Abstract
The aim of the present study was to determine the prevalence and related drug resistance of AmpC β-lactamases in Acinetobacter baumannii in tertiary-level hospitals in the Xuzhou region in China. A total of 134 clinical isolates of non-repetitive Acinetobacter baumannii were collected from different hospitals in the Xuzhou region, and multiplex polymerase chain reaction (PCR) was employed to determine the genotype of AmpC. The PCR products were purified and sequenced. The susceptibility to antibiotics was tested using the biometrics automated microbiological-assay system, VITEK-2. Amongst the 134 isolated strains, 96 strains were found to produce AmpC β-lactamases, and the positive rate was 72%, all of which carried acinetobacter-derived cephalosporinase (ADC) type AmpC resistance genes. The drug sensitivity tests indicated that the positive Acinetobacter baumannii strains were resistant to the majority of extended-spectrum β-lactam antibiotics, but were only sensitive to polymyxin. In conclusion, the incidence of AmpC enzymes in Acinetobacter baumannii strains in tertiary-level hospitals in the Xuzhou area is relatively high, and resistance to the majority of extended-spectrum β-lactam antibiotics may be related to the ADC type of AmpC.
Keywords: Acinetobacter baumannii, acinetobacter-derived cephalosporinase, AmpC β-lactamases, polymerase chain reaction
Introduction
Acinetobacter baumannii (A. baumannii), a non-fermenting bacteria, is the known conditioned pathogen leading to noso-comial infection, which exists widely in nature and the hospital environment and is the only Gram-negative bacillus which can survive for weeks on human skin (1). With the wide use of immunosuppressive drugs, the increase in invasion surgeries and the development of intensive care technology (2), an increasing number of A. baumannii infections have been detected. Multi-drug resistance and the amplification of resistance to A. baumannii have increased the mortality rate in the clinic (3). Resistance to antibiotics against A. baumannii mostly occurs due to the loss of membrane proteins, active efflux mechanisms, changes in penicillin-binding proteins (PBPs) and the production of various enzymes, amongst which the production of β-lactamases plays an important role in the resistance to β-lactam antibiotics (5). Both domestic and foreign studies have suggested the severe drug resistance to A. baumannii (4). The aim of the present study was to perform a genetic test and drug resistance analysis for the AmpC enzyme in A. baumannii isolated from tertiary-level hospitals in the Xuzhou region, China by polymerase chain reaction (PCR), to strengthen the study of drug resistance genes, which is of great significance ot understand the distribution of gene type to control and treat infections caused by A. baumannii. The findings of our study may facilitate and provide guidelines for the rational clinical use of antibiotics and the prevention and epidemic control of the drug-resistant bacterium, A. baumannii.
Materials and methods
Materials
A total of 134 clinical isolates of non-repetitive A. baumannii were collected from patients at the First People's Hospital, Affiliated to Xuzhou Medical College, Central Hospital in Xuzhou, China, from August 2012 to November 2012. The Escherichia coli ATCC 25922 strain was employed as a negative quality control, while Enterobacter cloacae was employed as a positive quality control. The VITEK® 2 Compact system was obtained from bioMérieux, Inc., Craponne, France; the Gel Imaging System was from Tianneng, Shanghai, China; the GeneAmp PCR System was purchased from Biometra GmbH, Goettingen, Germany; the Electrophoresis System was from Beijing Liuyi Instrument Plant, Beijing, China; the Mueller-Hinton (MH) was from Oxoid, Basingstoke, UK; the Ex Taq enzyme, dNTPs and the DNA Marker 1200 were from Tiangen Biotech, Beijing, China (http://www.tiangen.com/); and agarose and ethidium bromide were from Sigma, St. Louis, MO, USA.
Methods
Clinical samples were cultured in blood agar culture medium at 35°C for 18–24 h. The identification of the bacteria was carried out according to the ‘National Clinical Laboratory procedures’. All strains were tested for drug sensitivity analysis with the Gram-negative bacteria GN and AST GN-13 identification of the French Merieux automatic bacteria Vitek-2 identification system.
According to the instructions provided with the bacteria DNA extracting kit (Tiangen Biochemical Technology Co., Ltd., Beijing, China), DNA samples of the clinical strains were extracted as a PCR template. The primers used for PCR amplification (7 pairs created by Shenggong Corp., Shanghai, China) were as previously reported (6,7) and are presented in Table I. PCR was performed with a final volume of 25 µl. The PCR program consisted of an initial denaturation step at 94°C for 3 min, followed by 28 cycles of DNA denaturation at 94°C for 30 sec, primer annealing at 56°C for 30 sec, and primer extension at 72°C for 1 min. After the final cycle, a final extension step at 72°C for 7 min was added. The PCR product was analyzed by gel electrophoresis on 1.5% agarose gels. The gels were stained with ethidium bromide at 0.5 µg/ml and visualized using a UV transillumination imaging system (Furi Technology Co., Ltd., Shanghai, China).
Table I.
PCR primer sequences and target genes.
| Genes | Primer sequences (5′→3′) | Expected amplification size (bp) |
|---|---|---|
| blaADC | P1: TAAACACCACATATGTTCCG | 663 |
| P2: ACTTACTTCAACTCGCGACG | ||
| blaMOX | P1: GCT GCT CAA GGA GCA CAG GAT | 520 |
| P2: CAC ATT GAC ATA GGT GTG GTG C | ||
| blaCIT | P1: TGG CCA GAA CTG ACA GGC AAA | 462 |
| P2: TTT CTC CTG AAC GTG GCT GGC | ||
| blaDHA | P1: AAC TTT CAC AGG TGT GCT GGG T | 405 |
| P2: CCG TAC GCT TAC TGG CTT TGC | ||
| blaACC | P1: AAC AGC CTC AGC AGC CGG TTA | 346 |
| P2: TTC GCC GCA ATC ATC CCT AGC | ||
| blaEBC | P1: TCG GTA AAG CCG ATG TTG CGG | 302 |
| P2: CTT CCA CTG CGG CTG CCA GTT | ||
| blaFOX | P1: AAC ATG GGG TAT CAG GGA GAT G | 190 |
| P2: CAA AGC GCG TAA CCG GAT TGG |
PCR, polymerase chain reaction.
Sequence analysis
Two strains with positive PCR amplication were selected, and were bidirectionally sequenced (by Shenggong Corp.) and searched using the BLAST program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequencing results were compared with data on the GenBank database.
Results
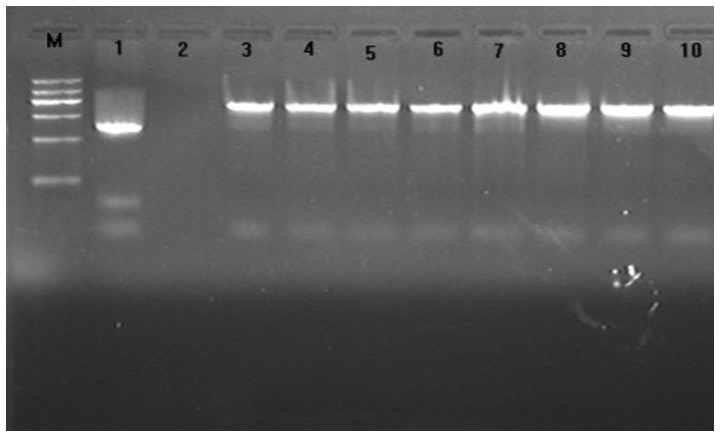
The results indicated that the majority of A. baumannii bacteria were present in the ICU deparments of the hospitals (Table II). The amplification products from 96 strains were observed for each template, and the size observed was consistent with the expected size (663 bp). All A. baumannii isolates were of the acinetobacter-derived cephalosporinase (ADC) type based on the size of the fragments amplified by the primers. The positive rate was 72% (96/134) (Fig. 1).
Table II.
Distribution of the 134 Acinetobacter baumannii strains in distinct departments.
| Wards | Strain | Percentage (%) |
|---|---|---|
| ICU | 80 | 59.70 |
| Department of Respiratory Care | 6 | 4.48 |
| Department of Neurology | 4 | 11.19 |
| Department of Geriatrics | 3 | 2.24 |
| Department of Surgery | 15 | 19.40 |
| Others | 26 | 2.99 |
All the departments mentioned above belong to First People's Hospital Affiliated to Zuzhou Medical College, Central Hospital in Xuzhou, where we obtained the isolates from.
Figure 1.
Electrophoretogram of PCR amplification products for the ampC gene. Lane M, DNA marker (from top to bottom: 1200/900/700/500/300/100 bp); lane 1, positive control; lane 2, negative control; lanes 3–10, heterogeneous clinically-isolated Acinetobacter baumannii.
Results of sequencing analysis
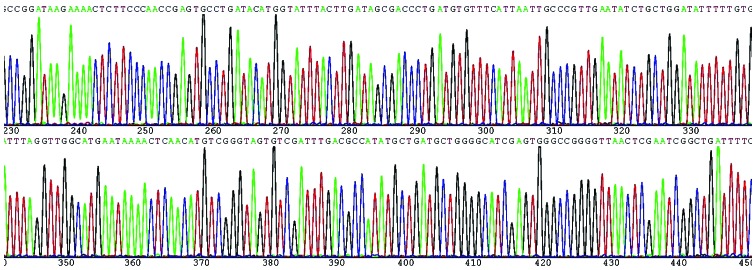
Based on the GenBank database (http://www.ncbi.nlm.nih.gov/Entrez/), the purification sequencing results revealed that the positive products of PCR amplication were the ampC gene of the ADC type. Partial sequencing results were shown in Fig. 2.
Figure 2.
Partial sequences of the ampC gene of ADC type.
Results of the drug sensitivity test
AmpC of A. baumannii was found to be significantly resistant to cephalosporin, quinolone, a synthetic compound combined with sulbactam and carbapenem antibiotics, but were sensitive to polymyxin (Table III).
Table III.
Results of the drug sensivity test of the 96 Acinetobacter baumannii strains positive for AmpC.
| Sensivity | Medium sensivity | Resistance | ||||
|---|---|---|---|---|---|---|
| Strain | Rate | Strain | Rate | Strain | Rate | |
| Cefoxitin | 0 | 0 | 0 | 0 | 96 | 100 |
| Ceftazidime | 0 | 0 | 0 | 0 | 96 | 100 |
| Cefepime | 0 | 0 | 0 | 0 | 96 | 100 |
| Levofloxacin | 0 | 0 | 0 | 0 | 96 | 100 |
| Gentamicin | 0 | 0 | 2 | 2.1 | 94 | 97.9 |
| Ampicillin/sulbactam | 1 | 1.0 | 7 | 7.3 | 88 | 91.7 |
| Piperacillin/tazobactam | 1 | 1.0 | 8 | 8.3 | 87 | 90.6 |
| Tienam | 7 | 7.3 | 1 | 1.0 | 88 | 91.7 |
| Polymyxin | 96 | 100 | 0 | 0 | 0 | 0 |
Discussion
A. baumannii is a clinically opportunistic pathogen, particularly for hospital-acquired pneumonia (8), and infections are associated with invasive medical procedures (9). According to the monitoring data of nosocomial infection in 2003 in the USA, the prevalence of A. baumannii infection, ranks fourth in nosocomial infection. With the widespread application of extended-spectrum antibiotics, the contribution of the multidrug and pandrug resistance pattern of A. baumannii has increased over the years, making the treatment of clinical infections more difficult (10). For Gram-negative bacteria, the production of β-lactamases plays an important role in the resistance to β-lactam antibiotics (5).
AmpC β-lactamases (AmpC enzymes) are produced by some bacteria and their production is mediated either by chromosomes or by plasmids of Gram-negative bacteria (5). As a ‘serine’ cephalosporinase, AmpC β-lactamases cannot be inhibited by clavulanic acid, but can be inhibited by cloxacillin (11). The enzymes belong to the functional group I [according to the Bush-Jacoby-Medeiros (B-J-M) classification] and the molecular class C. The A. baumannii bacterium is equipped with the chromosome encoded enzyme of class C, and ampC genes from heterogeneous A. baumannii strains highly correlate with each other, but differ from those from other types of strains. Thus, these enzymes are termed as the ADC family (7).
In this study, we genotyped A. baumannii isolates by PCR. Amplification products from 96 strains were observed for each template, with the expected size (663 bp). All A. baumannii isolates were of the ADC type based on the size of the fragments amplified by the primers. The absence of amplified products for the other 6 pairs of primers suggested that the assoication of the ampC gene with plasmid-mediated phenomenon did not occur in A. baumannii and in the other strains in this study. Two strains with positive PCR amplification were selected, the products of which were bidirectionally sequenced and searched using the BLAST program. The results revealed 100% homology with ADC-1. In accordance with previous literature (12), our results demonstrated a total positive rate of 72% for AmpC β-lactamases in A. baumannii in tertiary-level hospitals in the Xuzhou region in China.
In this study, 134 strains of A. baumannii were found to be extensively distributed in a range of departments, amongst which the ICU departmet was found to be the major source. The results of the drug sensitivity test revealed a multidrug, and even a pandrug resistance pattern of AmpC A. baumannii. Carbapenem is the priority drug used for the treatment of infections with AmpC G-bacillus. In this study, the resistance rate of ADC type AmpC A. baumannii to Tienam was as high as 91.7%, which is possibly associated with the spread of carbapenem of OXA type in Acinetobacter (13). Some lines of evidence indicate that sulbactam can irreversibly bind Acinetobacter PBP, which contributes to its inherent antimicrobial activity (14). However, this study demonstrated a drug resistance rate of approximately 90% to ampicillin-salbactam, complicating its drug-resistant mechanisms. We did not identify any drug-resistant strains for polymyxin, which can be a priority drug for treatment. It has been reported that aztreonam in combination with polymyxin may improve the therapeutic effect (14). Nonetheless, a kidney function test is necessary before the combined application of these two drugs, given that polymyxin may induce ototoxicity and renal toxicity. It has also been suggested that antibacterial peptides and vaccination may also be potential choices for therapeutic strategies (15).
In conclusion, the drug resistance condition of AmpC A. baumannii in tertiary-level hospitals in the Xuzhou region is relatively severe and warrants intensive monitoring.
References
- 1.Bayuga S, Zeana C, Sahni J, Della-Latta P, el-Sadr W, Larson E. Prevalence and antimicrobial patterns of Acinetobacter baumannii on hands and nares of hospital personnel and patients: the iceberg phenomenon again. Heart Lung. 2002;31:382–390. doi: 10.1067/mhl.2002.126103. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, Randhawa VS, Nirupam N, Rai Y, Saili A. Risk factors for carbapenem-resistant Acinetobacter baumanii blood stream infections in a neonatal intensive care unit, Delhi, India. J Infect Dev Ctries. 2014;8:1049–1054. doi: 10.3855/jidc.4248. [DOI] [PubMed] [Google Scholar]
- 3.Chopra T, Marchaim D, Awali RA, Krishna A, Johnson P, Tansek R, Chaudary K, Lephart P, Slim J, Hothi J, et al. Epidemiology of bloodstream infections caused by Acinetobacter baumannii and impact of drug resistance to both carbapenems and ampicillin-sulbactam on clinical outcomes. Antimicrob Agents Chemother. 2013;57:6270–6275. doi: 10.1128/AAC.01520-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dettori M, Piana A, Deriu MG, Lo Curto P, Cossu A, Musumeci R, Cocuzza C, Astone V, Contu MA, Sotgiu G. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol. 2014;37:185–191. [PubMed] [Google Scholar]
- 5.Poirel L, Lebessi E, Castro M, Fèvre C, Foustoukou M, Nordmann P. Nosocomial outbreak of extended-spectrum beta-lactamase SHV-5-producing isolates of Pseudomonas aeruginosa in Athens, Greece. Antimicrob Agents Chemother. 2004;48:2277–2279. doi: 10.1128/AAC.48.6.2277-2279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bou G, Martínez-Beltrán J. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2000;44:428–432. doi: 10.1128/AAC.44.2.428-432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu YN, Cao B, Wang H, Chen LA, She DY, Zhao TM, Liang ZX, Sun TY, Li YM, Tong ZH, et al. Adult hospital acquired pneumonia: A multicenter study on microbiology and clinical characteristics of patients from 9 Chinese cities. Zhonghua Jie He He Hu Xi Za Zhi. 2012;35:739–746. (In Chinese) [PubMed] [Google Scholar]
- 9.Trottier V, Namias N, Pust DG, Nuwayhid Z, Manning R, Marttos AC, Jr, Dunham MB, Schulman CI, McKenney MG. Outcomes of Acinetobacter baumannii infection in critically ill surgical patients. Surg Infect (Larchmt) 2007;8:437–443. doi: 10.1089/sur.2006.029. [DOI] [PubMed] [Google Scholar]
- 10.Lee YL, Chen YS, Toh HS, Huang CC, Liu YM, Ho CM, Lu PL, Ko WC, Chen YH, Wang JH, et al. Antimicrobial susceptibility of pathogens isolated from patients with complicated intra-abdominal infections at five medical centers in Taiwan that continuously participated in the Study for Monitoring Antimicrobial Resistance Trends (SMART) from 2006 to 2010. Int J Antimicrob Agents. 2012;40(Suppl):S29–S36. doi: 10.1016/S0924-8579(12)70007-9. [DOI] [PubMed] [Google Scholar]
- 11.Seral C, Gude MJ, Castillo FJ. Emergence of plasmid mediated AmpC β-lactamasas: Origin, importance, detection and therapeutical options. Rev Esp Quimioter. 2012;25:89–99. (In Spanish) [PubMed] [Google Scholar]
- 12.Zhu JM, Jiang RJ, Wu JL, Wu KL, Wang JM, Kong HS. Study on the AmpC gene of Multiple drug resistant Acinetobacter baumannii type ADC. Chin J Clin Infect Dis. 2008;1:222–226. (In Chinese) [Google Scholar]
- 13.Niumsup PR, Boonkerd N, Tansawai U, Tiloklurs M. Carbapenem-resistant Acinetobacter baumannii producing OXA-23 in Thailand. Jpn J Infect Dis. 2009;62:152–154. [PubMed] [Google Scholar]
- 14.Malone L, Kwon DH. Carbapenem-associated multidrug-resistant Acinetobacter baumannii are sensitised by aztreonam in combination with polyamines. Int J Antimicrob Agents. 2013;41:70–74. doi: 10.1016/j.ijantimicag.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Quintanilla M, Pulido MR, López-Rojas R, Pachón J, McConnell MJ. Emerging therapies for multidrug resistant Acinetobacter baumannii. Trends Microbiol. 2013;21:157–163. doi: 10.1016/j.tim.2012.12.002. [DOI] [PubMed] [Google Scholar]