Abstract
Background:
The role of retinoblastoma (Rb) protein in cell cycle regulation prompted us to take up this study with the aim of assessing its role in the progression of oral cancer and to correlate with various clinicopathological parameters, including habits such as smoking, Paan chewing, and alcoholism.
Materials and Methods:
This observational study included surgical specimens from 10 apparently normal oral mucosa, 14 oral reactive lesions (ORL), 29 precancerous lesions and 43 oral cancers. The expression of Rb protein in tissue samples were evaluated by immunohistochemistry and correlated with clinicopathological data. The percentage and mean expression of Rb protein were statistically analyzed using Student's t-test and P < 0.05 was considered as statistically significant difference.
Results:
The expression of Rb protein was found to increase from normal, ORL, precancerous lesions to cancers. A consistently high expression of Rb protein was seen in oral cancers, with an increase in well-differentiated and moderately differentiated tumors. Patients with combined habits of Paan chewing, smoking, and alcohol consumption had a higher expression compared with those without habits.
Conclusion:
Within the limitations of this study, it seems that overexpression of Rb protein noted in oral cancer, with an increase in well and moderately differentiated tumors suggest a possible role of Rb in differentiation. The high expression of Rb in patients with combined habits of Paan chewing, smoking and alcohol consumption indicates that Rb pathway may be altered in habit-related oral malignancies.
Keywords: Oral cancer, preneoplastic conditions, retinoblastoma protein, tumor suppressor gene, tumor suppressor proteins
INTRODUCTION
Head and neck cancer accounts for 10% of all malignancies worldwide, of which 40% are oral cancers.[1] The highest incidence is observed in India, with 83,000 new cases and 46,000 deaths from oral cancer every year.[2] The age standardized incidence rate of oral cancer in Trivandrum, India is 16.3/100,000 in males and 7.7 in females.[3] The high incidence of oral cancer in Trivandrum is related to smoking, Paan chewing, and alcohol use.[4] Adults above 40 years with the habit of smoking are at a higher risk for developing oral cancer.[5]
Multiple molecular mechanisms, including tumor suppressor genes are involved in the pathogenesis of oral cancer. Tumor suppressor gene is a gene whose protein product can inhibit the transformation of a normal cell to tumor cell and therefore, whose loss of function contribute to the malignant transformation of the cell. The retinoblastoma (Rb) tumor suppressor gene plays a key role in the regulation of cell cycle and differentiation. Its protein product Retinoblastoma tumor suppressor protein (pRb) acts as a regulator at the G1-S restriction point, capable of arresting the growth in mid G1-S phase. Critical to pRb function is the regulation of its phosphorylation state throughout the cell cycle and its ability to interact with other proteins.[6] Mutations lead to functional pRb inactivation and failure of pRb mediated growth and tumor suppression.[7]
Cell cycle regulation is critical in tumorigenesis, and Rb gene inactivation has been reported in various solid cancers.[8] Previous reports[9] suggested that Rb protein function was absent in malignant oral epithelium, whereas others[10,11] have indicated raised Rb protein during oral cancer progression. Although the alteration in Rb protein expression in tumorigenesis has not been completely elucidated, there is increasing evidence that Rb protein pathway is rendered dysfunctional in oral cancer.[12] The strong role of pRb in cell cycle regulation prompted us to take up this study, with the aim of assessing the role of Rb protein in the progression of oral cancer, by indicating its expression in the spectrum of lesions ranging from normal to the different histological types of oral cancers. The study also correlates the expression of Rb protein with the clinicopathological parameters including habits like smoking, Paan chewing and alcoholism. The role of pRb in the differentiation of tumor was also assessed.
MATERIALS AND METHODS
Case selection
This study was conducted in the Department of Oral Medicine and Radiology, Dental College, Trivandrum, with the collaboration of the Division of Cancer Research, Regional Cancer Centre, Trivandrum, India. Patients were screened and 4 groups namely normal, oral reactive lesions (ORL), precancerous lesions and oral cancers were selected for the study. After assessment of routine blood and urine investigations, incision biopsy under local anesthesia was taken from the selected groups, with their informed consent. Institutional Ethical Committee clearance was obtained.
Clinicopathological characteristics of patients
The study consisted of 96 patients with 40 males and 56 females. The clinicopathological characteristics of the patients included the data on age, sex, family history and habits like smoking, Paan chewing and alcoholism. The surgical specimens included 10 apparently normal oral mucosa, 14 ORL, 29 precancerous lesions and 43 oral cancers. The apparently normal tissue was taken from the buccal mucosa adjacent to impacted third molar, with the informed consent of patients who reported for extraction. The ORL included fibroma and mucocele. Among the 29 precancers, there were 19 cases of leukoplakia and 10 cases of oral submucous fibrosis (OSF). White lesions detected clinically as leukoplakia, with histological evidence of epithelial dysplasia (9 mild, 8 moderate and 2 severe dysplasia) were taken for immunohistochemical analysis. The precancerous lesions were considered as a single group in this study and mucosal dysplasia with different severity was not compared. Clinically diagnosed cases of leukoplakia, with histopathological features of severe dysplasia and invasion of tumor cells into the superficial connective tissue was included as early invasive squamous cell carcinoma (EISCC). The 43 cancers included 39 squamous cell carcinoma (SCC) and 4 verrucous carcinoma (VerrucousCa). Histologically, the tumors were graded into well differentiated (WDSCC), moderately differentiated (MDSCC) and poorly differentiated (PDSCC). Histological grading was based on the degree of resemblance of the invading carcinoma to the normal epithelium and its ability to form keratinizing islands, and follows the description in World Health Organization classification.[13]
Immunohistochemistry
Of the sections cut from formalin fixed, paraffin embedded specimens, one was used for routine hematoxylin and eosin staining and the other was used for immunohistochemistry staining by Avidin and Biotin methods. For antigen unmasking, the deparaffinized slides were placed in a rack in boiling citrate buffer in a pressure cooker containing 10 mM citrate buffer (pH-6) for at least 5 min. The slides were allowed to cool in buffer for 20 min and were washed twice in phosphate buffered saline (PBS) and used for the standard staining procedure. Endogenous peroxidase activity was blocked by incubating slides for 30 min in 0.3% H2O2 in methanol, washed with excess water and stabilized with PBS. The sections were incubated with 3% bovine serum albumin in humidified chamber for 30 min and removed. The sections were incubated with primary antibody Rb (Rb, Dako, Denmark) overnight at 4°C in the humid chamber. Washed extensively with PBS and then incubated with secondary biotinylated antibody for 30 min at 37°C. Washed in PBS thrice and incubated with Streptavidin Horse Radish Peroxidase reagent (Dako AS, Denmark) for 30 min. Washed extensively with PBS and substrate diaminobenzidine was added and incubated for 10 min in the dark, and a brown precipitate was formed. Washed with distilled water, and counter stained with hematoxylin for 1 min.
Evaluation of slides
The immunoreactivity was examined under light microscope (Leica-DMLB, Germany) (×400 magnification). The percentage of cells showing positive nucleus was counted by 2 pathologists who were equally experienced and both were blinded. Staining for Rb protein was considered as positive or negative based on the presence or absence of brown staining. Only nuclear staining was considered as evidence of protein. The Rb percentage 1-10% showing weak staining for Rb protein was considered as Rb negative and >10% was considered as Rb positive.[12] As the mean expression of Rb in the various groups of oral lesions was recorded, the Rb positive group was not graded percentage wise.
Statistical analysis
The expression of Rb protein in the tissue samples was statistically analyzed using the Student's t-test. For small sample size, the F-test (Levene's test)[14] was used. The results were expressed as mean ± standard error and P < 0.05 was considered to be statistically significant difference. The association between different variables was analyzed using Bivariate analysis (Spearman Rank Sum Test).
RESULTS
Among the 96 patients in the study, there were 40 males and 56 females. The mean age of patients included in normal was 41, ORL were 43, precancerous lesions were 49 and in oral cancer was 57 years.
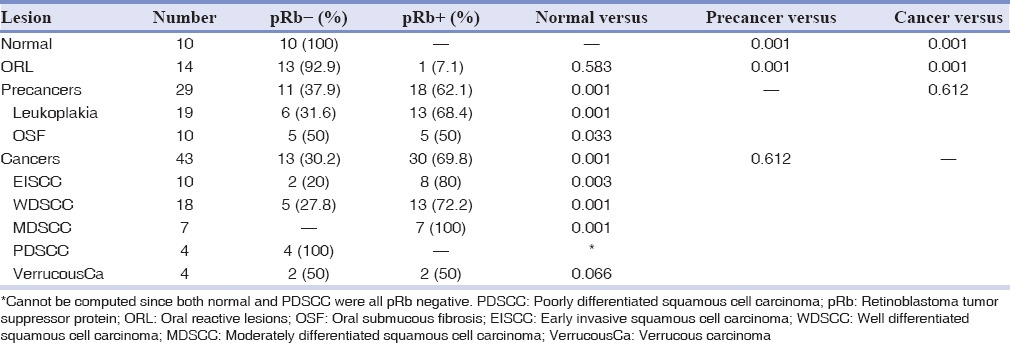
In this study, both percentage and mean expression of Rb protein were evaluated by immunohistochemistry. The percentage of Rb protein studied was found to increase from normal, ORL, precancerous lesions to oral cancers. All the 10 normal cases included in the study were pRb negative, with a percentage nuclear staining of 1%. The immunoreactivity of ORL was comparable to that in normal with 13 cases (92.9%) pRb negative showing a nuclear staining of 1%. The percentage of Rb protein expression in 29 precancerous lesions showed 18 (62.1%) pRb positive cases. Among the precancerous lesions studied, 68.4% of leukoplakia was pRb positive, while 50% of OSF was pRb negative. When the group of cancers were analyzed, there was a significant increase in the percentage expression with 30 cases (69.8%) pRb positive. 13 (72.2%) of the WDSCC and 7 (100%) of the MDSCC cases showed pRb positivity, which was significantly higher when compared to that in normal (P = 0.001). All the 4 cases (100%) of PDSCC was negative for pRb nuclear staining, which was comparable to that seen in normal. 50% of the VerrucousCa showed a positive pRb expression [Table 1].
Table 1.
Percentage of pRb in the oral lesions
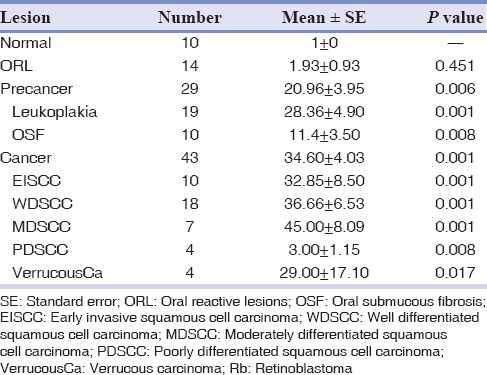
The mean expression of pRb in ORL (1.93 ± 0.93) though slightly increased, was comparable to that in normal (1 ± 0). The mean staining score of Rb protein was high in both precancers (20.96 ± 3.95) and cancers (34.60 ± 4.03) and the expression was significantly higher when compared to normal oral mucosa (P = 0.006 and P = 0.001 respectively). Among the precancerous lesions, the expression of pRb in leukoplakia was high (28.36 ± 4.90) and was comparable to that seen in cancers. However, in the case of OSF, there was a significant decrease in the mean pRb expression (11.4 ± 3.50). Among the group of cancers studied, there was an increase in the expression of pRb from WDSCC (36.66 ± 6.53) to MDSCC (45.00 ± 8.09) and down regulation in PDSCC (3.00 ± 1.15). A high expression was also noted in VerrucousCa [Table 2].
Table 2.
Mean expression of Rb protein
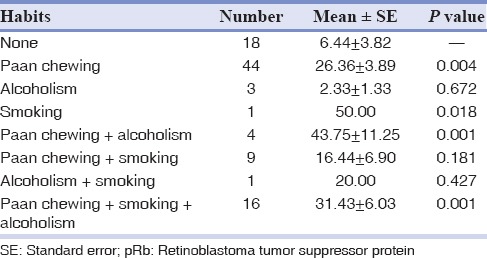
The expression of Rb protein did not show any statistically significant correlation with regard to age, sex and family history. When the habits of the patients were correlated for the mean expression of pRb, it was noted that patients with combined habits of Paan chewing, smoking and alcohol consumption had a significantly higher expression of pRb (31.43 ± 6.03) compared to those without any habits (6.44 ± 3.82). One patient with the habit of smoking alone had a high expression of pRb (50.00). In alcohol users, there was a down regulation in the expression of pRb (2.33 ± 1.33). However, alcoholism when combined with smoking and Paan chewing, the expression of pRb was high (20.00 and 43.75 ± 11.25, respectively) [Table 3].
Table 3.
Correlation of habits with mean pRb expression
The immunohistochemical expression of Rb protein in the various oral lesions studied is shown in Figures 1a–e.
Figure 1a.
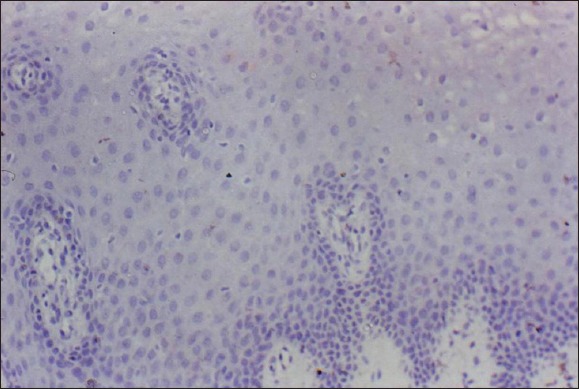
Normal oral epithelium: Negative retinoblastoma tumor suppressor protein expression (×400).
Figure 1b.
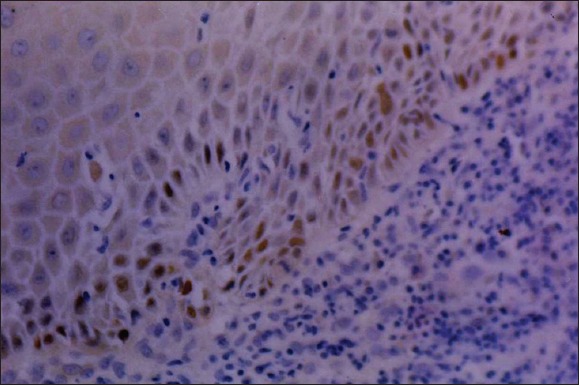
Leukoplakia: High retinoblastoma tumor suppressor protein expression (×400).
Figure 1c.
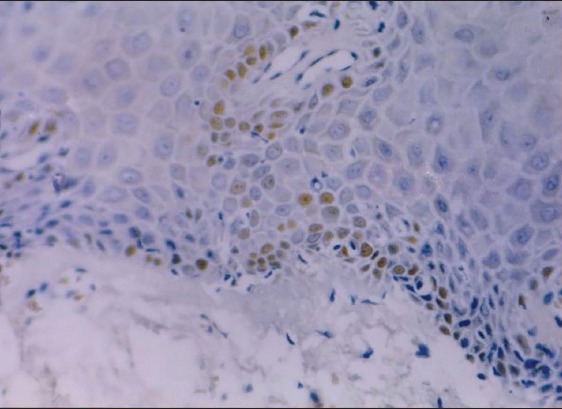
Oral submucous fibrosis: Moderate retinoblastoma tumor suppressor protein expression (×400).
Figure 1d.
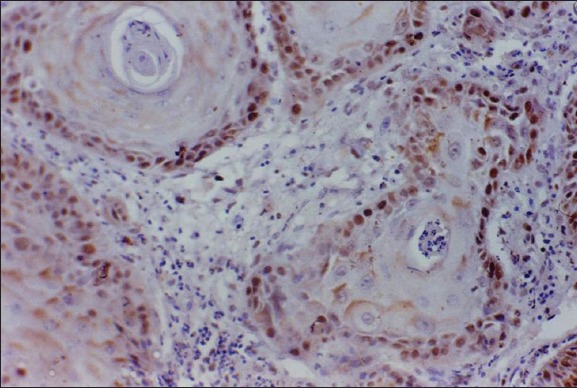
Well differentiated squamous cell carcinoma: Very high retinoblastoma tumor suppressor protein expression (×400).
Figure 1e.
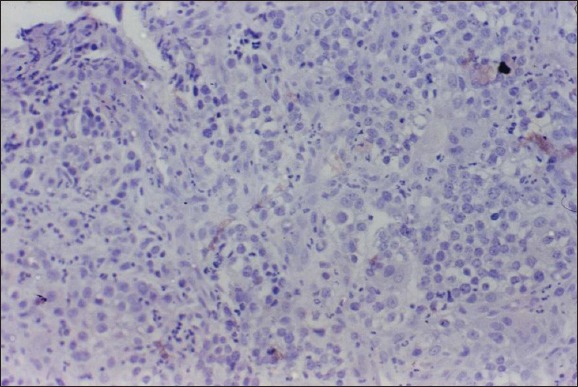
Poorly differentiated squamous cell carcinoma: Negative retinoblastoma tumor suppressor protein expression (×400).
DISCUSSION
In this study, the mean expression of Rb protein in the different stages from normal oral mucosa to the cancerous stage was evaluated by immunohistochemistry and correlated with clinicopathological data. The results indicate that, both the percentage and mean expression of Rb protein demonstrated an increase from normal, ORL, precancerous lesions to oral cancers. All normal cases in the study were negative for the expression of Rb protein. Pande et al.[6] showed 93% of normal tissues to be pRb positive, when purely negative cases (0% nuclear staining) were only considered as negative, with the range of positivity being 1-10%. The mean expression of pRb in ORL was comparable to that seen in normal oral mucosa.
The percentage and mean expression of Rb protein in precancers and cancers were significantly higher when compared to that in normal and ORL. Among the precancerous lesions, the expression of pRb in leukoplakia was high, which was comparable to that seen in cancers, suggesting an overexpression of Rb protein in comparison to that in normal. Schoelch et al.[10] demonstrated a high expression of Rb protein in oral premalignant lesions, while Pande et al.[6] found that there was a significant decrease in the expression. However, in OSF, the Rb protein expression did not appear to be affected to such an extent as that in leukoplakia. This could be due to the fact that, leukoplakia is an alteration affecting the proliferation of epithelial cells, while the abnormality in OSF lies with collagen synthesis. This suggests that Rb protein has probably no role to play in the pathogenesis of malignancies in OSF and its alteration could be a later step leading to the progression to oral cancer.
A consistently high expression of Rb protein, much above that in normal and ORL, was seen in oral cancers. Previous reports[9] suggest that Rb protein function is absent in malignant oral epithelium and others[10,11] have shown raised protein levels during oral cancer progression. In a study conducted by Muirhead et al.[8]87% of oral cancer patients showed expression of Rb protein. Regezi et al.[11] detected an overexpression of pRb in lateral tongue cancers, while Schoelch et al.[10] found that there was an increase in the expression of Rb protein with progression of the disease. Of note, in recent studies, oral SCC presented higher expression of Rb protein compared with premalignant lesions, 75 versus 25%.[12] The increase of nuclear pRb levels in the development of malignancy appears inconsistent with previous reports focusing on loss of Rb function in head and neck malignancies.[15,16,17,18,19] Increased pRb expression has also been demonstrated in laryngeal carcinoma,[20] colorectal cancer[21] and in ovarian epithelial tumors.[22,23] Trudel et al.[24] observed intense Rb positivity or “overexpression” in some grade 3 breast cancers, but they could not clearly explain the reason for this and considered this finding paradoxical.
It has been suggested by previous workers[25] that overexpression of pRb reflect an alteration in the Rb pathway resulting in loss of tumor suppressor function. Although the mechanism for increased pRb expression and possible alterations is not clear, a number of possibilities exist. The overexpressed pRb may represent a hyperphosphorylated form of the protein. Rb acts to control cell proliferation through regulation of the cell cycle at the G1-S phase transition. The growth suppressor activity of pRb is controlled at the level of phosphorylation. When pRb is hypophosphorylated, it is capable of binding to E2F transcription factor.[26] However, when pRb is phosphorylated by cyclin dependent kinase complexes, E2F is released, and cell can initiate DNA synthesis.[27] An alternate explanation concerns the possible role of pRb in the regulation of apoptosis. Overexpression of Rb protein has been shown to block p53 mediated apoptosis.[28] The role of transcription regulator E2F has been postulated as a possible mechanism for this apoptotic function, as overexpression of E2F alone can lead to apoptosis.[29] These findings suggest a possible role of overexpressed pRb in binding E2F and inhibiting normal apoptotic function. Finally, binding of pRb to other proteins such as MDM2 or certain DNA viral oncoproteins may also abrogate the function of pRb, while preserving its expression.[25]
An overexpression of pRb was noted in early invasive squamous cell carcinoma (EISCC) which was consistent with previous reports.[10] In the early stages of oral cancer, it cannot be excluded that Rb gene might contain genetic abnormalities, which could lead to functionally inactivated protein with a subsequent longer degradation resulting in high protein levels.[19]
When the different histological types of oral cancers were analyzed separately, Rb protein expression was related to differentiation of tumor, with an increase in the expression of pRb from WDSCC to MDSCC. However, it was surprising to note that, there was a down regulation in PDSCC, where all the cases were negative for the expression of Rb protein, which was comparable to that seen in normal. Tanaka et al.[30] found a similar result of increased expression of Rb protein in WDSCC than in PDSCC. This could be that, in the case of oral cancer, the Rb protein produced is an abnormal type, while in PDSCC there could be a down regulation of Rb production, both leading to low availability of functional pRb. In neoplasia, nonfunctional mutant Rb, low Rb expression and maximally phosphorylated Rb have been described, indicating the potential mechanisms by which neoplastic cells can escape from cell cycle control in various types of tumors.[11] Terminal differentiation is characterized by permanent cell cycle withdrawal, and is on the molecular level, accompanied by the modulation of numerous genes relevant for the expression of the differentiated phenotype. Rb gene was found to play an important role in it.[25] Muirhead et al. found that 39 of 45 tumors that expressed Rb displayed marked keratinization and 5 out of 6 poorly differentiated tumors were negative for the expression of Rb. In their study, an inverse relationship between p16 and Rb expression was noted, and they hypothesize that p16 or Rb has an effect on cell differentiation. The cells are probably arrested at a stage within the process of differentiation, leading to the tumors comprising of poorly differentiated areas.[8] An inverse relationship between p16 and Rb indicate that the alteration in multiple tumor suppressor pathways needs to be studied for assessing the role of Rb in differentiation.[6,8] Inactivation of pRb by oncoviral proteins interferes with differentiation and can reactivate the cell cycle machinery even in terminally differentiated cells.[31] This hypothesis is supported by the fact that, pRb was recognized mainly in the cells of well-differentiated carcinomas and not in poorly differentiated ones. The high mean expression of Rb protein in VerrucousCa suggests that, pRb may be functionally inactivated by other mechanisms, including human papilloma virus infection.
The expression of Rb protein in the various groups of oral lesions, when correlated clinicopathologically, did not show any statistical significance with regard to age, sex, and family history that was consistent with previous reports.[8] When the habits of patients were analyzed, it was found that, in patients with a combination of smoking, Paan chewing and alcoholism, there was a higher expression of pRb when compared to those without habits and a down regulation of pRb in alcohol users. Pande et al.[6,32] have indicated that alterations in p16/pRb pathway may be involved in the development of betel and tobacco related oral malignancies. However, alcoholism when combined with smoking and Paan chewing, the expression of pRb was high suggesting a synergistic role of these habits in the expression of Rb protein.
CONCLUSION
It was evident from this study that, the expression of Rb protein was found to increase from normal through the various groups ORL, precancers to cancers. The overexpression seen in oral cancer, with an increase in well-differentiated and moderately differentiated tumors suggest the possible role of Rb in differentiation. It can be postulated that the gross down regulation in poorly differentiated tumors, probably due to loss or inactivation of the gene or protein, could be the mechanism driving the epithelial cells to the poorly differentiated phenotype. This has to be confirmed with the help of mutational and loss of heterozygosity studies. The overexpression of pRb in patients with a combination of habits like smoking, Paan chewing and alcohol consumption, and the gross down regulation of pRb in alcohol users indicates that the alteration of Rb pathway may be involved in the development of the habit related oral malignancies.
However, due to the limited sample size of poorly differentiated tumors in the present study, further studies based on larger series of patients and involvement of multiple tumor suppressor pathways are needed to confirm the role of Rb in differentiation. The results indicate that, the level of pRb expression, and not simply its presence or absence, should be determined when assessing the impact of Rb on cancer progression.
Footnotes
Source of Support: Nil.
Conflicts of Interest: The authors of this manuscript declare that they have no conflicts of interest, real or perceived, financial or nonfinancial in this article.
REFERENCES
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Whelan SL, Ferlay J, Teppo L, Thomas DB. Lyon: IARC Press; 2002. Cancer Incidence in Five Continents. [Google Scholar]
- 3.Krishnan Nair M, Varghese C, Mathew A, Gangadharan P. Cancer incidence in Trivandrum, India. In: Parkin DM, Whelan SL, Ferlay J, Young J Jr, Raymond L, editors. Cancer Incidence in Five Continents. VII. Lyon: International Agency for Research on Cancer; 1997. pp. 358–67. IARC Scientific Publications No.: 143. [Google Scholar]
- 4.Muwonge R, Ramadas K, Sankila R, Thara S, Thomas G, Vinoda J, et al. Role of tobacco smoking, chewing and alcohol drinking in the risk of oral cancer in Trivandrum, India: A nested case-control design using incident cancer cases. Oral Oncol. 2008;44:446–54. doi: 10.1016/j.oraloncology.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Jadhav K, Singh D. Assessment of psychological dependence among tobacco users: A survey held among the rural population of India to call for attention of tobacco cessation centers. Dent Res J (Isfahan) 2013;10:467–73. [PMC free article] [PubMed] [Google Scholar]
- 6.Pande P, Mathur M, Shukla NK, Ralhan R. pRb and p16 protein alterations in human oral tumorigenesis. Oral Oncol. 1998;34:396–403. doi: 10.1016/s1368-8375(98)00024-4. [DOI] [PubMed] [Google Scholar]
- 7.Kim MS, Li SL, Bertolami CN, Cherrick HM, Park NH. State of p53, Rb and DCC tumor suppressor genes in human oral cancer cell lines. Anticancer Res. 1993;13:1405–13. [PubMed] [Google Scholar]
- 8.Muirhead DM, Hoffman HT, Robinson RA. Correlation of clinicopathological features with immunohistochemical expression of cell cycle regulatory proteins p16 and retinoblastoma: Distinct association with keratinisation and differentiation in oral cavity squamous cell carcinoma. J Clin Pathol. 2006;59:711–5. doi: 10.1136/jcp.2005.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Todd R, Hinds PW, Munger K, Rustgi AK, Opitz OG, Suliman Y, et al. Cell cycle dysregulation in oral cancer. Crit Rev Oral Biol Med. 2002;13:51–61. doi: 10.1177/154411130201300106. [DOI] [PubMed] [Google Scholar]
- 10.Schoelch ML, Regezi JA, Dekker NP, Ng IO, McMillan A, Ziober BL, et al. Cell cycle proteins and the development of oral squamous cell carcinoma. Oral Oncol. 1999;35:333–42. doi: 10.1016/s1368-8375(98)00098-0. [DOI] [PubMed] [Google Scholar]
- 11.Regezi JA, Dekker NP, McMillan A, Ramirez-Amador V, Meneses-Garcia A, Ruiz-Godoy Rivera LM, et al. P53, p21, Rb, and MDM2 proteins in tongue carcinoma from patients <35 versus >75 years. Oral Oncol. 1999;35:379–83. doi: 10.1016/s1368-8375(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 12.de Oliveira MG, Ramalho LM, Gaião L, Pozza DH, de Mello RA. Retinoblastoma and p53 protein expression in pre-malignant oral lesions and oral squamous cell carcinoma. Mol Med Rep. 2012;6:163–6. doi: 10.3892/mmr.2012.876. [DOI] [PubMed] [Google Scholar]
- 13.Barnes L, Eveson JW, Reichart P, Sidransky D. Lyon: IARC Press; 2005. World Health Organization Classification of Tumours. Pathology and Genetics of Head and Neck Tumors; p. 119. [Google Scholar]
- 14.Levene H, Olkin I, Hotelling H, editors. Stanford: Stanford University Press; 1960. Contributions to Probability and Statistics: Essays in Honour of Harold Hotelling; pp. 278–92. [Google Scholar]
- 15.Pavelic ZP, Lasmar M, Pavelic L, Sorensen C, Stambrook PJ, Zimmermann N, et al. Absence of retinoblastoma gene product in human primary oral cavity carcinomas. Eur J Cancer B Oral Oncol. 1996;32B:347–51. doi: 10.1016/0964-1955(96)00025-5. [DOI] [PubMed] [Google Scholar]
- 16.Yokoyama J, Shiga K, Sasano H, Suzuki M, Takasaka T. Abnormalities and the implication of retinoblastoma locus and its protein product in head and neck cancers. Anticancer Res. 1996;16:641–4. [PubMed] [Google Scholar]
- 17.Maestro R, Piccinin S, Doglioni C, Gasparotto D, Vukosavljevic T, Sulfaro S, et al. Chromosome 13q deletion mapping in head and neck squamous cell carcinomas: Identification of two distinct regions of preferential loss. Cancer Res. 1996;56:1146–50. [PubMed] [Google Scholar]
- 18.Gimenez-Conti IB, Collet AM, Lanfranchi H, Itoiz ME, Luna M, Xu HJ, et al. P53, Rb, and cyclin D1 expression in human oral verrucous carcinomas. Cancer. 1996;78:17–23. doi: 10.1002/(SICI)1097-0142(19960701)78:1<17::AID-CNCR4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 19.Andl T, Kahn T, Pfuhl A, Nicola T, Erber R, Conradt C, et al. Etiological involvement of oncogenic human papillomavirus in tonsillar squamous cell carcinomas lacking retinoblastoma cell cycle control. Cancer Res. 1998;58:5–13. [PubMed] [Google Scholar]
- 20.Krecicki T, Smigiel R, Fraczek M, Kowalczyk M, Sasiadek MM. Studies of the cell cycle regulatory proteins P16, cyclin D1 and retinoblastoma protein in laryngeal carcinoma tissue. J Laryngol Otol. 2004;118:676–80. doi: 10.1258/0022215042244769. [DOI] [PubMed] [Google Scholar]
- 21.Ali AA, Marcus JN, Harvey JP, Roll R, Hodgson CP, Wildrick DM, et al. RB1 protein in normal and malignant human colorectal tissue and colon cancer cell lines. FASEB J. 1993;7:931–7. doi: 10.1096/fasebj.7.10.8344490. [DOI] [PubMed] [Google Scholar]
- 22.Dodson MK, Cliby WA, Xu HJ, DeLacey KA, Hu SX, Keeney GL, et al. Evidence of functional RB protein in epithelial ovarian carcinomas despite loss of heterozygosity at the RB locus. Cancer Res. 1994;54:610–3. [PubMed] [Google Scholar]
- 23.Kim TM, Benedict WF, Xu HJ, Hu SX, Gosewehr J, Velicescu M, et al. Loss of heterozygosity on chromosome 13 is common only in the biologically more aggressive subtypes of ovarian epithelial tumors and is associated with normal retinoblastoma gene expression. Cancer Res. 1994;54:605–9. [PubMed] [Google Scholar]
- 24.Trudel M, Mulligan L, Cavenee W, Margolese R, Côté J, Gariépy G. Retinoblastoma and p53 gene product expression in breast carcinoma: Immunohistochemical analysis and clinicopathologic correlation. Hum Pathol. 1992;23:1388–94. doi: 10.1016/0046-8177(92)90059-c. [DOI] [PubMed] [Google Scholar]
- 25.Cote RJ, Dunn MD, Chatterjee SJ, Stein JP, Shi SR, Tran QC, et al. Elevated and absent pRb expression is associated with bladder cancer progression and has cooperative effects with p53. Cancer Res. 1998;15(58):1090–4. [PubMed] [Google Scholar]
- 26.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 27.Stone JG, Siedlak SL, Tabaton M, Hirano A, Castellani RJ, Santocanale C, et al. The cell cycle regulator phosphorylated retinoblastoma protein is associated with tau pathology in several tauopathies. J Neuropathol Exp Neurol. 2011;70:578–87. doi: 10.1097/NEN.0b013e3182204414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haupt Y, Rowan S, Shaulian E, Vousden KH, Oren M. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 1995;9:2170–83. doi: 10.1101/gad.9.17.2170. [DOI] [PubMed] [Google Scholar]
- 29.Qin XQ, Livingston DM, Kaelin WG, Jr, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci U S A. 1994;91:10918–22. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka N, Odajima T, Nakano T, Kimijima Y, Yamada S, Ogi K, et al. Immunohistochemical investigation of new suppressor oncogene p130 in oral squamous cell carcinoma. Oral Oncol. 1999;35:321–5. doi: 10.1016/s1368-8375(98)00089-x. [DOI] [PubMed] [Google Scholar]
- 31.Herwig S, Strauss M. The retinoblastoma protein: A master regulator of cell cycle, differentiation and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 32.Pande P, Soni S, Kaur J, Agarwal S, Mathur M, Shukla NK, et al. Prognostic factors in betel and tobacco related oral cancer. Oral Oncol. 2002;38:491–9. doi: 10.1016/s1368-8375(01)00090-2. [DOI] [PubMed] [Google Scholar]


