Abstract
The endothelial PAS domain protein 1 (EPAS1) gene functions to sense the blood oxygen level by regulating the hypoxia-inducible transcription factor pathway, and single nucleotide polymorphisms (SNPs) of EPAS1 have been found to have a strong and positive selection in the adaptation of the native Tibetan highland population to high-altitude hypoxia. The aim of the present study was to investigate the effect of EPAS1 SNPs on the risk of acute mountain sickness (AMS) and the physiological responses to acute high-altitude hypoxia in lowland humans. Three tag SNPs (rs6756667, rs13419896 and rs4953354; minor allele frequency, ≥5%) were selected and genotyped in 603 unrelated Han Chinese men, who had traveled to Lhasa (a high-altitude hypoxia environment) by plane, using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry method. The data showed that the EPAS1 rs6756667 wild-type GG homozygous genotype was associated with elevated AMS risk compared with the AA and AG genotypes (odds ratio, 1.815; 95% confidence interval, 1.233–2.666; P=0.0023) using the dominant-model analysis. EPAS1 rs6756667 GG genotypes were also associated with higher levels of hemoglobin, red blood cells and hematocrit than those carrying the AG heterozygote during AMS development. These findings indicate that EPAS1 SNPs play a role in the physiological effects of AMS, and these effects could be further evaluated as a therapeutic strategy to control acute hypoxia-related human diseases.
Keywords: acute mountain sickness, endothelial PAS domain protein 1, hypoxia, single nucleotide polymorphisms
Introduction
Acute mountain sickness (AMS) is a medical condition, resulting from acute exposure to high-altitude environments (generally ≥2,500 m) and is characterized by headache, dizziness, vomiting, fatigue and sleep disturbance (1,2). The incidence of AMS ranges from 9 to 84% with variable altitudes, ascent rates and latitudes (3). AMS can progress to life-threatening high-altitude cerebral or pulmonary edema (4), which seriously impacts the health of the individual and reduces physical capability. With increasing numbers of lowland individuals travelling to high plateaus for a variety of reasons, AMS has become a public health concern (5). To date, the pathophysiology of AMS has yet to be fully elucidated, although such a condition is believed to manifest neurological complications linked to hypoxia (2). It appears that, under the same acute hypoxic conditions, certain lowland individuals are more susceptible to AMS than others (6); therefore, it is possible that genetic factors, such as the expression and variants of genes or single nucleotide polymorphisms (SNPs), play a role in AMS susceptibility.
Numerous genome-wide studies (7–11) and other reports (12–14) of genetic variations in native Tibetan highland populations have revealed that SNPs of endothelial PAS domain protein 1 (EPAS1), which is also known as hypoxia-inducible factor-2α (HIF-2α), have a strong and positive selection in high-altitude hypoxia adaptation and are associated with lower hemoglobin (Hb) concentrations (8). EPAS1 encodes the oxygen-sensitive subunit of the HIF-2 transcription factor and plays a role in regulating erythropoietin (15) and vascular endothelial growth factor expression (16) under hypoxic conditions. Genetic variants of EPAS1 that are shown to be more common in highlanders may be worth investigating in lowlanders with AMS, as the advantage conferred by the variants may be beneficial under both acute and chronic hypoxia and represent the migrational selection for AMS resistance in the population at high altitude. Of the AMS genetic association studies that have been performed, each has been limited to a few individuals or other gene variants for AMS association (17,18). Furthermore, little is known about the association between the EPAS1 gene polymorphisms and the risk of AMS in lowlanders following acute exposure to high-altitude hypoxia (19). Since the Han Chinese populate a lowland region, they are considered to represent an appropriate population for investigations into the association between genetic factors and susceptibility or resistance to AMS.
In the present study, three tag SNPs [rs6756667, rs13419896 and rs4953354; minor allele frequency (MAF), ≥5%] were selected and genotyped in 603 unrelated Han Chinese men, who had traveled to Lhasa by plane, using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) method. The physiological relevance of the three SNPs in the development of AMS was then assessed, in order to enhance the understanding of the molecular mechanisms associated with AMS risk and facilitate the development of a novel strategy to control AMS.
Materials and methods
Study population and AMS evaluation
A total of 603 unrelated male Chinese Han subjects were recruited from Chongqing, China. These individuals were physical labors who traveled from Chengdu (500 m above sea level) to Lhasa, Tibet (3,700 m above sea level) by plane. One week before the trip, all the participants underwent health examinations in Xinqiao Hospital (Chongqing, China); the participants were enrolled in this study if they met the following inclusion criteria: i) Aged between 18 and 45 years; ii) of Han Chinese ethnicity and with Han Chinese parents; and iii) no history of high-altitude residency. The exclusion criteria were as follows: i) Individuals with diseases with similar clinical manifestations to AMS, such as migraine and insomnia; and ii) individuals with respiratory or cardiovascular system diseases, neuropsychosis, cerebrovascular disease, cancer or dysfunction of the liver or kidneys. The individuals were assessed using the current consensus of the Lake Louise scoring system (LLss) (20) 18–24 h after their flight to Lhasa, which took place between June 21 and 25, 2012. The individuals with an LLss score of ≥3 points and a headache were assigned to the AMS group (n=369, 61.2% of the participants), while individuals with a score of <3 points or a score of ≥3 points but no headache were assigned as a non-AMS group (n=234, 38.8% of the participants).
Ethics statement
All subjects gave their informed consent for inclusion prior to participation in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Xinqiao Hospital, Third Military Medical University (identification code, 2012014; approved May 9, 2012; Chinese Clinical Trial Register no. ChiCTR-OCS-11002213).
Data and blood sample collection
The demographic data were collected in Chongqing, China, and physiological variables were measured within 18–24 h of acute exposure to hypoxia (Lhasa, Tibet, China). Demographic data [age, body mass index (BMI), tobacco smoking and alcohol consumption] and AMS symptoms (dizziness, headache, gastrointestinal disturbance, fatigue and difficulty sleeping) were recorded using structured case report form questionnaires. The physiological variables, including blood pressure (BP), arterial oxygen saturation (SaO2) and heart rate (HR), were measured once the participants had rested in the supine position for 10 min. BP was recorded using a mercury sphygmomanometer, and SaO2 and HR were monitored using a Finger-Pulse Oximeter 503 (Criticare Systems, Inc., Waukesha, WI, USA). BMI was calculated following the measurement of the weight and height of the participants.
For blood sampling, ~5 ml venous blood was collected from each subject into EDTA tubes. Following collection, the sample was centrifuged at 492 × g for 10 min at 4°C in order to separate the blood cells and plasma. The plasma samples were stored at −80°C until required for serological analysis, and the isolated blood cells were immediately processed in order to extract DNA for genotyping.
Selection of EPAS1 SNPs
The human EPAS1 gene (accession no. NC_000002.12) is localized on chromosome 2 with nucleic acid positions between 46,297,402 and 46,386,703 (National Center for Biotechnology Information GenBank data; http://www.ncbi.nlm.nih.gov/genbank/). Genetic variation data for the EPAS1 gene were obtained from the HapMap project (21) for 82 healthy Chinese Han Beijing (CHB) individuals (www.hapmap.org). In this study, three SNPs (rs6756667, rs13419896 and rs4953354) with an MAF of ≥0.05 were selected. EPAS1 rs13419896 and rs6756667 were selected based on two genome-wide studies (8,14), while rs4953354 was selected according to data from the Kunming Primate Research Center (Kunming, China) (9). A maximally informative tag of each SNP was selected using Tagger software (http://www.broad.mit.edu/mpg/haploview), which captures all known common genetic variations in a gene based on a linkage disequilibrium (LD) threshold of r2 ≥0.8 (22). The inverse of r2 represents the ratio of sample size required to detect an indirect association with a non-assayed SNP to direct an association at the same statistical power.
DNA extraction and genotyping
Genomic DNA from each individual was extracted using the Ezup Column Blood Genomic DNA Extraction kit (Shanghai Sangon Biotechnology Co., Ltd., Shanghai, China) according to the manufacturer's instructions and stored at −80°C until use. To genotype these three SNPs, polymerase chain reaction primers were first designed using Assay Design software (version 3.1; Sequenom, Inc., San Diego, CA, USA) (Table I) and then synthesized by Shanghai Sangon Biotechnology Co., Ltd. MALDI-TOF (Sequenom Inc., San Diego, CA, USA) was subsequently performed for genotyping. All genotyping was performed in a blinded fashion without any knowledge of the identity of the individual, and 10% of the DNA samples were repeatedly genotyped to monitor the genotyping quality.
Table I.
Primers used in this study to detect EPAS1 SNPs.
| SNP | Forward primer | Reverse primer | Sequencing primer |
|---|---|---|---|
| rs6756667 | 5′-ACGTTGGATGTCGGAATCGACAGACTGGTG-3′ | 5′-ACGTTGGATGCTGACCAAGAGTTGATGCTG-3′ | 5′-AAACTGCTGTAAGGTGA-3′ |
| rs13419896 | 5′-ACGTTGGATGCTGAACCAGAGTCAGTAACC-3′ | 5′-ACGTTGGATGAACCCTTCCTGGTTGAGTAG-3′ | 5′-GAAAAGAGTCAGTAACCAATCCTAG-3′ |
| rs4953354 | 5′-ACGTTGGATGTCTGCCATTGTCTTGCATTG-3′ | 5′-ACGTTGGATGAAGAGGCGAAATGTGCAGAC-3′ | 5′-TGAAAGTATTTATGGAGCATATCT-3′ |
SNP, single nucleotide polymorphism.
Statistical analysis
The statistical analyses were performed with SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). P≤0.05 was considered to indicate a statistically significant difference, or a P-value of <0.017 following Bonferroni correction for multiple testing. All continuous variables are expressed as the mean ± standard deviation. Differences between AMS and non-AMS subjects were evaluated using the Student's t-test for continuous variables and the χ2 test for categorical variables. The Student's t-test was therefore performed to compare the number of red blood cells (RBCs), hematocrit (HCT), Hb, SaO2, HR and BP between different genotypes of AMS individuals (a two-tailed P≤0.05 indicated statistical significance). The allele frequencies of each SNP were calculated by gene counting, and the genotype distribution of each SNP was tested for Hardy-Weinberg equilibrium (HWE) using χ2 analyses. Three genetic models (allele-dose, dominant and recessive) were subsequently used to assess association of these SNPs with AMS risk. The association between the genotypes and the AMS morbidity rate was calculated using χ2 analysis. Odds ratios (ORs) with 95% confidence intervals (CIs) were adjusted for age and BMI and calculated using multiple logistic regression analyses to estimate the relative risk of AMS.
Results
Subject characteristics
The characteristics of this study cohort are summarized in Table II. In brief, there were 369 AMS cases (61.2%) and 234 non-AMS controls (38.8%) in this unmatched, nested case-control study. All participants were young Han Chinese men with a median age of 22.96±3.74 years (range, 18–45 years). No significant differences were found between the AMS and non-AMS individuals in terms of age, BMI, tobacco smoking or alcohol consumption (all P>0.05). The baseline characteristics were thus considered comparable between the two groups. The comparison of clinical characteristics between the two groups revealed that the AMS individuals had a significantly higher HR and lower SaO2 than the non-AMS individuals (both P<0.01); however, the two groups showed similar levels of Hb, RBCs and HCT, as well as systolic and diastolic BPs (all P>0.05).
Table II.
Characteristics of the study subjects (n=603).
| Characteristics | AMS | Non-AMS | P-value |
|---|---|---|---|
| No. of patients, n (%) | 369 (61.19) | 234 (38.81) | |
| Baseline demographic data | |||
| Agea, yearsa | 23.18±3.85 | 22.64±3.56 | 0.088 |
| BMIa, kg/m2 | 21.83±2.31 | 21.63±2.16 | 0.289 |
| Tobacco smoking status, % | 0.964 | ||
| Never | 36.05 | 33.18 | |
| Former | 59.01 | 61.14 | |
| Current | 4.94 | 5.68 | |
| Alcohol consumption, % | 0.102 | ||
| Never | 31.43 | 25.58 | |
| Former | 22.00 | 18.60 | |
| Current | 46.57 | 55.82 | |
| Clinical indexesa | |||
| SBP, mm/Hg | 118.95±12.14 | 117.61±10.66 | 0.107 |
| DBP, mm/Hg | 78.47±10.01 | 78.70±10.14 | 0.781 |
| HR, bpm | 86.29±12.61 | 83.49±12.60 | 0.008 |
| SaO2, % | 87.93±3.38 | 88.91±2.79 | <0.001 |
| Hb, g/1 | 145.62±0.89 | 145.14±1.65 | 0.206 |
| RBCs, x1012/1 | 4.56±0.03 | 4.61±0.05 | 0.893 |
| HCT, % | 41.27±0.23 | 41.22±0.41 | 0.263 |
| LLss-related symptoms, n (%) | |||
| Headache | 369 (100.00) | 74 (31.62) | |
| Dizziness | 340 (92.14) | 102 (43.59) | <0.001 |
| Fatigue | 333 (90.24) | 84 (36.32) | <0.001 |
| Gastrointestinal symptoms | 123 (33.33) | 14 (5.60) | <0.001 |
| Difficulty sleeping | 303 (82.11) | 80 (34.19) | <0.001 |
| LLssa | 5.05±1.89 | 1.56±1.17 | <0.001 |
| Degree of AMS, n (%) | |||
| Mild: LLss score between 3 and 5 | 175 (47.43) | ||
| Severe: LLss score ≥5 | 194 (52.57) |
Data are presented as the mean ± standard deviation. Significant P-values (<0.05) are shown in bold print. AMS, acute mountain sickness; BMI, body mass index; DBP, diastolic blood pressure; SBP, systolic blood pressure; HR, heart rate; SaO2, arterial oxygen saturation; Hb, hemoglobin; RBCs, red blood cells; HCT, hematocrit; LLss, Lake Louise scoring system.
Using the LLss, which included measurements of headache, dizziness, gastrointestinal symptoms, difficulty sleeping and fatigue (23), it was found that the incidence of these five symptoms was significantly different between the two groups, with the AMS group exhibiting significantly higher incidences than the controls (all P<0.001), particularly with regard to the incidence of headache. The total LLss score of the AMS group was 5.05±1.89, which was significantly (P<0.001) higher than that of the non-AMS group (1.56±1.17). Specifically, there were 175 individuals (47.43%) with mild AMS (LLss, 3–5) and 194 individuals (52.57%) with severe AMS (LLss, ≥5).
Association of EPAS1 SNPs with AMS risk
In this study, three EPAS1 SNPs were genotyped in the individuals, and it was found that the overall MAF of the three SNPs (including AMS and non-AMS subjects) was similar to that reported in the HapMap III database (Tables III and IV). The distribution of these three SNP genotypes was within HWE (P>0.05) between the AMS and non-AMS individuals.
Table III.
Distribution of the three endothelial PAS domain protein 1 tag SNPs among the subjects.
| MAF, n (%) | Genotype, n (%) | HWE P-value | ||||||
|---|---|---|---|---|---|---|---|---|
| SNPs | N | Minor allele | Subjects | Database | Wild | Heterozygous | Variant | |
| rs6756667 | 603 | A | 13.3 | 5.6 | 467 (77.45) | 130 (21.56) | 6 (0.99) | 0.35 |
| rs13419896 | 602 | A | 31.5 | 27.8 | 281 (47.47) | 251 (43.40) | 60 (10.13) | 0.72 |
| rs4953354 | 602 | G | 10.0 | 13.3 | 451 (74.92) | 141 (23.42) | 10 (1.66) | 0.79 |
SNP, single nucleotide polymorphism; MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium.
Table IV.
Association between endothelial PAS domain protein 1 tag single nucleotide polymorphisms and the risk of AMS.
| A, rs6756667 | |||||
|---|---|---|---|---|---|
| Factor | Genotype/allele | AMS, n (%) | Non-AMS, n (%) | OR (95% CI) | P-value |
| Genotype | GG | 301 (81.6) | 166 (70.9) | 0.002 | |
| AG | 67 (18.2) | 63 (26.9) | |||
| AA | 1 (0.2) | 5 (2.2) | |||
| Allele | G | 669 (90.7) | 395 (84.4) | 1.701 (1.261–2.545) | 0.001 |
| A | 69 (9.3) | 73 (15.6) | |||
| Dominant model | GG | 301 (81.6) | 166 (70.9) | 1.815 (1.233–2.666) | 0.002 |
| AG+AA | 68 (18.4) | 68 (29.1) | |||
| Recessive model | AG+GG | 368 (99.7) | 229 (97.9) | 8.035 (1.007–69.214) | 0.025 |
| AA | 1 (0.3) | 5 (2.1) | |||
| B, rs13419896 | |||||
| Factor | Genotype/allele | AMS, n (%) | Non-AMS, n (%) | OR (95% CI) | P-value |
| Genotype | AA | 34 (9.2) | 26 (11.1) | 0.452 | |
| AG | 154 (42.0) | 105 (45.1) | |||
| GG | 179 (48.8) | 102 (43.8) | |||
| Allele | G | 512 (69.8) | 309 (66.3) | 1.172 (0.914–1.502) | 0.211 |
| A | 222 (30.2) | 157 (33.7) | |||
| Dominant model | GG | 179 (48.8) | 102 (43.8) | 1.222 (0.879–1.701) | 0.232 |
| AG+AA | 188 (51.2) | 131 (56.2) | |||
| Recessive model | AG+GG | 333 (90.7) | 207 (88.8) | 1.230 (0.717–2.110) | 0.451 |
| AA | 34 (9.3) | 26 (11.2) | |||
| C, rs4953354 | |||||
| Factor | Genotype/allele | AMS, n (%) | Non-AMS, n (%) | OR (95% CI) | P-value |
| Genotype | AA | 276 (75.0) | 175 (74.8) | 0.372 | |
| AG | 88 (23.9) | 53 (22.6) | |||
| GG | 4 (1.1) | 6 (2.6) | |||
| Allele | A | 640 (87.0) | 403 (86.1) | 1.075 (0.766–1.509) | 0.674 |
| G | 96 (13.0) | 65 (13.9) | |||
| Dominant model | AA | 301 (81.6) | 166 (70.9) | 1.011 (0.693–1.476) | 0.953 |
| AG+GG | 68 (18.4) | 68 (29.1) | |||
| Recessive model | AG+AA | 364 (98.9) | 228 (97.4) | 2.395 (0.669–8.578) | 0.167 |
| GG | 4 (1.1) | 6 (2.6) | |||
The OR was adjusted for age and body mass index using the multiple logistic regression analyses. Significant P-values of <0.017 following Bonferroni correction are shown in bold print. Allele-dose (A allele vs. G allele), dominant effect (variant homozygotes + heterozygotes vs. wild homozygotes) and the recessive effect (variant homozygotes vs. heterozygotes + wild homozygotes) were analyzed using the χ2 test. AMS, acute mountain sickness; OR, odds ratio; CI, confidence interval.
Under an additive model and Bonferroni correction, and following adjustments for age and BMI, it was found that the SNP rs6756667 was associated with the risk of developing AMS (Table IV). By contrast, no statistically significant association was found between rs13419896 or rs4953354 and the risk of AMS in the allele-dose, dominant and recessive models (all P>0.1).
The genotype frequencies of EPAS1 rs6756667 GG, AG and AA were 81.60% (n=301), 18.2% (n=67) and 0.3% (n=1), respectively, in the AMS group and 70.9% (n=166), 26.9% (n=63) and 2.1% (n=5), respectively, in the non-AMS group. The distribution of the overall genotype frequency was significantly different between the AMS and non-AMS groups (P=0.002). The allele frequency of G (AMS, 90.7% vs. non-AMS, 84.4%) and A (AMS, 9.3% vs. non-AMS, 15.6%) was also significantly different between these two groups. The individuals carrying the rs6756667 G allele had a 1.701-fold greater risk of developing AMS (OR, 1.701; 95% CI, 1.261–2.545; P=0.001) compared with those carrying the rs6756667 A allele. The rs6756667 GG homozygous genotype was also significantly associated with elevated AMS risk compared with the rs6756667 homozygous AA and heterozygous AG genotypes (OR, 1.815; 95% CI, 1.233–2.666; P=0.002) following the dominant model analysis; however, the recessive model analysis did not show a statistically significant association between rs6756667 and the AMS risk (OR, 8.035; 95% CI, 1.233–2.666; P=0.025).
Association between EPAS1 rs6756667 and AMS severity
The individuals carrying the variant G allele had a significantly higher rate of mild AMS when compared with those carrying the A allele (P=0.004 and 0.018 for mild AMS using the allele-dose and dominant models, respectively). For severe AMS, the subjects carrying the variant G allele also exhibited a significantly higher rate when compared with those carrying the A allele (P=0.014, 0.008 and 0.041 for severe AMS using the allele-dose, dominant and recessive models, respectively) (Table V).
Table V.
Association between the endothelial PAS domain protein 1 rs6756667 single nucleotide polymorphism and AMS severity.
| P-value | |||||
|---|---|---|---|---|---|
| Allele/genotype | Mild AMS, n (%) | Severe AMS, n (%) | Non-AMS, n (%) | Pa | Pa |
| A | 34 (9.7) | 35 (9.0) | 73 (15.6) | – | – |
| G | 316 (90.3) | 353 (91.0) | 395 (84.4) | 0.004 | 0.014 |
| GG | 142 (81.1) | 159 (82.0) | 166 (70.9) | 0.018 | 0.008 |
| AG+AA | 33 (18.9) | 35 (18.0 | 68 (29.1) | ||
| AG+GG | 174 (99.4) | 194 (100.0) | 229 (97.9) | – | 0.041 |
| AA | 1 (0.6) | 0 (0.000) | 5 (2.1) | ||
Mild AMS vs. non-AMS
Severe AMS vs. non-AMS. Significant P-values (<0.05) are shown in bold print. Mild AMS vs. non-AMS and severe AMS vs. non-AMS in allele-dose, dominant and recessive models were analyzed using the χ2 test. AMS, acute mountain sickness.
Association between EPAS1 rs6756667 and clinicophysiological characteristics
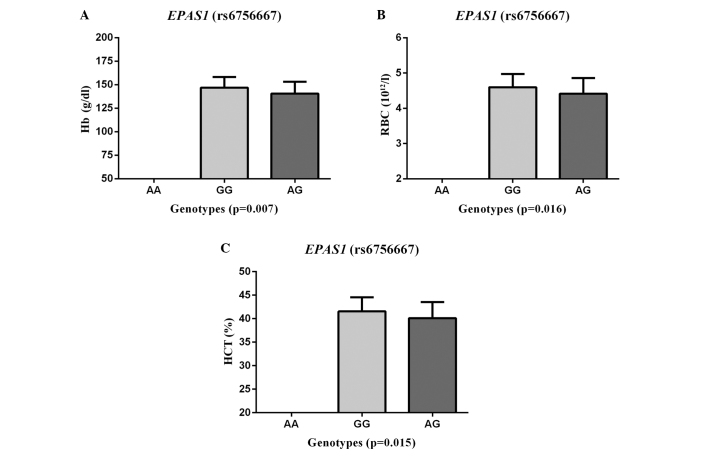
The SNP rs6756667 was next assessed for associations with certain clinicophysiological characteristics (RBCs, HCT, Hb, SaO2, HR and BP) in the AMS group. A significant association was found between the SNP rs6756667 and Hb (P=0.007), RBCs (P=0.016) and HCT (P=0.015) (Fig. 1), but there was no evident association with SaO2, HR or BP (all P>0.05). The AMS patients with GG genotypes had significantly higher levels of Hb, RBCs and HCT than those with the rs6756667 AG heterozygous genotype.
Figure 1.
Analysis of genotypes of EPAS1 rs6756667 single nucelotide polymorphism for association with (A) Hb, (B) number of RBCs and (C) HCT in AMS patients. A significant correlation was found between EPAS1 (rs6756667) and (A) Hb, (B) RBC levels and (C) HCT in AMS patients from the male Han Chinese population. Data for the EPAS1 GG/AG genotypes (x-axis) vs. (A) Hb concentration (A), (B) RBC count and (C) HCT (y-axis) are shown. P-values refer to the comparison between the GG and AG genotypes. The physiological data are expressed as the mean ± standard deviation. Differences between genotypes were evaluated with a Student's t-test. EPAS1, endothelial PAS domain protein 1; AMS, acute mountain sickness; Hb, hemoglobin; RBC, red blood cell; HCT, hematocrit.
Discussion
In this study, the association between three EPAS1 SNPs and the risk of AMS in a male Chinese Han population was assessed. It was found that the EPAS1 rs6756667 G allele was associated with a risk of developing AMS and that the EPAS1 rs6756667 SNP was also associated with Hb, RBC and HCT levels; AMS patients carrying the rs6756667 GG genotype exhibited significantly higher Hb, RBC and HCT levels than those with the AG genotype. These data indicate that EPAS1 SNPs play a role in the physiological effects of AMS development in this Chinese population; however, further study is required to evaluate and confirm whether targeting EPAS1 could be a therapeutic strategy to control acute hypoxia-related human diseases.
With regard to the association between AMS and genetic variants in humans, this study was the first, to the best of our knowledge, to investigate the potential clinical relevance of EPAS1 rs6756667, rs13419896 and rs4953354 polymorphisms in a large Chinese population. The major attraction of the use of the LD tag SNPs was that it enabled a reduction in the genotyping cost while maintaining a powerful detection system of non-assayed causal SNPs. These tag SNPs are a subset of all variants within a chromosome region for the study of an association with a disease. In the current study, the approach to select SNPs was different from that used in other studies, such as in the study by Buroker et al (19). In their study, one SNP (rs46441523), which contained five base pairs of nucleic acids from the start of exon 6 that had a 78% frequency difference between the highland Tibetans and the lowland Han Chinese, was assessed.
EPAS1 has 16 exons extending over 90 kb on chromosome 2, with a large first intron (50 kb) (24). EPAS1 rs6756667, rs13419896 and rs4953354 are localized in the EPAS1 intron region and can affect the binding to various transcription factors, as indicated by bioinformatics analysis (http://www.gene-regulation.com/pub/programs.html; http://motif.genome.ad.jp). In the present study, the exonic SNPs were not selected, as a result of the lack of information on SNPs in the exons of EPAS1 in Tibetans (8–10). Furthermore, previous studies have shown that the majority of the EPAS1 SNPs that are highly differentiated between Tibetans on the Tibetan Plateau and non-Tibetan CHB individuals only occur within the introns of EPAS1 (9,25). At the protein level, exposure to acute hypoxia induces rapid but short-lived responses, which often occur as a result of the modification of existing proteins; by contrast, chronic hypoxia induces responses that are delayed but durable and that depend on changes in mRNA and protein expression (26).
A major limitation of a traditional case-control study is the potential for population stratification when inappropriate patient-control matching occurs, such as the use of healthy blood donors as the control group. To avoid a confounding association, an unmatched, nested case-control study design was selected (27); in the study, male lowland Han individuals were enrolled and prospectively followed, in order to determine whether those who had genetic variants had a lesser or higher risk of AMS. It was found that the rs6756667 GG genotype significantly increased the risk of developing AMS compared with the AA and heterozygous AG genotypes. The present results are consistent with those of a previous study involving individuals of a Tibetan ethnicity, including Sherpas, that was performed by Hanaoka et al (28). Previous genome-wide studies have revealed that the rs6756667 polymorphism is significantly associated with high-altitude adaptation in Tibetans (8,14). Furthermore, Beall et al (8) investigated 31 SNPs, including rs6756667, and the data showed a significant association with the blood levels of Hb in Tibetans. Results from Hanaoka et al (28) indicated that EPAS1 was under selection for adaptation to the high-altitude life of the Sherpa population. Bioinformatics analysis (http://www.gene-regulation.com/pub/programs.html; http://motif.genome.ad.jp) has revealed that the rs6756667 site contains a non-functional domain, but cis-element activity regulates EPAS1 expression by binding certain transcription factors (CREB, CRB-BP or v-ErbA); however, additional functional studies are necessary to clarify this theory. These findings underscore a potentially important role of EPAS1 in influencing the development of life-threatening high-altitude cerebral or pulmonary edema from AMS. The analysis of patients with cerebral and pulmonary edema is likely to provide further validation.
The present study additionally assessed physiological data (Hb, HCT and RBC levels), in order to investigate their association with EPAS1 rs6756667 SNP. At sea level, Hb, HCT and RBCs are important physiological requirements for the Hb-oxygen transport system to efficiently carry and deliver oxygen to the tissues, cells and molecules of the organism (29). The ascent to high altitude is associated with a reduction in the atmospheric O2 level; therefore, individuals ascending to high altitude undergo the physiological stimulus of systemic hypoxia (27), which evokes an adaptive (homeostatic) response. EPAS1 plays a central role in the response to hypoxia (30), and hypoxia-sensing genes, including the EPAS1 pathways, are the key regulators of RBC production (erythropoiesis), development, energy metabolism, vasculogenesis, iron metabolism, cardiopulmonary regulation and tumor promotion in mammals (13,31). In the present study, it was found that the EPAS1 rs6756667 SNP was significantly associated with levels of Hb, RBCs and HCT in the AMS group, indicating that EPAS1 SNPs are associated with AMS development and that GG homozygotes may have an advantage over heterozygotes during this process. A previous study showed that EPAS1 was associated with certain clinical phenotypes in high-altitude illness (19). In the study, the EPAS1 rs46441523 (C/G) SNP was assessed for an association with age, RBCs, HCT, Hb, SaO2, HR and systolic/diastolic BP among the chronic mountain sickness (CMS) and AMS groups. An association was found between age and CMS, but no association was found among the difference parameters and AMS (19). The results may thus be complicated by different sites and subjects (including males and females).
The present study does have certain limitations. First, only three, and not all, of the EPAS1 tag SNPs were selected. Furthermore, no verification study was performed using another set of samples, and the effect of the SNP that was positively associated with AMS on the expression or function of EPAS1 protein was not assessed. In addition, the complexity and difficulties in obtaining additional blood samples did not enable the plasma index to be determined in all subjects at high altitude, and the subjects were randomly selected for the routine blood examination (such as levels of Hb, RBCs and HCT). Another limitation was that the proportion of individuals with the AA genotype in the database was very low (5.6%); therefore, patients with the rs6756667 AA homozygous genotype were not included for the analysis of physiological features (Hb, RBCs and HCT) in the AMS population in the present study. Despite the limited power of the clinical association study, however, the results have shown for the first time, to the best of our knowledge, the presence of an association between the rs6756667 polymorphism and the risk of developing AMS in this male Chinese Han population.
Acknowledgements
This study was supported in part by a grant from the Special Health Research Project, Chinese Ministry of Health (grant no. 201002012). The authors would like to thank all the individuals that participated in the study and are grateful to Dr En-Hao Zhang, Dr Xiao-Han Ding, Dr Ming-Yue Rao and Mr. Can Chen of the research team for their continued support of this study. The authors would additionally like to thank Medjaden Bioscience Ltd. for assisting in the preparation of this manuscript.
References
- 1.Basnyat B, Murdoch DR. High-altitude illness. Lancet. 2003;361:1967–1974. doi: 10.1016/S0140-6736(03)13591-X. [DOI] [PubMed] [Google Scholar]
- 2.MacInnis MJ, Wang P, Koehle MS, Rupert JL. The genetics of altitude tolerance: The evidence for inherited susceptibility to acute mountain sickness. J Occup Environ Med. 2011;53:159–168. doi: 10.1097/JOM.0b013e318206b112. [DOI] [PubMed] [Google Scholar]
- 3.Barry PW, Pollard AJ. Altitude illness. BMJ. 2003;326:915–919. doi: 10.1136/bmj.326.7395.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartsch P, Swenson ER. Clinical practice: Acute high-altitude illnesses. N Engl J Med. 2013;368:2294–2302. doi: 10.1056/NEJMcp1214870. [DOI] [PubMed] [Google Scholar]
- 5.Schommer K, Bartsch P, Maeder MB. High Altitude Illness. Notarzt. 2013;29:175–183. (In German) [Google Scholar]
- 6.Schneider M, Bernasch D, Weymann J, Holle R, Bartsch P. Acute mountain sickness: Influence of susceptibility, preexposure, and ascent rate. Med Sci Sports Exerc. 2002;34:1886–1891. doi: 10.1097/00005768-200212000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, Scherer SW, Julian CG, Wilson MJ, López Herráez D, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, Li C, Li JC, Liang Y, McCormack M, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y, Yang ZH, Zhang H, Cui C, Qi X, Luo X, Tao X, Wu T, Ouzhuluobu Basang, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 10.Simonson TS, Yang YZ, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 11.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji LD, Qiu YQ, Xu J, Irwin DM, Tam SC, Tang NL, Zhang YP. Genetic adaptation of the hypoxia-inducible factor pathway to oxygen pressure among eurasian human populations. Mol Biol Evol. 2012;29:3359–3370. doi: 10.1093/molbev/mss144. [DOI] [PubMed] [Google Scholar]
- 13.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu S, Li S, Yang Y, Tan J, Lou H, Jin W, Yang L, Pan X, Wang J, Shen Y, et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 2011;28:1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- 15.Percy MJ, Furlow PW, Lucas GS, Li X, Lappin TR, McMullin MF, Lee FS. A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N Engl J Med. 2008;358:162–168. doi: 10.1056/NEJMoa073123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bougatef F, Quemener C, Kellouche S, Naïmi B, Podgorniak MP, Millot G, Gabison EE, Calvo F, Dosquet C, Lebbé C, et al. EMMPRIN promotes angiogenesis through hypoxia-inducible factor-2alpha-mediated regulation of soluble VEGF isoforms and their receptor VEGFR-2. Blood. 2009;114:5547–5556. doi: 10.1182/blood-2009-04-217380. [DOI] [PubMed] [Google Scholar]
- 17.Buroker NE, Ning XH, Zhou ZN, Li K, Cen WJ, Wu XF, Zhu WZ, Scott CR, Chen SH. AKT3, ANGPTL4, eNOS3 and VEGFA associations with high altitude sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau. Int J Hematol. 2012;96:200–213. doi: 10.1007/s12185-012-1117-7. [DOI] [PubMed] [Google Scholar]
- 18.Ding H, Liu Q, Hua M, Ding M, Du H, Zhang W, Li Z, Zhang J. Polymorphisms of hypoxia-related genes in subjects susceptible to acute mountain sickness. Respiration. 2011;81:236–241. doi: 10.1159/000322850. [DOI] [PubMed] [Google Scholar]
- 19.Buroker NE, Ning XH, Zhou ZN, Li K, Cen WJ, Wu XF, Zhu WZ, Scott CR, Chen SH. EPAS1 and EGLN1 associations with high altitude sickness in Han and Tibetan Chinese at the Qinghai-Tibetan Plateau. Blood Cells Mol Dis. 2012;49:67–73. doi: 10.1016/j.bcmd.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Roach RC, Bärtsch P, Oelz O, Hachett P. (Lake Louise AMS Scoring Consensus Committee). The Lake Louise acute mountain sickness scoring system. In: Sutton JR, Houston CS, Coates G, editors. Hypoxia and Molecular Medicine. Queen City Printers; Burlington, VT: 1993. pp. 272–274. [Google Scholar]
- 21.International HapMap Consortium, corp-author. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Imray C, Booth A, Wright A, Bradwell A. Acute altitude illnesses. Brit Med J. 2011;343:d4943. doi: 10.1136/bmj.d4943. [DOI] [PubMed] [Google Scholar]
- 24.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 25.Wang BB, Zhang YB, Zhang F, Lin H, Wang X, Wan N, Ye Z, Weng H, Zhang L, Li X, et al. On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLoS One. 2011;6:e170022. doi: 10.1371/journal.pone.0017002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92:967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai RJ, Glynn RJ, Gagne JJ. Performance of disease risk score matching in nested case-control studies. Pharmacoepidemiol Drug Saf. 2014;23(Suppl):148. [Google Scholar]
- 28.Hanaoka M, Droma Y, Basnyat B, Ito M, Kobayashi N, Katsuyama Y, Kubo K, Ota M. Genetic variants in EPAS1 contribute to adaptation to high-altitude hypoxia in Sherpas. PLoS One. 2012;7:e50566. doi: 10.1371/journal.pone.0050566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen FB. The dual roles of red blood cells in tissue oxygen delivery: Oxygen carriers and regulators of local blood flow. J Exp Biol. 2009;212:3387–3393. doi: 10.1242/jeb.023697. [DOI] [PubMed] [Google Scholar]
- 30.Hirota K, Semenza GL. Regulation of angiogenesis by hypoxia-inducible factor 1. Crit Rev Oncol Hematol. 2006;59:15–26. doi: 10.1016/j.critrevonc.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Semenza GL. Hypoxia-inducible factors: Mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33:207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]