Summary
Centromeres are the chromosomal regions promoting kinetochore assembly for chromosome segregation. In many eukaryotes, the centromere consists of up to mega base pairs of DNA. On such “regional centromeres,” kinetochore assembly is mainly defined by epigenetic regulation [1]. By contrast, a clade of budding yeasts (Saccharomycetaceae) has a “point centromere” of 120–200 base pairs of DNA, on which kinetochore assembly is defined by the consensus DNA sequence [2, 3]. During evolution, budding yeasts acquired point centromeres, which replaced ancestral, regional centromeres [4]. All known point centromeres among different yeast species share common consensus DNA elements (CDEs) [5, 6], implying that they evolved only once and stayed essentially unchanged throughout evolution. Here, we identify a yeast centromere that challenges this view: that of the budding yeast Naumovozyma castellii is the first unconventional point centromere with unique CDEs. The N. castellii centromere CDEs are essential for centromere function but have different DNA sequences from CDEs in other point centromeres. Gene order analyses around N. castellii centromeres indicate their unique, and separate, evolutionary origin. Nevertheless, they are still bound by the ortholog of the CBF3 complex, which recognizes CDEs in other point centromeres. The new type of point centromere originated prior to the divergence between N. castellii and its close relative Naumovozyma dairenensis and disseminated to all N. castellii chromosomes through extensive genome rearrangement. Thus, contrary to the conventional view, point centromeres can undergo rapid evolutionary changes. These findings give new insights into the evolution of point centromeres.
Graphical Abstract
Highlights
-
•
A new type of point centromere has been identified in budding yeast N. castellii
-
•
Its DNA sequence and evolutionary origin are different from other point centromeres
-
•
N. castellii centromeres are bound by CBF3 that recognizes other point centromeres
-
•
Contrary to the conventional view, point centromeres can change rapidly in evolution
All known point centromeres share common DNA sequences and a single evolutionary origin. Kobayashi et al. have identified a new type of point centromere in budding yeast N. castellii. Its DNA sequence and evolutionary origin are different from other point centromeres. Discovery of the new centromere redefines the evolution of point centromeres.
Results and Discussion
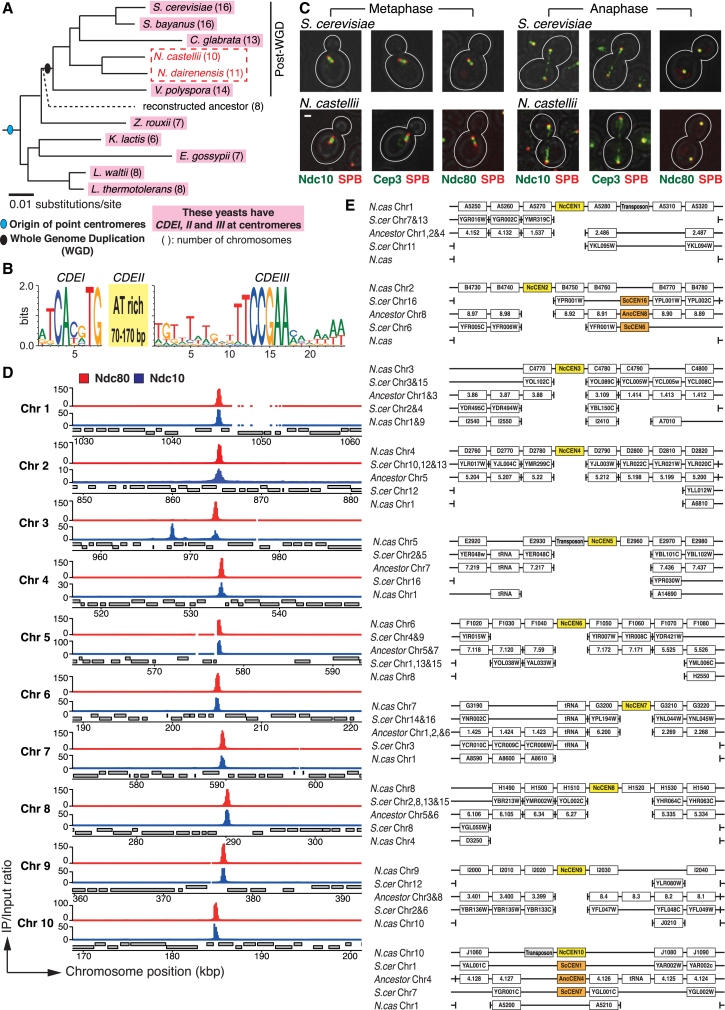
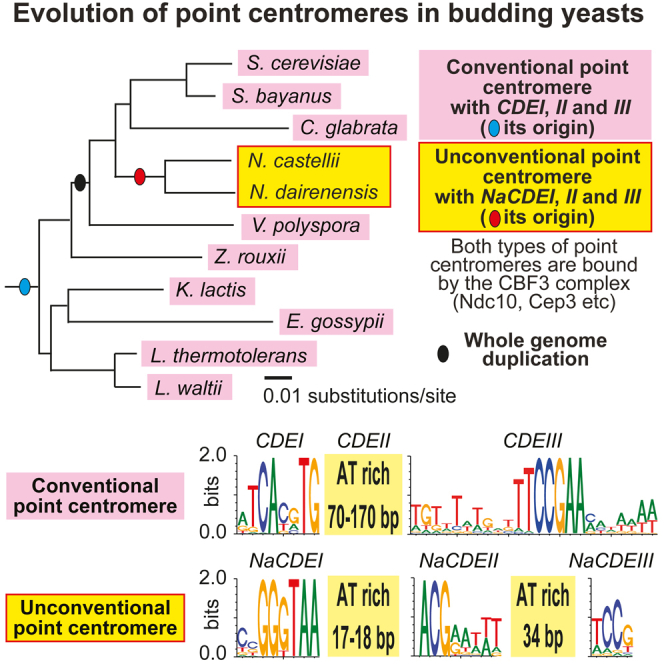
All known point centromeres have a common DNA sequence, known as CDEI, II, and III (Figures 1A and 1B) [5, 6]. Among different budding yeast species, the order of orthologous genes is generally conserved around the centromeres (i.e., in synteny) [11]. These indicate that the point centromeres themselves are orthologous. However, there are peculiar exceptions to this view: Naumovozyma castellii (N. castellii) (previously called Saccharomyces castellii) and its close relative N. dairenensis belong to the clade of budding yeasts expected to have point centromeres [3, 7], but no CDEI,II,III-like sequences are found at the loci expected from synteny (Figure 1A; see Figure S1D) [6, 12]. They may use a novel type of point centromere or have re-acquired regional centromeres during their evolution. Here, we aim to identify the centromere in N. castellii.
Figure 1.
Kinetochore Proteins Show Single-Peak Localization on Each Chromosome of N. castellii
(A) A clade of budding yeast species carries point centromeres with common consensus DNA sequences CDEI, II, and III (pink), except for N. castellii and N. dairenensis (red rectangle). Phylogenetic tree of budding yeast species is taken from [6, 7] with modification. Horizontal length in the tree is proportional to the change in genomic DNA sequences. The WGD (black oval) is estimated to have occurred approximately 100 million years ago [8], and the N. castellii/N. dairenensis divergence is about half this age.
(B) Nucleotide sequence of CDEI, II, and III [5, 6]. Logos of nucleotides graphically represent their frequency at individual positions. CDEII is 70–170 bp DNA sequence with >79% AT content.
(C) Localization of Ndc10, Cep3, and Ndc80 in representative S. cerevisiae and N. castellii cells (in metaphase and anaphase). Ndc10, Cep3, and Ndc80 were tagged with three tandem copies of GFP (3× GFP) in N. castellii (T11587, T11584, and T11586, respectively) and with 1× GFP in S. cerevisiae (T11520, T11522, and T11521, respectively). Spindle pole body (SPB) components, Spc42 and Spc110, were tagged with 4× mCherry and 1× mCherry in N. castellii and S. cerevisiae, respectively. Cell shapes are outlined in white. The scale bar represents 1 μm. Metaphase and anaphase are defined by the distance between two SPBs (<2.5 μm and >3 μm, respectively). Note that, during anaphase, CBF3 also localizes on the spindle as well as at kinetochores [9].
(D) Ndc80 (red) shows single-peak localization on each chromosome (Chr) of N. castellii. Ndc10 (blue) also shows peaks at the same regions. NDC80-6xHA (T9328) and NDC10-6xHA (T9326) cells were processed for ChIP-seq. Gray bars represent open reading frames of genes; top on Watson strand, bottom on Crick strand. Chromosome positions are shown in length (kilo base pairs [kbp]) from the left telomere. In addition to the positions of Ndc80 peaks, Ndc10 showed two extra peaks (Figure S1A); one of them is at 968 kbp on chromosome 3, as shown here.
(E) Gene order in S. cerevisiae and the reconstructed ancestor, aligned around N. castellii CENs (yellow box). Gene orders were analyzed using YGOB [10]. Vertical tick bars represent gaps; i.e., genes to the right and left are not neighbors on a chromosome. Two chromosome series of N. castellii and S. cerevisiae are aligned with one series of the ancestor because N. castellii and S. cerevisiae are post-WGD yeasts (see A). Orange boxes represent centromeres in the ancestor (AncCEN) and S. cerevisiae (ScCEN).
See also Figure S1.
Kinetochore Components Show Single-Peak Localization on Each Chromosome of N. castellii by Chromatin Immunoprecipitation
In Saccharomyces cerevisiae (S. cerevisiae) and other budding yeasts, their point centromeres are recognized by the CBF3 complex, which binds the CDEIII DNA consensus [5, 13]. The CBF3, consisting of Ndc10, Cep3, Ctf13, and Skp1 proteins, is exclusively found in budding yeasts with point centromeres. In S. cerevisiae, the CBF3 and other kinetochore components show a bi-lobed pattern on the metaphase spindle and segregate following movement of the spindle poles during anaphase [14] (Figure 1C, top). Despite an apparent lack of CDEI,II,III-containing centromeres, the N. castellii genome encodes orthologs of the CBF3 components [10, 15]. In N. castellii, Ndc10 and Cep3 proteins showed the kinetochore-like localization pattern, similar to S. cerevisiae (Figure 1C, bottom). The same localization pattern was found for Ndc80 (Figure 1C, bottom), an outer kinetochore component [5]. Thus, Ndc10, Cep3, and Ndc80 might indeed be N. castellii kinetochore components.
To identify N. castellii centromeres, we added epitope tags to Ndc10, Cep3, and Ndc80 at their original loci, carried out chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-seq), and analyzed in reference to the annotated N. castellii genome sequence [15, 16]. We also carried out ChIP-seq for Cse4, a centromere-specific histone H3 variant. Crucially, Ndc80 ChIP-seq gave a distinct single peak at an intergenic region on each of ten chromosomes (Figures 1D and S1A). Cse4, Ndc10, and Cep3 gave peaks at the same ten intergenic regions as Ndc80 and gave one, two, and six additional peaks, respectively (Figures 1D and S1A–S1C). The chromosomal regions, where Ndc80 showed accumulation (together with Ndc10, Cep3, and Cse4), may serve as the centromeres in N. castellii. On this assumption, we tentatively named them N. castellii CEN1–10, or NcCEN1–10 for short, on chromosomes 1–10, respectively.
Most N. castellii CENs Are Not at Conserved Syntenic Locations on Chromosomes, Compared with Locations of Other Point Centromeres
We compared the order of orthologous genes between S. cerevisiae, N. castellii, and an ancestor. This ancestor is evolutionarily positioned prior to the whole-genome duplication (WGD), which occurred during the evolution of budding yeasts [3] (Figure 1A), and its genome was constructed using bioinformatics [17]. Between S. cerevisiae and the ancestor, the gene order across the centromeres is conserved, including the centromeres themselves [11, 17] (Figure S1D). Thus, chromosomal positions of the centromeres did not change during evolution from the ancestor to S. cerevisiae. The gene order across the majority of the ancestral centromeric regions (excluding the centromeres themselves) is also conserved without rearrangement on N. castellii chromosomes (Figure S1D). However, N. castellii CEN locations are not syntenic to the CENs in the ancestor (Figures S1D and S1E). This suggests that centromeres “disappeared” from these ancestral centromeric loci during the N. castellii evolution. In fact, ancestral CEN4 and N. castellii CEN10 make the only centromere pair whose surrounding orthologs show complete synteny (Figures S1D and S1E). Conversely, N. castellii CEN1–9 are not located in regions of conserved gene order when compared to S. cerevisiae or the ancestor (Figure 1E). More specifically, the synteny along N. castellii chromosomes (relative to those of S. cerevisiae and the ancestor) is disrupted at the positions of NcCENs. It is therefore likely that N. castellii CEN1–9 have been positioned, at least partly, by genome rearrangement during evolution, rather than by de novo centromere formation between the existing genes.
Candidate Centromere Regions Show Dynamic Behaviors Expected for Functional Centromeres in N. castellii Cells
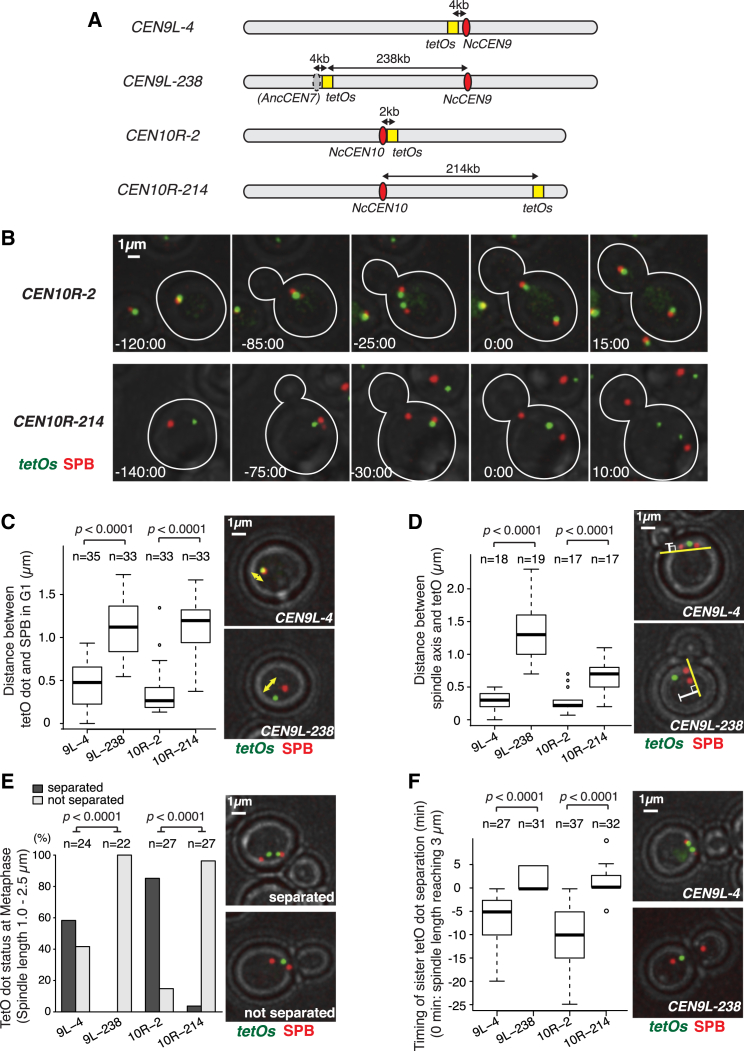
We addressed whether N. castellii CENs show expected localizations of functional centromeres in cells. If N. castellii CENs promote kinetochore assembly, spindle microtubules should attach and apply forces on these chromosome regions. In S. cerevisiae, such forces cause separation of sister chromatids up to 10 kb from centromeres on the metaphase spindle [18–21]. To investigate this in N. castellii, we inserted tet operators at 2–4 kb from NcCEN9 (CEN9L-4) and NcCEN10 (CEN10R-2; Figure 2A). As controls, we also inserted the tet operators on chromosome arms (CEN9L-238 and CEN10R-214). CEN9L-238 is also positioned at 4 kb right of the locus corresponding to the ancestor CEN7, based on synteny. The tet operators were bound by TetR-GFP fusion proteins [22] and visualized as small GFP dots (Figure 2B). In G1 phase, GFP dots at CEN9L-4 and CEN10R-2 localized in the vicinity of the spindle pole body (SPB), whereas those at CEN9L-238 and CEN10R-214 were at a larger distance from the SPB (Figure 2C). In metaphase, GFP dots at CEN9L-4 and CEN10R-2 located near the axis defined by two SPBs (Figure 2D) and often showed two signals, indicative of sister chromatid separation (Figure 2E). In contrast, the GFP dots at CEN9L-238 or CEN10R-214 did not separate until early anaphase (Figures 2E and 2F), whereas the GFP dots at CEN9L-4 and CEN10R-2 moved immediately after SPB segregation during anaphase (Figure 2B, top). These behaviors of GFP dots at CEN9L-4 and CEN10R-2 are similar to those at the S. cerevisiae centromeres [18–21] and consistent with NcCENs indeed being functional centromeres in N. castellii.
Figure 2.
N. castellii Candidate Centromeres Show Dynamic Localizations during the Cell Cycle, which Are Consistent with Those of Functional Centromeres
(A) Diagram showing positions of the insertion of tetO arrays (112 tandem repeats; yellow box) on chromosomes 9 and 10. NcCEN9 and NcCEN10 are shown as red ovals. The corresponding position of the ancestor CEN7 (AncCEN7), based on synteny, is shown as a gray oval.
(B) Live-cell images of tetO arrays shown in (A). Images of SPC42-4xmCherry TetR-GFP cells with tetOx112 at CEN10R-2 (T11466) or at CEN10R-214 (T11467) were acquired every 5 min. Spc42 is an SPB component. Time (min: s) is shown relative to anaphase onset (when the distance between SPBs exceeded 3 μm). The scale bar represents 1 μm.
(C–F) Analyses of tetOs localization in cells. Live-cell images of SPC42-4xmCherry TetR-GFP cells with tetOx112 at CEN9L-4 (T11501), CEN9L-238 (T11500), CEN10R-2 (T11466), or CEN10R-214 (T11467) were acquired every 5 min. (C) Distance between the SPB and the tetO dot in G1 phase is shown (in cells with one SPB but no bud). In box plots, a thick line represents a median; a box shows the range of the first to third quartile (interquartile range: IQR); the upper and lower whiskers show the maximum and minimum values, respectively, which do not exceed 3/2 IQR beyond the box; and open circles show outliers. (D) Distance between the tetO dot and the spindle axis in metaphase is shown (cells with two SPBs less than 2 μm apart and with a single tetO dot). Box plots are as in (C). (E) Frequency of separation and non-separation of the tetO dot in metaphase is shown (cells with two SPBs 1.0–2.5 μm apart). (F) Timing of separation of the tetO dot, relative to anaphase onset, is shown (as defined in B). Representative cells show separation of sister tetO dots (at CEN9L-4) before anaphase onset (top) and no separation of them (at CEN9-L-238) after anaphase onset (bottom). Box plots are as in (C).
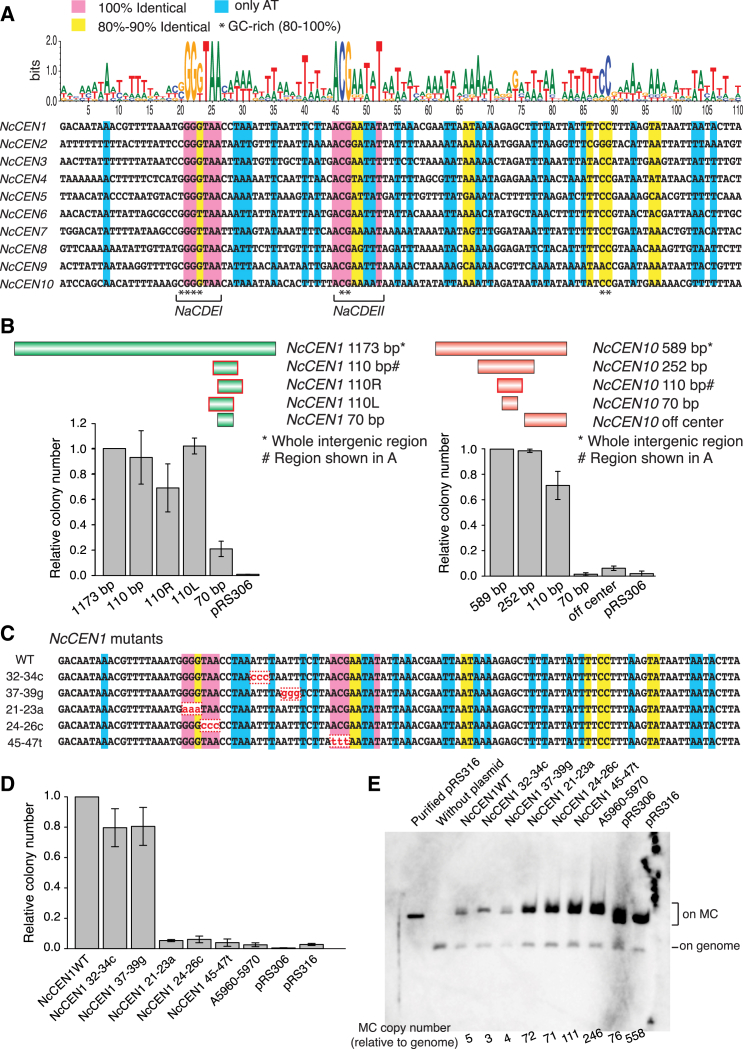
N. castellii CENs Include Unique Consensus DNA Elements, which Are Crucial for Minichromosome Propagation
We compared DNA sequences at NcCEN1–10, which include Ndc80-enriched regions in ChIP-seq (Figure 1D). Approximately in the middle of the enriched regions, we identified NcCEN consensus DNA sequences (Figures 3A and S2A). In particular, two short DNA elements showed very high similarity in all ten NcCENs at positions 20–26 and 45–52 in Figure 3A, which we name NaCDEI and NaCDEII (Naumovozyma consensus DNA element), respectively. In addition, other regions showed similarity or common AT- or GC-rich areas among NcCENs. The overall consensus within the 70-bp sequence at positions 20–89 in Figure 3A is unique to NcCENs and found at no other regions in the N. castellii genome. Remarkably, the consensus found at N. castellii CENs is very different from the consensus of other point centromeres (CDEI, II, and III; Figure 1B).
Figure 3.
N. castellii CENs Have Unconventional Consensus DNA Elements, which Are Important for Minichromosome Propagation
(A) Consensus DNA sequence found at N. castellii CENs. Logos of nucleotides (top) graphically represent their frequency at individual positions. Nucleotide positions, highlighted in pink and yellow, represent those identical in 100% and 80%–90% NcCENs, respectively. Blue shows positions only with A and T, whereas an asterisk shows GC-rich (80%–100%) positions. Two highly conserved elements were named NaCDEI (position 20–26) and NaCDEII (position 45–52), which are SGGKTAA (S: G or C; K: G or T) and ACGDDWWT (D: not C; W: A or T), respectively.
(B) Consensus DNA sequence of 110 bp shows full centromere activity for minichromosome (MC) propagation. The DNA fragments shown in the diagram (top) were inserted into the pRS306 plasmid, which carries S. cerevisiae URA3 [23] that works as a selection marker in N. castellii cells [24]. These DNA constructs were introduced into N. castellii haploid cells with ura3-1 (T11421) for colony formation assay. NcCEN1 1,173 bp and NcCEN10 589 bp cover the whole intergenic region containing a Ndc80-enriched region (Figure 1D). NcCEN1 110 bp and NcCEN10 110 bp are shown in (A), whereas NcCEN1 70 bp and NcCEN10 70 bp correspond to positions 20–89 in (A). NcCEN1 110 bp DNA sequence was shifted to right and left by 20 bp along the chromosome, making NcCEN1 110R and 110L, respectively. Colony numbers are normalized to that with NcCEN1 1,173 bp and NcCEN10 589 bp. Error bars represent SEM (n = 3).
(C) Mutations introduced to NcCEN1 DNA sequence (110 bp shown in A). WT, wild-type.
(D) Colony formation assay using NcCEN1 mutants shown in (C). The number of N. castellii colonies with a MC carrying each mutant (on 1,173 bp wild-type NcCEN1; see B) was normalized to that with wild-type NcCEN1 (1,173 bp; see B). The A5690–A5970 intergenic DNA fragment (1,071 bp) from a chromosome 1 arm was also integrated into a MC and used as a negative control. Error bars represent SEM (n = 3).
(E) Copy number (per cell) of each MC was evaluated using Southern blots. Genomic and MC DNA was digested by Not I, separated by electrophoresis, blotted, and probed with the ampicillin resistance gene (the host strain has one copy of it at hoΔ allele on the genome). The number at the bottom shows a ratio of each MC to the genome (ratio of intensity of hybridized bands).
See also Figure S2.
We evaluated the activity of candidate centromeres on minichromosomes in N. castellii cells. If minichromosomes were able to undergo both DNA replication and mitotic segregation, they are stably propagated during cell proliferation. DNA replication origins have not yet been identified in N. castellii, and we investigated the propagation of pRS306 and pRS316 plasmids, on which an S. cerevisiae centromere and replication origin are absent and present, respectively [23]. pRS306 and pRS316 were maintained at high copy number in N. castellii cells (>70 per cell; see Figure 3E). Yeast cells accumulate minichromosomes with high copy number if their replication occurs normally but segregation is inefficient [25]. We reasoned that, in N. castellii cells, an S. cerevisiae centromere does not promote minichromosome segregation efficiently, but its replication is supported, even without an S. cerevisiae replication origin (Figures S2B–S2D). Notably, addition of NcCEN1 to pRS306 caused a marked reduction in its copy number to 3–5 per cell (see Figure 3E; NcCEN1 WT) and the formation of many more yeast colonies (Figure 3B; NcCEN1 1,173 bp). Addition of NcCEN10 showed similar effects (Figure 3B; NcCEN10 589 bp), whereas addition of a control, chromosome arm DNA fragment (A5960–5970) had no such effect (see Figures 3D and 3E). Thus, N. castellii CENs are able to facilitate minichromosome propagation and yeast colony formation, presumably by promoting mitotic segregation.
To determine the minimum DNA sequence carrying a centromere activity, we examined the ability of several DNA fragments within NcCEN1 and NcCEN10 to support minichromosome propagation (Figure 3B). The 70-bp sequence at position 20–89 in Figure 3A was essential for centromere activity, and additional 20- to 40-bp sequences around it facilitated the activity. The 20- to 40-bp sequences are not particularly similar among the NcCENs (Figure 3A), but their AT richness may contribute to their centromere activity (Table S1) as does the AT-rich CDEII in other budding yeasts [13]. Subsequently, we addressed whether the consensus DNA elements NaCDEI and NaCDEII are important for centromere activity. Three base-pair mutations within the NaCDEI (21–23a and 24–26c) and NaCDEII (45–47t) of NcCEN1 (Figure 3C) showed substantial decreases in yeast colony formation (Figure 3D) and high copy numbers of minichromosomes (>70 per cell), indicative of inefficient segregation (Figure 3E). Three base-pair control mutations between NaCDEI and NaCDEII (32–34c and 37–39 g) showed similar numbers of yeast colonies to wild-type NcCEN1 (Figure 3D) and maintained low copy numbers of minichromosomes (3–5 per cell; Figure 3E). Thus, NaCDEI and NaCDEII are important for the centromere activity.
Using these assays, we next evaluated requirement of RNAi for centromere activity in N. castellii. This pathway is present in some budding yeasts, including N. castellii [26], and required for centromere activity in fission yeast [27]. The centromere activity for minichromosome propagation was still normal without the RNAi pathway in N. castellii (Figures S2E and S2F).
Consensus DNA Elements in N. castellii CENs Promote CBF3 Binding and Facilitate Centromere Activity on Authentic Chromosomes
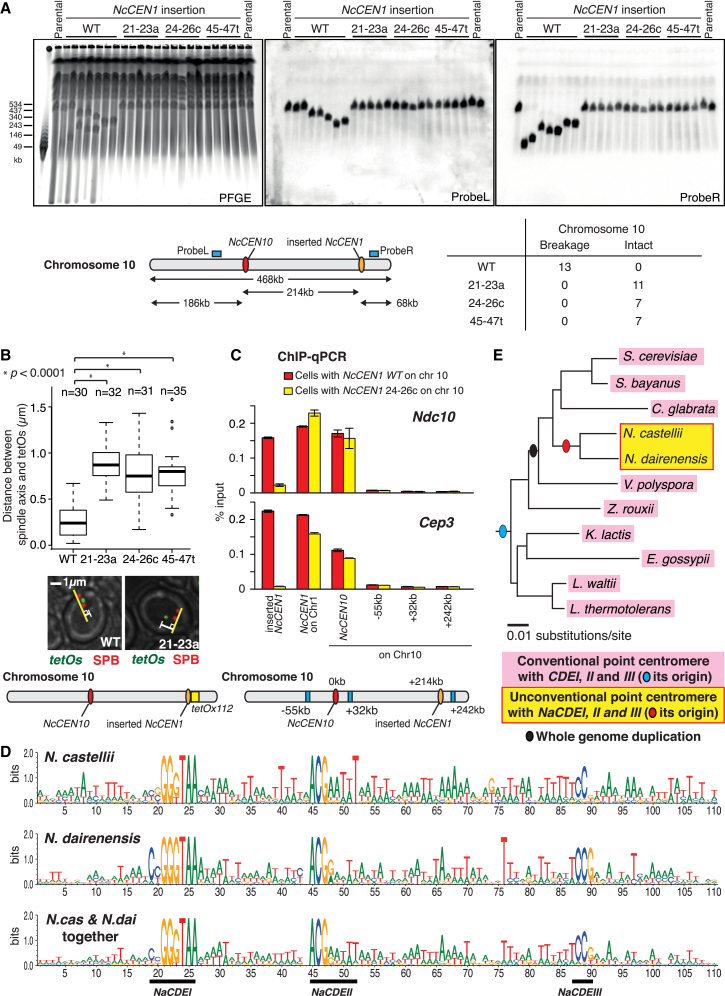
We next addressed whether the centromere DNA elements identified above are also crucial for centromere activity on authentic chromosomes. When an additional active centromere is inserted into a yeast chromosome, it causes chromosome breakage between the original and the newly inserted centromere [28, 29]. We employed this procedure to assess the centromere activity in N. castellii. We inserted wild-type and mutated NcCEN1s on the chromosome 10 arm (Figure 4A, diagram) and analyzed breakage of this chromosome using pulsed field gel electrophoresis (PFGE), followed by Southern blotting (Figure 4A). After insertion of a wild-type NcCEN1, all of 13 randomly chosen clones showed breakage of chromosome 10 (Figures 4A and S3A). By contrast, no such breakage was observed after insertion of NcCEN1 carrying mutations at NaCDEI and NaCDEII (21–23a, 24–26c, and 45–47t). Next, we visualized the intracellular localization of wild-type and mutated NcCEN1s, inserted on chromosome 10. Wild-type NcCEN1 on chromosome 10 was near SPBs during telophase to G1 (Figure S3B, left) and on the metaphase spindle with frequent sister separation (Figures 4B and S3B, right). However, mutated NcCEN1 (21–23a, 24–26c, and 45–47t) did not show such behavior. Thus, NaCDEI and NaCDEII are crucial for centromere activity on authentic chromosomes.
Figure 4.
Consensus DNA Elements in N. castellii CENs Are Important for the Centromere Function in the Context of Authentic Chromosomes
(A) NcCEN1 wild-type (WT; 1,173 bp; Figure 3B) and its mutants (1,173 bp with 21–23a, 24–26c, and 45–47t; Figure 3C) were inserted at 214 kb right of NcCEN10 on chromosome 10 (orange oval in diagram) in haploid N. castellii cells (T11605). Karyotypes of individual clones were analyzed (representative examples are shown here) by pulsed field gel electrophoresis (PFGE), followed by Southern blots with ProbeL and ProbeR; see positions of the probes in diagram. Table shows the number of clones that did, or did not, show breakage of chromosome 10 (shortening detected by ProbeL and/or ProbeR; Figure S3A).
(B) NcCEN1 wild-type (WT; T11632), 21–23a (Figure 3C; T11842), 24–26c (T11843), and 45–47t (T11844), each marked with tetOx112, were inserted at 214 kb right of NcCEN10 on chromosome 10 in SPC42-4xmCherry TetR-GFP cells. Cells were imaged, and the distance between the spindle axis and inserted NcCEN1 was analyzed as in Figure 2D. Box plots are as in Figure 2C.
(C) NDC10-3xFLAG cells with inserted NcCEN1 wild-type (WT; T11845) and 24–26c (T11846), as well as CEP3-3xFLAG cells with inserted NcCEN1 wild-type (WT; T11847) and 24–26c (T11848), were processed for ChIP-qPCR. Diagram shows chromosome loci for quantification by PCR (blue box). Primers for qPCR were designed to distinguish the original NcCEN1 on chromosome 1 and NcCEN1 inserted on chromosome 10. Error bars represent SEM (n = 3).
(D) N. dairenensis candidate centromeres (Figure S4B) have consensus DNA elements, NaCDEI, II, and III, which are also found at N. castellii CENs (Figure 3A). Logos of nucleotides graphically represent their frequency at individual positions, in aligned N. castellii centromeres (top), N. dairenensis candidate centromeres (middle), and both together (bottom).
(E) Conclusion of this study: budding yeasts, highlighted in pink, have point centromeres with CDEI, II, and III, which are conserved across several species. In contrast, N. castellii and N. dairenensis, highlighted in yellow, carry unconventional point centromeres with NaCDEI, II, and III, which have very different DNA sequences from conventional CDEI, II, and III. Blue and red ovals show the origins of the two types of point centromere in the yeast phylogenetic tree.
See also Figures S3 and S4.
As shown earlier, CBF3 components Ndc10 and Cep3 bind NcCENs (Figures 1D and S1C). To address whether this binding requires NaCDEs, we used ChIP followed by qPCR (ChIP-qPCR; Figure 4C). Both Ndc10 and Cep3 showed enrichment at the original NcCEN1 and wild-type NcCEN1 inserted on chromosome 10, but not at the mutated NcCEN1 on chromosome 10 (Figure 4C). Thus, the consensus DNA elements within NcCEN are required for CBF3 binding. Furthermore, we found both Ndc10 and Cep3 are essential genes in N. castellii (Figure S3C), as is expected if they have central roles in recognizing NcCENs.
How can the CBF3 complex, which recognizes standard CDE I,II,III-type CENs, also bind NcCENs despite the different DNA sequences? We investigated the evolutionary conservation of Ndc10 and Cep3, the CBF3 components recognizing consensus DNA elements in budding yeasts [5, 13]. A putative DNA-binding domain of Cep3 is conserved between N. castellii and other budding yeasts [30]. By contrast, the core DNA-binding domain of Ndc10 showed a more-rapid change during evolution of N. castellii, compared with other budding yeasts with standard CENs (Figures S3D–S3F). Such a rapid change may have happened to adapt to the new type of point centromere.
N. dairenensis, a Close Relative of N. castellii, Has N. castellii-like Consensus DNA Elements at Its Candidate Centromere Regions
We next aimed to identify candidate centromeres in N. dairenensis, a close relative to N. castellii (Figure 1A). Based on the annotated N. dairenensis genome [16], we found that the orders of orthologous genes around most N. castellii CENs are conserved on N. dairenensis chromosomes (Figure S4A). Crucially, at the corresponding intergenic regions, we identified CDEs that are very similar to NaCDEI, II found at N. castellii CENs (Figures 4D and S4B). It is likely that these regions serve as N. dairenensis centromeres, but we could not test this prediction because of a lack of molecular genetics methods in N. dairenensis. This analysis revealed a third sequence element with evolutionary conservation (NaCDEIII; Figure 4D), but mutagenesis showed that it is not essential for centromere function in N. castellii (Figure S4B legend). N. dairenensis Ndc10 showed evolutionary changes, similarly to N. castellii Ndc10 (Figures S3D–S3F). In conclusion, N. castellii CENs and N. dairenensis candidate centromeres have very similar consensus DNA elements (Figure 4D). The new type of centromere CDEs (NaCDEI, II, and III) originated prior to the branching point of N. castellii and N. dairenensis in evolution (Figure 4E).
Conclusions
We have identified centromeres in the budding yeast N. castellii. We conclude that they make point centromeres, because (1) consensus DNA elements are found among all ten centromeres, (2) these DNA elements are important for the centromere activity, and (3) a short DNA fragment (110 bp) containing the consensus DNA elements is sufficient for centromere function. Crucially, the consensus centromere DNA elements are very different from those in other known point centromeres, highlighting the N. castellii centromere as the first unconventional, i.e., non-CDE I,II,III-type, point centromere (Figure 4E). The gene order analyses give the following insights: first, most N. castellii centromeres are not located in intergenic regions orthologous to those containing standard CDEI,II,III-type point centromeres in other species (Figure S1D), although these two are often in close proximity (Figure S4C). This indicates that these N. castellii centromeres did not descend from standard point centromeres at their individual chromosome regions. Second, at most N. castellii centromeres, synteny is disrupted when compared with the ancestral budding yeast genome (Figure 1E). This can be explained if N. castellii centromeres were propagated to all chromosomes during evolution through extensive genome rearrangement. The origin of the N. castellii centromere is still elusive, but it may have been propagated and superseded the conventional point centromeres.
Acknowledgments
We thank M. Gierlinski and members of the T.U.T., Y.S., C.N., and K.H.W. groups for helpful discussion; L. Clayton for editing the manuscript; J. Piskur, M. Cohn, R. Ciosk, K. Nasmyth, K.E. Sawin, and R.Y. Tsien for reagents; S. Swift for technical help; K.P. Byrne for bioinformatics assistance; and D.P. Bartel for his generous support. This work was supported by the Wellcome Trust (096535 and 097945), European Research Council (322682 and 268893), grants-in-aid from MEXT (KAKENHI; 221S0002), and BBSRC (BB/E023754/1 and BB/K007211/1). T.U.T. is a Wellcome Trust Principal Research Fellow.
Published: July 9, 2015
Footnotes
This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with the article online at http://dx.doi.org/10.1016/j.cub.2015.06.023.
Accession Numbers
DNA sequences have been deposited under the following accession codes: ChIP-seq of Ndc80-6xHA, DDBJ: DRA002836; Ndc10-6xHA, DDBJ: DRA003502; Cep3-3xFLAG, DDBJ: DRA002836; Cse4-6xHA, DDBJ: DRA003067; N. castellii CEN1-10, DDBJ: LC029901–LC029910; and N. dairenensis CEN1-11, DDBJ: BR001264–BR001274.
Supplemental Information
References
- 1.Verdaasdonk J.S., Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pluta A.F., Mackay A.M., Ainsztein A.M., Goldberg I.G., Earnshaw W.C. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 3.Dujon B. Yeast evolutionary genomics. Nat. Rev. Genet. 2010;11:512–524. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 4.Malik H.S., Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 5.Meraldi P., McAinsh A.D., Rheinbay E., Sorger P.K. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon J.L., Byrne K.P., Wolfe K.H. Mechanisms of chromosome number evolution in yeast. PLoS Genet. 2011;7:e1002190. doi: 10.1371/journal.pgen.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedtke S.M., Townsend T.M., Hillis D.M. Resolution of phylogenetic conflict in large data sets by increased taxon sampling. Syst. Biol. 2006;55:522–529. doi: 10.1080/10635150600697358. [DOI] [PubMed] [Google Scholar]
- 8.Wolfe K.H., Shields D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 9.Zeng X., Kahana J.A., Silver P.A., Morphew M.K., McIntosh J.R., Fitch I.T., Carbon J., Saunders W.S. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 1999;146:415–425. doi: 10.1083/jcb.146.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne K.P., Wolfe K.H. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfe K.H. Comparative genomics and genome evolution in yeasts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:403–412. doi: 10.1098/rstb.2005.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cliften P.F., Fulton R.S., Wilson R.K., Johnston M. After the duplication: gene loss and adaptation in Saccharomyces genomes. Genetics. 2006;172:863–872. doi: 10.1534/genetics.105.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman A.A., Sorger P.K. Structure and function of kinetochores in budding yeast. Annu. Rev. Cell Dev. Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- 14.He X., Rines D.R., Espelin C.W., Sorger P.K. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 15.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 16.Gordon J.L., Armisén D., Proux-Wéra E., ÓhÉigeartaigh S.S., Byrne K.P., Wolfe K.H. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc. Natl. Acad. Sci. USA. 2011;108:20024–20029. doi: 10.1073/pnas.1112808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon J.L., Byrne K.P., Wolfe K.H. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He X., Asthana S., Sorger P.K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 19.Goshima G., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka T., Fuchs J., Loidl J., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 21.Pearson C.G., Maddox P.S., Salmon E.D., Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaelis C., Ciosk R., Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 23.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Astromskas E., Cohn M. Tools and methods for genetic analysis of Saccharomyces castellii. Yeast. 2007;24:499–509. doi: 10.1002/yea.1488. [DOI] [PubMed] [Google Scholar]
- 25.Hieter P., Mann C., Snyder M., Davis R.W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 26.Drinnenberg I.A., Weinberg D.E., Xie K.T., Mower J.P., Wolfe K.H., Fink G.R., Bartel D.P. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lejeune E., Allshire R.C. Common ground: small RNA programming and chromatin modifications. Curr. Opin. Cell Biol. 2011;23:258–265. doi: 10.1016/j.ceb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Haber J.E., Thorburn P.C. Healing of broken linear dicentric chromosomes in yeast. Genetics. 1984;106:207–226. doi: 10.1093/genetics/106.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brock J.A., Bloom K. A chromosome breakage assay to monitor mitotic forces in budding yeast. J. Cell Sci. 1994;107:891–902. doi: 10.1242/jcs.107.4.891. [DOI] [PubMed] [Google Scholar]
- 30.Bellizzi J.J., 3rd, Sorger P.K., Harrison S.C. Crystal structure of the yeast inner kinetochore subunit Cep3p. Structure. 2007;15:1422–1430. doi: 10.1016/j.str.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.