Highlights
-
•
We compared current viral respiratory diagnostic techniques with NGS.
-
•
NGS is able to detect respiratory viruses in clinical diagnostic samples.
-
•
With the current sample preparation method, NGS is less sensitive than RT-PCR.
-
•
NGS provided additional sequence and typing information compared with RT-PCR.
Abbreviations: RT-PCR, real-time polymerase chain reaction; NGS, next generation sequencing; NPS, nasopharyngeal swab; VTM, viral transport medium; HRV, human rhinovirus; IFA, influenza A; IFB, influenza B; RSV, respiratory syncytial virus; ADV, adenovirus; hMPV, human metapneumovirus; PIV-1-4, parainfluenza virus 1-4; HCoV, human coronavirus; WoSSVC, West of Scotland Specialist Virology Center; HEV, human enterovirus; Ct, cycle threshold; BLAST, basic local alignment search tool; TRT, turn-around time
Keywords: Next generation sequencing, Virus detection;, Viral respiratory infection, Diagnostics
Abstract
Background
Molecular assays are the gold standard methods used to diagnose viral respiratory pathogens. Pitfalls associated with this technique include limits to the number of targeted pathogens, the requirement for continuous monitoring to ensure sensitivity/specificity is maintained and the need to evolve to include emerging pathogens. Introducing target independent next generation sequencing (NGS) could resolve these issues and revolutionise respiratory viral diagnostics.
Objectives
To compare the sensitivity and specificity of target independent NGS against the current standard diagnostic test.
Study design
Diagnostic RT-PCR of clinical samples was carried out in parallel with target independent NGS. NGS sequences were analyzed to determine the proportion with viral origin and consensus sequences were used to establish viral genotypes and serotypes where applicable.
Results
89 nasopharyngeal swabs were tested. A viral pathogen was detected in 43% of samples by NGS and 54% by RT-PCR. All NGS viral detections were confirmed by RT-PCR.
Conclusions
Target independent NGS can detect viral pathogens in clinical samples. Where viruses were detected by RT-PCR alone the Ct value was higher than those detected by both assays, suggesting an NGS detection cut-off – Ct = 32. The sensitivity and specificity of NGS compared with RT-PCR was 78% and 80% respectively. This is lower than current diagnostic assays but NGS provided full genome sequences in some cases, allowing determination of viral subtype and serotype. Sequencing technology is improving rapidly and it is likely that within a short period of time sequencing depth will increase in-turn improving test sensitivity.
1. Background
Virus specific molecular assays such as real-time PCR (RT-PCR) are now considered the gold standard in the diagnosis of viral respiratory tract infections. They are rapid, relatively inexpensive and offer increased sensitivity and specificity over prior techniques such as virus culture and direct immunofluorescence. Assays can be developed quickly to detect novel/emerging pathogens and can be combined to identify multiple microbiological pathogens in a single test. Yet there is a limit to the number of targets, usually up to four, which can be included in an in-house test before compromising test sensitivity. As a result, diagnostic laboratories must develop a panel of multiplex tests in order to detect the whole range of pathogens. Also, as for all PCR based assays, detection is based on targeting conserved regions of the pathogen genome and mutations can lead to reduced sensitivity or false negative results. Furthermore, only the targeted pathogens included in the assay will be identified, therefore atypical or emerging pathogens will generally evade detection by PCR. Although commercial PCR based tests [1] are available that overcome some of the pitfalls associated with in-house tests, they remain PCR based technologies and as a result suffer from the same sequence based pitfalls outlined above.
Introducing NGS into a diagnostic setting may revolutionize the investigation of respiratory infections. Combining sequence independent amplification with NGS will potentially detect viral and non-viral pathogens within a clinical specimen without actively targeting them, while simultaneously analyzing the genetic sequence. NGS is established in virus discovery, whole genome studies and metagenome studies [2], [3], [4] thus the simultaneous detection of multiple different pathogens with this technique is possible. However the efficacy and feasibility of employing such techniques in a diagnostic setting requires further study.
2. Objectives
Here we present a pilot study that compares current diagnostic techniques, namely RT-PCR with NGS in the detection of RNA viruses in respiratory samples from individuals symptomatic of a respiratory illness.
3. Study design
3.1. Samples
Eighty nine nasopharyngeal swabs (NPS) were collected from adults with upper respiratory tract infections between May 2010 and October 2011. Samples were collected as part of the VIDARIS trial, a random subset of which were used in this study. It should be noted that over half of the participants in this trial were vaccinated against influenza. Ethical approval was provided by the Upper South B Regional Ethics Committee. All participants provided written informed consent [5]. Swabs were stored in viral transport media (VTM) at −80 °C until testing. The VTM was thawed at 37 °C and centrifuged at 1500 × g for 10 min to remove debris. Total nucleic acids were extracted from 200 μl of the supernatant (MagJet Viral DNA and RNA kit, Thermo Scientific) and eluted in 100 μl of water.
3.2. Next generation sequencing method
A 20 μl aliquot of the extract was treated with DNAse 4U (Turbo DNAse 2U/μl, Life technologies) for 30 min at 37 °C. RNA was purified from the reaction using RNAClean XP beads (Agencourt), eluted in 15 μl of water and reverse transcription carried out using Maxima Minus H (Thermofisher) at 50 °C for 60 min with 0.2pM primer FR26RV-N (5′ GCC GGA GCT CTG CAG ATA TCN NNN NN 3′). Second strand cDNA was synthesised (NEBNext mRNA 2nd Strand Synthesis, New England Biolabs) and the reaction purified with Ampure XP beads (Agencourt). Sequence-independent single primer amplification (SISPA) was carried out with the Advantage 2 PCR kit (Clontech) and 0.2pM primer FR20RV (5′ GCC GGA GCT CTG CAG ATA TC 3′). The PCR product was purified with Ampure XP beads (Agencourt) and quantified (Qubit HS DNA, Life Technologies). 1 ng of cDNA was used to prepare barcoded sequencing libraries with the Nextera XT DNA Sample Prep kit (Illumina) and indices from the Nextera XT Index Kit as per the manufacturer’s instructions. Up to 24 sample libraries were pooled per sequencing run and 151 bp paired-end reads were generated on the Illumina MiSeq.
3.3. Bioinformatic analysis
Sequencing adapters and low quality sequencing reads were removed (Trim Galore!, Babraham Bioinformatics) and low–complexity reads filtered out (PrinSeq [6]). High quality paired-end sequences were retained for downstream analyses. These sequences were mapped to a database containing a human genome and cDNA references, to remove host sequences. Unmapped sequences were entered into the Metamos pipeline [7] which employs multiple de novo assemblers with k-mer optimisation to assemble contigs. The contigs from the most effective assembly were then taxonomically classified using the Basic Local Alignment Search Tool (BLAST) against the GenBank nucleotide and non-redundant databases (cut off E value 0.001). Identical sequences between samples were removed using BedTools and unique sequences were retained for further analysis. Sequenced reads were then mapped back to the top taxonomic hit for each sample and visualized using Tablet [8], to quantify viral reads within each sample and generate a consensus sequence. Where appropriate, greater than 90% of reference genome coverage, the consensus sequences were aligned with known reference sequences and phylogenetic analysis carried out using MEGA6 [9]. Taxonomic hits were compared with the results of the diagnostic qRT-PCR (Table 1 ).
Table 1.
A summary of sequenced reads, real-time PCR and NGS data, including RT-PCR results for all positive samples and the accession number of top taxonomic hits from BLAST along with reference assembly details.
| Sample | Total Reads | % mapping to human reference | PCR | Ct | Taxonomy | Accession | % reads mapping | % genome covered | Nucleotide mismatch |
|---|---|---|---|---|---|---|---|---|---|
| 1C2 | 1107488 | 81.67 | HRV | 26.75 | Enterovirus D-68 | AB601885 | 1.46 | 14.9 | 1.4 |
| 2A3 | 1110727 | 88.23 | HCoV 229E | 18.14 | HCoV 229E | JX503060 | 9.63 | 99.8 | 0.6 |
| 2A4 | 311141 | 98.39 | HCoV 229E | 20.00 | HCoV 229E | JX503060 | 0.16 | 55.3 | 0.6 |
| 2C3 | 470840 | 93.93 | HCoV 229E | 25.83 | HCoV 229E | JX503060 | 0.48 | 20.0 | 0.7 |
| 2D2 | 743683 | 94.07 | HCoV 229E | 19.74 | HCoV 229E | JX503060 | 2.50 | 73.9 | 0.6 |
| 1D3 | 801021 | 96.66 | HCoV NL63 | 27.17 | HCoV NL63 | KF530112 | 0.38 | 11.7 | 0.5 |
| 1E7 | 426736 | 92.10 | HCoV NL63 | 24.36 | HCoV NL63 | JQ765569 | 0.97 | 23.3 | 0.5 |
| 1F8 | 644660 | 93.24 | HCoV NL63 | 25.22 | HCoV NL63 | JQ765567 | 0.03 | 3.7 | 0.4 |
| 1H1 | 608153 | 94.92 | HCoV OC43 | 17.21 | HCoV OC43 | KF530099 | 3.48 | 99.9 | 1.6 |
| 2D5 | 992040 | 93.04 | HCoV OC43 | 14.63 | HCoV OC43 | JN129835 | 2.70 | 97.6 | 0.7 |
| 1G6 | 81749 | 37.68 | hMPV | 26.84 | hMPV-B | KJ627397 | 1.41 | 18.5 | 2.0 |
| 2A9 | 626886 | 84.12 | hMPV | 25.68 | hMPV-B | KF530171 | 6.35 | 48.4 | 3.1 |
| 1C7 | 538412 | 69.88 | HRV | 24.93 | HRV-A1 | JN837694 | 4.76 | 87.4 | 1.2 |
| 1D1 | 1010455 | 59.82 | HRV | 20.68 | HRV-A1 | JN837694 | 32.03 | 100 | 1.9 |
| 1D4 | 1089553 | 23.19 | HRV | 19.81 | HRV-A1 | JN837694 | 74.67 | 99.6 | 2.2 |
| 1I5 | 728425 | 92.82 | HRV | 33.66 | HRV-A1 | JN837694 | 0.01 | 39.0 | 5.6 |
| 1H3 | 413108 | 83.13 | HRV | 20.44 | HRV-A2 | X02316 | 6.65 | 96.0 | 8.1 |
| 2C1 | 816200 | 91.89 | HRV | 20.24 | HRV-A21 | JN837693 | 5.32 | 97.9 | 2.5 |
| 1B1 | 546282 | 20.17 | HRV | 18.73 | HRV-A49 | JN798589 | 75.48 | 100 | 2.5 |
| 2B7 | 596971 | 95.94 | HRV | 17.24 | HRV-A60 | JN798590 | 1.82 | 95.3 | 3.7 |
| 2D6 | 1016390 | 82.29 | HRV | 23.07 | HRV-A8 | FJ445113 | 6.23 | 97.0 | 8.1 |
| 1G1 | 265505 | 78.73 | HRV | 28.49 | HRV-A90 | FJ445167 | 0.02 | 2.6 | 10.8 |
| 1C3 | 1278307 | 65.56 | HRV | 28.15 | HRV-A97 | FJ445172 | 0.31 | 20.9 | 8.7 |
| 1I8 | 1132106 | 99.02 | HRV | 27.85 | HRV-B27 | JF285309 | 0.01 | 6.7 | 3.7 |
| 1I4 | 641357 | 98.15 | HRV | 26.01 | HRV-B3 | JF285331 | 0.64 | 97.2 | 2.4 |
| 1H7 | 657627 | 56.80 | HRV | 19.39 | HRV-B4 | JN798573 | 39.08 | 99.7 | 1.4 |
| 1B2 | 638257 | 93.07 | HRV | 26.78 | HRV-B92 | FJ445169 | 2.53 | 56.4 | 7.5 |
| 1I6 | 1063930 | 57.27 | HRV | 29.11 | HRV-C (UTR) | JX129433 | 2.92 | N/A | 2.3 |
| 2A6 | 1056405 | 83.90 | HRV | 23.12 | HRV-C11 | EU840952 | 0.29 | 27.2 | 8.9 |
| 1E1 | 758165 | 61.76 | HRV | 20.1 | HRV-C15 | GU219984 | 33.93 | 100 | 2.0 |
| 1E8 | 887501 | 96.50 | HRV | 25.09 | HRV-C15 | GU219984 | 0.06 | 29.7 | 2.5 |
| 1I2 | 710612 | 60.67 | HRV | 19.80 | HRV-C17 | JN815240 | 33.29 | 99.1 | 3.8 |
| 1B8 | 818055 | 96.78 | Influenza A | 22.34 | Influenza A H3N2 | KJ942712 | 0.03 | 30.6–60.3 | various |
| 1B5 | 696929 | 50.67 | PIV-3 | 27.57 | PIV-3 | KJ672618 | 5.62 | 5.0 | 0.5 |
| 1G2 | 309778 | 78.95 | PIV-3 | 28.09 | PIV-3 | KJ672606 | 0.04 | 2.8 | 0.6 |
| 1E3 | 700028 | 93.78 | RSV | 20.84 | RSVA | KJ627329 | 3.71 | 91.0 | 0.7 |
| 1F1 | 322516 | 85.54 | RSV | 25.08 | RSVA | KF826849 | 3.79 | 54.6 | 0.5 |
| 2B4 | 612887 | 55.78 | RSV | 19.32 | RSVB | JX576741 | 40.52 | 99.3 | 2.0 |
| 1G1 | 265505 | 78.73 | Adeno | 35.1 | N/A | ||||
| 2A2 | 745105 | 98.00 | HCoV 229E | 32.35 | N/A | ||||
| 2C4 | 759226 | 98.06 | HCoV 229E | 33.84 | N/A | ||||
| 2D3 | 765913 | 89.42 | HCoV OC43 | 24.05 | N/A | ||||
| 1C9 | 837310 | 47.23 | hMPV | 35.51 | N/A | ||||
| 1B7 | 537522 | 62.21 | HRV | 28.94 | N/A | ||||
| 1F7 | 236596 | 70.59 | HRV | 28.03 | N/A | ||||
| 2B9 | 569654 | 76.13 | HRV | 33.31 | N/A | ||||
| 2D4 | 1034235 | 96.30 | PIV-2 | 36.07 | N/A | ||||
| 1E5 | 714509 | 96.87 | RSV | 30.01 | N/A | ||||
| 2B6 | 854593 | 77.09 | RSV | 33.96 | N/A |
3.4. Diagnostic in-house RT-PCR methods
40 μl of the nucleic acid extract was then screened for human rhinovirus (HRV), influenza A/B (IFA/IFB), respiratory syncytial virus (RSV), adenovirus (ADV), human metapneumovirus (hMPV), parainfluenzavirus 1–4 (PIV 1–4), coronaviruses (HCoV) HKU1, NL63, OC43 and 229E and Mycoplasma pneumonia using the routine diagnostic qRT-PCR at the West of Scotland Specialist Virology Centre (WoSSVC) as previously described [10].
4. Results
4.1. Next generation sequencing
The average number of sequences generated per sample was ∼660,640 (range 30,872–1,278,122) after quality trimming and filtering. Viral contigs were found in 53/89 samples but following removal of duplicate reads this was reduced to 46/89. In a subset of samples (n = 8), there were fewer than 10 unique viral reads detected by the NGS assay alone. Due to the low number of reads we deemed these to be negative by NGS. The viral sequences detected in the remaining 38 samples belonged to the Picornaviridae, Coronaviridae, Paramyxoviridae and Orthomyxoviridae (Table 1). No mixed infections were detected by NGS.
Picornaviruses were most frequently detected (n = 21) and classified as HRV in 20/21 and enterovirus (HEV) in 1/21 cases. These could be subdivided into 3 rhinovirus species, A (11/21), B (4/21), C (5/21) and HEV D. Picornavirus sequences generated by NGS showed high similarity at the nucleotide level to reference genomes available in the NCBI database, allowing us to assign a serotype in all but one cases (Table 1). The extent of reference genome coverage and phylogenetic similarity to reference sequences are shown in Fig. 1, Fig. 2 , respectively. Numerous HRV serotypes and an HEV-D68 were detected. Human coronaviruses (HCoV) were detected in nine samples and were found to belong to the following types: HCoV 229E (4/9), HCoV NL63 (3/9) and HCoV OC43 (2/9). Paramyxoviruses were detected in seven samples. These included hMPV-B (2/7), PIV-3 (2/7), RSV-A (2/7) and RSV-B (1/7). An Orthomyxovirus was detected in one sample and typed as Influenza A H3N2.
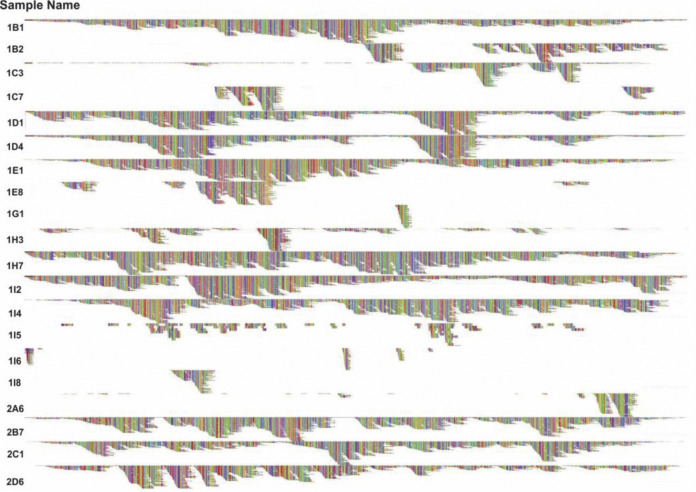
Fig. 1.
HRV genome coverage – a graphic visualization of the reference genome coverage following Bowtie2 alignment of sequencing reads to the top BLAST hit for each HRV positive sample.
Fig. 2.
Neighbor-joining (NJ) phylogenetic tree of full-genome reference sequences and a subset of the NGS consensus sequences with full or near-full reference genome coverage. The branch annotations represent the bootstrap values (percentage of 1000 samples trees). Sequences generated in this study are shown as circles and squares.
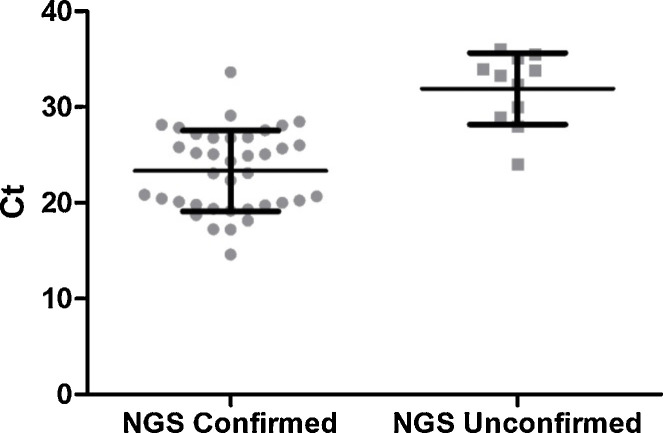
All viruses identified by NGS were confirmed by qRT-PCR. However, in eleven cases, virus was identified by RT-PCR only. This included the following viruses, ADV (1/11), PIV-2 (1/11), hMPV (1/11), RSV (2/11), HCoV (3/11) and HRV (3/11). One sample was found by RT-PCR to contain a mixture of both ADV and HRV. The NGS method failed to detect the ADV in this sample. Where NGS confirmed the findings of the RT-PCR assay the Ct values were significantly lower than the group where a virus was detected by the RT-PCR assay alone (p < 0.0001, unpaired t-test), shown in Fig. 3 . No samples contained viral pathogens detected by NGS only.
Fig. 3.
Threshold cycle (Ct) values respiratory of samples positive by RT-PCR and either confirmed or unconfirmed by the NGS assay. The bars indicate the mean and one standard deviation. An unpaired t-test demonstrates a significant difference between the two groups (p < 0.0001).
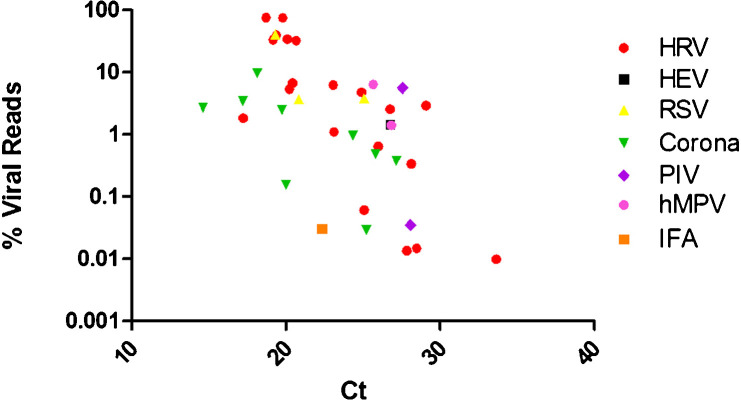
Further examination of the relationship between the numbers of viral sequenced reads and the threshold cycle (Ct) value provided by the RT-PCR assay is shown in Fig. 4 . The log of the percentage of sequenced reads correlates with the Ct value indicating that a higher viral load is associated with a greater proportion of viral reads.
Fig. 4.
The percentage of sequenced reads, after quality trimming, mapping to the taxonomic reference genome determined by BLAST. Linear regression of data, R2 value = 0.19 (p = 0.0057). This is suggestive of a log relationship between the proportion of reads mapping and the Ct value.
Despite sample treatment to enrich for viral RNA, a substantial number of bacterial transcript sequences were generated, however partial genome amplification of bacteria only allows classification to the level of order/family (data not shown).
5. Discussion
As a pilot study designed to assess the utility of NGS as a method for the detection of respiratory RNA viruses, we compared an in-house NGS method with an established in-house RT-PCR test using 89 respiratory samples. A viral pathogen was detected by the NGS method in 38 samples with the RT-PCR confirming these and detecting a further 11 viruses. Overall, based on these results, the NGS assay had a sensitivity of 77.55% (95% CI 63.37–88.21%) and specificity of 80.49% (95% CI 65.13–91.15%) or 100% (95% CI 91.31–100%) if considering samples with fewer than 10 unique reads are negative, compared to RT-PCR. It is possible that the sensitivity of NGS detection could vary between virus groups however the number of detections in this cohort was not powered to assess each individually.
The 11 NGS negative/RT-PCR positive samples contained virus at lower Cts compared to those positive by both assays, suggesting the current NGS method has a cut off in the region of Ct 32, approximately 1–2 logs less sensitive than the qRT-PCR method (data not shown). This finding is similar to that outlined by Prachayangprecha et al. At present, this level of sensitivity is not appropriate for diagnostic services to replace qRT-PCR. Increasing the depth of sequencing could improve the sensitivity of NGS, either by reducing the multiplexing of samples (i.e., reducing the number of samples processed per sequencing run) or using an alternative platform with greater capacity such as the Illumina NextSeq or HiSeq. Reducing the contamination of host DNA through host DNA depletion holds the greatest promise [11] to increase the depth of viral and microbial DNA as we found up to 99% of reads obtained per sample were derived from the host.
One of the 11 pathogens not detected by the NGS method was adenovirus. It is unclear whether this was due to the sensitivity cut off discussed above (Ct of 35.1) or due to the initial DNAse step outlined in the method. In future, if this is to be adapted for the detection of DNA viruses and other non viral causes of respiratory infection then other ways of enrichment will need to be sought. Perhaps target enrichment via hybridization, but again this may encounter the issue of missing novel or changed viruses.
Although the NGS method failed to detect all the positive samples, it did offer several advantages over the RT-PCR method. For example, the NGS provided more detailed typing information for the detected viruses including subtype data for RSV and hMPV as well as serotype data for the HRV and HEV.
Such information is highly informative. As well as enabling real-time diagnostic data to be produced, NGS could simultaneously provide resistance testing (e.g., H275Y mutation of influenza A, conferring oseltamivir resistance) and typing data, as demonstrated here and previously [12], [13]. This could be used to inform public health of circulating strains and could allow rapid detection of the emergence of novel subtypes (e.g., EV68 and ADV 14) or highlight potential outbreaks. In future, it may also detect viral polymorphisms associated with disease severity [14], [15].
Similar to other studies we also found a correlation between NGS sequence reads and RT-PCR cycle thresholds [16], [17]. This was particularly strong for rhinoviruses – mainly because these represented the majority of the pathogens detected. This suggests that sequence reads might be a suitable proxy for viral/target copies. This would enable laboratories/clinicians to interpret the clinical relevance of results [18]. Such data can also be used to infer prognosis or treatment response.
Viral co-infection detection by NGS could not be assessed in this cohort as only a single co-infection episode was detected by RT-PCR. The population under study were normally healthy adults who did not require health care interventions. There is little information on co-infections in such individuals but evidence suggests that viral co-infections occur more commonly in hospitalised individuals [19]. The detection of influenza was also lower than may have been expected which probably relates to the level vaccination amongst the cohort. Further studies including children and unvaccinated individuals would be of use.
The NGS approach did not detect positive results that were missed by the RT-PCR. Although this panel was small, our data supports other studies which have concluded that current respiratory panels are appropriate for the detection of the main causes of viral respiratory disease.
A major barrier to the introduction of NGS as a routine diagnostic test is its cost and turnaround time. The current cost of sequencing is prohibitive as a diagnostic test when compared to RT-PCR but this has reduced in recent years, a trend which will probably continue in the future. Although not designed to detect bacteria the current method still detected bacterial genomes in nearly all samples, this finding shows that syndromic testing is a possibility. If viral, bacterial and fungal diagnostic tests can be combined into a single assay this would benefit the cost effectiveness of developing NGS as a diagnostic tool.
The turnaround-time of the above process was in the region of seven days, including preliminary data analysis whereas the turnaround time for RT-PCR methods is usually just a few hours. As a result, NGS is unlikely to become a routine diagnostic test in the near term. However there are many steps that could be accelerated with automation. The development of kit based library preparation methods has also resulted in a condensed process, reducing hands-on time. Technical advances that allow a greater depth of sequencing could obviate the need for enrichment processes. Furthermore third generation sequencers, such as the PacBio RS II, offer the potential for even more rapid sequencing. Taken together, these advances are likely to improve the TRT of NGS significantly.
Competing interests
None declared.
Funding
This study was funded by the UK Medical Research Council (Grant reference number G0801822).
Ethical approval
The study was approved by the Upper South B Regional Ethics Committee, New Zealand (URB/09/10/050) and all participants provided written informed consent.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jcv.2015.06.082.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Gadsby N.J. Comparison of the Luminex Respiratory Virus Panel fast assay with in-house real-time PCR for respiratory viral infection diagnosis. J. Clin. Microbiol. 2010;48(6):2213–2216. doi: 10.1128/JCM.02446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker K.S. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441(2):95–106. doi: 10.1016/j.virol.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ip H.S. Identification and characterization of Highlands J virus from a Mississippi sandhill crane using unbiased next-generation sequencing. J. Virol. Methods. 2014;206:42–45. doi: 10.1016/j.jviromet.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Van den Hoecke S. Analysis of the genetic diversity of influenza A viruses using next-generation DNA sequencing. BMC Genomics. 2015;16(1):79. doi: 10.1186/s12864-015-1284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murdoch D.R. Effect of vitamin D3 supplementation on upper respiratory tract infections in healthy adults: the VIDARIS randomized controlled trial. JAMA. 2012;308(13):1333–1339. doi: 10.1001/jama.2012.12505. [DOI] [PubMed] [Google Scholar]
- 6.Schmieder R., R, Edwards Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Treangen T.J. MetAMOS: a modular and open source metagenomic assembly and analysis pipeline. Genome Biol. 2013;14(1):R2. doi: 10.1186/gb-2013-14-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milne I. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14(2):193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 9.Tamura K. MEGA6: Molecular evolutionary genetics analysis version 6. 0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunson R.N., Carman W.F. During the summer 2009 outbreak of swine flu in Scotland what respiratory pathogens were diagnosed as H1N1/2009? BMC Infect. Dis. 2011;11:192. doi: 10.1186/1471-2334-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oyola S.O. Efficient depletion of host DNA contamination in malaria clinical sequencing. J. Clin. Microbiol. 2013;(3):745–751. doi: 10.1128/JCM.02507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petty T.J. Comprehensive human virus screening using high-throughput sequencing with a user-friendly representation of bioinformatics analysis: a pilot study. J. Clin. Microbiol. 2014;52(9):3351–3361. doi: 10.1128/JCM.01389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao G.et al. Identification of novel viruses using VirusHunter–an automated data analysis pipeline. PLoS ONE. 2013;8(10):e78470. doi: 10.1371/journal.pone.0078470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houng H.S. Emergent 2009 influenza A(H1N1) viruses containing HA D222N mutation associated with severe clinical outcomes in the Americas. J. Clin. Virol. 2012;53(1):12–15. doi: 10.1016/j.jcv.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Selleri M. Detection of haemagglutinin D222 polymorphisms in influenza A(H1N1) pdm09-infected patients by ultra-deep pyrosequencing. Clin. Microbiol. Infect. 2013;19(7):668–673. doi: 10.1111/j.1469-0691.2012.03984.x. [DOI] [PubMed] [Google Scholar]
- 16.Prachayangprecha S. Exploring the potential of next-generation sequencing in detection of respiratory viruses. J. Clin. Microbiol. 2014;52(10):3722–3730. doi: 10.1128/JCM.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosseel T. DNase SISPA-next generation sequencing confirms Schmallenberg virus in Belgian field samples and identifies genetic variation in Europe. PLoS ONE. 2012;7(7):e41967. doi: 10.1371/journal.pone.0041967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeVincenzo J.P. Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 2010;182(10):1305–1314. doi: 10.1164/rccm.201002-0221OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goka E.A. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol. Infect. 2015;143(1):37–47. doi: 10.1017/S0950268814000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.