berendsen H. J. C., Postma J. P. M., Vangunsteren W. F., Dinola A. & Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690, 10.1063/1.448118 (1984). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.