Abstract
Arteriovenous malformations (AVMs) of the central nervous system are considered as congenital disorders. They are composed of abnormally developed dilated arteries and veins and are characterized microscopically by the absence of a capillary network. We previously reported DNA fragmentation and increased expression of apoptosis-related factors in AVM lesions. In this article, we used microarray analysis to examine differential gene expression in relation to clinical manifestations in 11 AVM samples from Japanese patients. We categorized the genes with altered expression into four groups: death-related, neuron-related, inflammation-related, and other. The death-related differentially expressed genes were MMP9, LIF, SOD2, BCL2A1, MMP12, and HSPA6. The neuron-related genes were NPY, S100A9, NeuroD2, S100Abeta, CAMK2A, SYNPR, CHRM2, and CAMKV. The inflammation-related genes were PTX3, IL8, IL6, CXCL10, GBP1, CHRM3, CXCL1, IL1R2, CCL18, and CCL13. In addition, we compared gene expression in those with or without clinical characteristics including deep drainer, embolization, and high-flow nidus. We identified a small number of genes. Using these microarray data we are able to generate and test new hypotheses to explore AVM pathophysiology. Microarray analysis is a useful technique to study clinical specimens from patients with brain vascular malformations.
Keywords: arteriovenous malformations, DNA microarray, clinical characteristics
Introduction
Arteriovenous malformations (AVMs) of the central nervous system are generally considered as congenital disorders that result from aberrant differentiation of the mesoderm during embryonic development. AVMs are composed of abnormally developed dilated arteries and veins and are characterized microscopically by the absence of a capillary network.1–4) Although many studies have addressed the epidemiological characteristics, natural history, radiological features, and clinical behavior of AVMs, less is known about the molecular properties of these lesions.1–4) Recent studies have revealed abnormal expression of angiogenic growth factors and their receptors compared with that in normal brain tissue.5–8) Moreover, we have reported that AVM lesions display DNA fragmentation and increased expression of apoptosis-related factors.9–11) In this study, we examined differential gene expression in AVMs and analyzed this expression in relation to clinical manifestations in Japanese patients.
Materials and Methods
I. Patients
Eleven specimens from patients with cerebral AVMs were used in this study. All samples were obtained during surgery and were snap-frozen in liquid nitrogen. The relevant clinical and lesion features of the cases are summarized in Table 1.
Table 1.
Clinical summary of the patients
| Case | Age | Sex | Hemorrhage | High flow | Deep | Embolization | Seizure | Size | S-M grade | Location |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 60 | M | No | Yes | No | No | No | 3 cm | 2 | Occipital |
| 2 | 2 | F | Yes | Yes | Yes | No | No | 5 cm | 4 | Frontal |
| 3 | 28 | F | No | No | No | No | Yes | 2 cm | 1 | Temporal |
| 4 | 32 | M | No | Yes | No | Yes | Yes | 2 cm | 1 | Frontal |
| 5 | 49 | M | Yes | Yes | Yes | No | No | 3 cm | 2 | Frontal |
| 6 | 25 | M | Yes | Yes | No | No | No | 2 cm | 2 | Occipital |
| 7 | 28 | M | No | Yes | No | No | Yes | 4 cm | 2 | Frontal |
| 8 | 17 | F | No | No | Yes | Yes | No | 5 cm | 4 | Cerebellum |
| 9 | 29 | F | Yes (op) | Yes | No | Yes | No | 4 cm | 2 | Temporooccipital |
| 10 | 45 | M | Yes | Yes | No | No | No | 3 cm | 2 | Parietal |
| 11 | 38 | M | No | Yes | No | No | No | 2 cm | 1 | Parietal |
F: female, M: male, op: intraoperative hemorrhage, S-M: Spetzler and Martin.
II. Preparation of RNA
RNA was isolated from the specimens which is nudus including brain parenchyma as follows. Briefly, RNAlater® (Life Technologies Inc., Carlsbad, California, USA) was added at a volume of 1 ml/100 mg sample. The samples were thawed and then homogenized three times for 20 sec on ice. After the addition of 0.1 vol 1-bromo-3-chloropropane, the homogenate was vortexed for 15 sec and incubated on ice for 1 hr. After centrifugation, the upper aqueous phase was transferred to a new tube and a half volume of isopropanol was added. The solution was then mixed and incubated on ice for 1 hr. After centrifugation, the supernatant was removed. The RNA pellet was washed with 80% ethanol and resuspended in diethylpyrocarbonate-treated water. The RNA was affinity column-purified using an RNeasy Mini Kit (Qiagen Inc., Valencia, California, USA) according to the manufacturer's protocol. Control RNA was extracted from a middle cerebral artery and a cortical tissue sample from a Caucasian male.
III. Microarray analysis
Microarray analysis was conducted by Hokkaido System Science Co. Ltd. (Sapporo). Total RNA was extracted from three biological replicates of each sample, and then it was used for cRNA synthesis. The resulting cRNA was subsequently labeled with Cyanin3 using a Quick Amp Labeling Kit (Agilent Technologies Inc., Santa Clara, California, USA), and purified using RNeasy mini spin columns (Qiagen) to generate the cRNA target solution. The cRNA target solution was then hybridized to the microarray (Arabidopsis Oligo DNA microarray ver. 4.0; Agilent Technologies). After washing and air-drying, the slide was scanned at a resolution of 5 μm using a microarray scanner (Agilent Technologies). The digitalized data were imported into software (GeneSpring GX 10; Agilent Technologies) and normalized to shift to the 75th percentile. The following flagged features were cut off: features that were not positive and significant, and features that were not above background levels. After filtering for flags, 32 348 probes remained. On the microarray, some genes are represented by several oligonucleotides that have distinct 60-mer sequences from different regions within the same gene.
Results
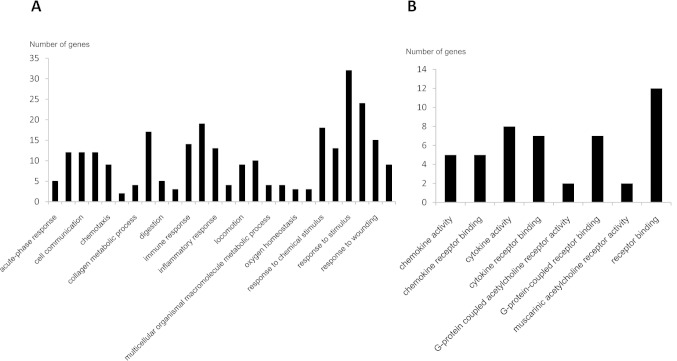
Tables 2 and 3 indicate the genes that displayed an absolute fold change of at least ± 300. We categorized these genes into four groups: death-related, neuron-related, inflammation-related, and others. The differentially expressed death-related genes were MMP9, LIF, SOD2, BCL2A1, MMP12, and HSPA6. The neuron-related genes were NPY, S100A9, NeuroD2, S100Abeta, CAMK2A, SYNPR, CHRM2, and CAMKV. The inflammation-related genes were PTX3, IL8, IL6, CXCL10, GBP1, CHRM3, CXCL1, IL1R2, CCL18, and CCL13. In addition, we classified significantly changed genes based on biological process and molecular function (Fig. 1).
Table 2.
Genes with altered expression in cerebral arteriovenous malformation Part 1
| ProbeName | Regulation | Common name | Category | Description |
|---|---|---|---|---|
| A_23_P166848 | Up | LTF | O | Homo sapiens lactotransferrin (LTF), mRNA |
| A_23_P40174 | Up | MMP9 | D | Homo sapiens matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (MMP9), mRNA |
| A_23_P207520 | Up | COL1A1 | O | Homo sapiens mRNA for prepro-alpha1(I) collagen |
| A_23_P212914 | Up | RUFY3 | O | Homo sapiens RUN and FYVE domain containing 3 (RUFY3), transcript variant 1, mRNA |
| A_23_P121064 | Up | PTX3 | I | Homo sapiens pentraxin-related gene, rapidly induced by IL-1 beta (PTX3), mRNA |
| A_24_P122137 | Up | LIF | D | Homo sapiens leukemia inhibitory factor (cholinergic differentiation factor) (LIF), mRNA |
| A_23_P53137 | Up | HBG1 | O | Homo sapiens hemoglobin, gamma A (HBG1), mRNA |
| A_32_P87013 | Up | IL8 | I | Homo sapiens interleukin 8 (IL8), mRNA |
| A_32_P70158 | Up | LILRB3 | O | Homo sapiens leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (LILRB3), transcript variant 2, mRNA |
| A_23_P142533 | Up | COL3A1 | O | Homo sapiens collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant) (COL3A1), mRNA |
| A_24_P24371 | Up | ENST00000390543 | O | Immunoglobulin heavy chain C gene segment [Source: IMGT/GENE_DB; Acc: IGHG4] |
| A_23_P71037 | Up | IL6 | A, I | Homo sapiens interleukin 6 (interferon, beta 2) (IL6), mRNA |
| A_23_P81898 | Up | UBD | O | Homo sapiens ubiquitin D (UBD), mRNA |
| A_23_P324384 | Up | RPS4Y2 | O | Homo sapiens ribosomal protein S4, Y-linked 2 (RPS4Y2), mRNA |
| A_32_P385587 | Up | ALAS2 | O | Homo sapiens aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) (ALAS2), nuclear gene encoding mitochondrial protein, transcript variant 1, mRNA |
| A_24_P935819 | Up | SOD2 | D | Homo sapiens superoxide dismutase 2, mitochondrial, mRNA (cDNA clone MGC: 21350 IMAGE: 4184203), complete cds |
| A_24_P303091 | Up | CXCL10 | I | Homo sapiens chemokine (C-X-C motif) ligand 10 (CXCL10), mRNA |
| A_23_P106602 | Up | CRISPLD2 | O | Homo sapiens cysteine-rich secretory protein LCCL domain containing 2 (CRISPLD2), mRNA |
| A_23_P170233 | Up | CSTA | O | Homo sapiens cystatin A (stefin A) (CSTA), mRNA |
| A_23_P158817 | Up | IGH@ | O | Homo sapiens cDNA FLJ27104 fis, clone SPL04981, highly similar to Ig gamma-2 chain C region |
| A_24_P169873 | Up | ENST00000390539 | O | Immunoglobulin heavy chain C gene segment [Source: IMGT/GENE_DB; Acc: IGHA2] |
| A_23_P62890 | Up | GBP1 | I | Homo sapiens guanylate binding protein 1, interferon-inducible, 67kDa (GBP1), mRNA |
| A_32_P22654 | Up | ALAS2 | O | Homo sapiens aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) (ALAS2), nuclear gene encoding mitochondrial protein, transcript variant 1, mRNA |
| A_23_P33723 | Up | CD163 | O | Homo sapiens CD163 molecule (CD163), transcript variant 1, mRNA |
| A_32_P39440 | Up | BC030813 | O | Homo sapiens cDNA clone MGC: 22645 IMAGE: 4700961, complete cds |
| A_23_P23048 | Up | S100A9 | N | Homo sapiens S100 calcium binding protein A9 (S100A9), mRNA |
| A_23_P256470 | Down | NPY | N | Homo sapiens neuropeptide Y (NPY), mRNA |
| A_23_P205428 | Down | FOXG1 | O | Homo sapiens forkhead box G1B (FOXG1B), mRNA [NM_005249] |
| A_24_P817236 | Down | ENST00000366569 | O | Muscarinic acetylcholine receptor M3 [Source: Uniprot/SWISSPROT; Acc: P20309] |
| A_24_P142343 | Down | HRNBP3 | O | Homo sapiens hypothetical protein LOC146713 (HRNBP3), mRNA |
| A_24_P500584 | Down | XIST | O | Homo sapiens X (inactive)-specific transcript (XIST) on chromosome X |
| A_32_P85360 | Down | THC2770932 | O | Unknown |
| A_24_P347319 | Down | KCNC2 | O | Homo sapiens potassium voltage-gated channel, Shaw-related subfamily, member 2 (KCNC2), transcript variant 1, mRNA |
| A_23_P401472 | Down | CHRM3 | I | Homo sapiens cholinergic receptor, muscarinic 3 (CHRM3), mRNA |
| A_32_P142818 | Down | DLX1 | O | Homo sapiens distal-less homeobox 1 (DLX1), transcript variant 1, mRNA |
| A_23_P67569 | Down | PRG2 | O | Homo sapiens plasticity-related gene 2 (PRG2), mRNA |
A: angiogenesis, D: death, I: inflammation, N: neuron, O: others.
Table 3.
Genes with altered expression in cerebral arteriovenous malformation Part 2
| ProbeName | Regulation | Common name | Category | Description |
|---|---|---|---|---|
| A_24_P335092 | Up | SAA1 | O | Homo sapiens serum amyloid A1 (SAA1), transcript variant 1, mRNA |
| A_23_P43979 | Up | M87790 | O | Human (hybridoma H210) anti-hepatitis A immunoglobulin lambda chain variable region, constant region, complementarity-determining regions mRNA, complete cds |
| A_23_P434809 | Up | S100A8 | N | Homo sapiens S100 calcium binding protein A8 (S100A8), mRNA |
| A_23_P7144 | Up | CXCL1 | I | Homo sapiens chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (CXCL1), mRNA |
| A_23_P64539 | Up | HBG1 | O | Homo sapiens hemoglobin, gamma A (HBG1), mRNA |
| A_23_P79398 | Up | IL1R2 | I | Homo sapiens interleukin 1 receptor, type II (IL1R2), transcript variant 1, mRNA |
| A_23_P99515 | Up | C13orf33 | O | Homo sapiens chromosome 13 open reading frame 33 (C13orf33), mRNA |
| A_24_P357847 | Up | BC030813 | O | Homo sapiens cDNA clone MGC: 22645 IMAGE: 4700961, complete cds |
| A_23_P431388 | Up | SPOCD1 | O | Homo sapiens SPOC domain containing 1 (SPOCD1), mRNA |
| A_23_P152002 | Up | BCL2A1 | D | Homo sapiens BCL2-related protein A1 (BCL2A1), mRNA |
| A_23_P160286 | Up | PRG4 | O | Homo sapiens proteoglycan 4 (PRG4), mRNA |
| A_23_P90710 | Up | DES | O | Homo sapiens desmin (DES), mRNA |
| A_23_P259071 | Up | AREG | O | Homo sapiens amphiregulin (schwannoma-derived growth factor) (AREG), mRNA |
| A_32_P116488 | Up | THC2677011 | O | Unknown |
| A_24_P605563 | Up | AY172962 | O | Homo sapiens anti-rabies SOJB immunoglobulin lambda light chain mRNA, complete cds |
| A_23_P55270 | Up | CCL18 | I | Homo sapiens chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated) (CCL18), mRNA |
| A_23_P4773 | Up | LILRB5 | O | Homo sapiens leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 5 (LILRB5), transcript variant 2, mRNA |
| A_23_P259314 | Up | RPS4Y1 | O | Homo sapiens ribosomal protein S4, Y-linked 1 (RPS4Y1), mRNA |
| A_23_P26965 | Up | CCL13 | I | Homo sapiens chemokine (C-C motif) ligand 13 (CCL13), mRNA |
| A_32_P192842 | Up | BM129308 | O | if20d02.x1 Melton Normalized Human Islet 4 N4-HIS 1 Homo sapiens cDNA clone IMAGE: 5677082 3′, mRNA sequence |
| A_23_P340698 | Up | MMP12 | D | Homo sapiens matrix metallopeptidase 12 (macrophage elastase) (MMP12), mRNA |
| A_23_P114903 | Up | HSPA6 | D | Homo sapiens heat shock 70kDa protein 6 (HSP70B′) (HSPA6), mRNA |
| A_32_P200144 | Up | IGH@ | O | Homo sapiens cDNA FLJ27104 fis, clone SPL04981, highly similar to Ig gamma-2 chain C region |
| A_32_P45738 | Down | PGAM1 | O | Homo sapiens phosphoglycerate mutase 1 (brain) (PGAM1), mRNA |
| A_23_P60130 | Down | MAL2 | O | Homo sapiens mal, T-cell differentiation protein 2 (MAL2), mRNA |
| A_23_P355377 | Down | SLC12A5 | O | Homo sapiens solute carrier family 12, (potassium-chloride transporter) member 5 (SLC12A5), mRNA |
| A_32_P25295 | Down | NEUROD2 | N | Homo sapiens neurogenic differentiation 2 (NEUROD2), mRNA |
| A_23_P2543 | Down | CUX2 | O | Homo sapiens cut-like 2 (Drosophila) (CUTL2), mRNA |
| A_24_P380311 | Down | CAMK2A | N | Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha (CAMK2A), transcript variant 1, mRNA |
| A_23_P302568 | Down | SLC30A3 | O | Homo sapiens solute carrier family 30 (zinc transporter), member 3 (SLC30A3), mRNA |
| A_23_P80718 | Down | SYNPR | N | Homo sapiens synaptoporin (SYNPR), mRNA |
| A_23_P145606 | Down | CHRM2 | N | Homo sapiens cholinergic receptor, muscarinic 2 (CHRM2), transcript variant 1, mRNA |
| A_23_P29680 | Down | CAMKV | N | Homo sapiens CaM kinase-like vesicle-associated (CAMKV), mRNA |
| A_23_P77731 | Down | CRYM | O | Homo sapiens crystallin, mu (CRYM), transcript variant 1, mRNA |
| A_23_P252817 | Down | SST | O | Homo sapiens somatostatin (SST), mRNA |
| A_23_P35725 | Down | ANO3 | O | Homo sapiens transmembrane protein 16C (TMEM16C), mRNA |
| A_23_P157926 | Down | LINGO2 | O | Homo sapiens leucine rich repeat and Ig domain containing 2 (LINGO2), mRNA |
| A_23_P408195 | Down | TMEM155 | O | Homo sapiens transmembrane protein 155 (TMEM155), mRNA |
A: angiogenesis, D: death, I: inflammation, N: neuron, O: others.
Fig. 1.
Classified genes with significantly altered expression based on biological process (A) and molecular function (B).
Next, we analyzed gene expression in relation to clinical characteristics. First, we analyzed gene expression in the samples that were or were not from deep-draining veins. We identified 32 genes that showed greater than 10-fold change in deep-draining samples (Table 4). Among them, FGF9, which is an angiogenesis-related gene, was upregulated. We next compared gene expression in those with or without preoperative embolization, and found 21 genes that showed a greater than 10-fold change in those with embolization (Table 5). Among them, PTX3, MMP3, and GDNF were downregulated in the samples with preoperative embolization. When we compared expression in the samples with or without a high-flow nidus, we identified 40 genes with a greater than 10-fold change in samples with high flow (Table 6). Neuron-related genes, including NPY and NeuroD, were downregulated in high-flow AVMs.
Table 4.
Clinical presentation and gene expression (deep-draining veins)
| ProbeName | Fold change | Regulation | Common name | Category | Description |
|---|---|---|---|---|---|
| A_23_P24294 | 17.487488 | Up | SLC17A6 | O | Homo sapiens solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (SLC17A6), mRNA |
| A_32_P164593 | 12.236315 | Up | ZMAT4 | O | Homo sapiens zinc finger, matrin type 4 (ZMAT4), mRNA |
| A_23_P334308 | 10.814596 | Up | MTUS2 | O | Homo sapiens KIAA0774 (KIAA0774), transcript variant 1, mRNA |
| A_23_P2283 | 14.029393 | Up | TAC3 | O | Homo sapiens tachykinin 3 (neuromedin K, neurokinin beta) (TAC3), transcript variant 1, mRNA |
| A_23_P92860 | 12.137709 | Up | CCNO | O | Homo sapiens cyclin U (CCNU), mRNA |
| A_24_P142343 | 21.59073 | Up | O | Homo sapiens hypothetical protein LOC146713 (HRNBP3), mRNA | |
| A_24_P25137 | 10.262987 | Up | CHRM3 | O | Homo sapiens cholinergic receptor, muscarinic 3 (CHRM3), mRNA |
| A_32_P166733 | 13.562277 | Up | BU686948 | O | UI-CF-DU1-ado-e-06-0-UI.s1 UI-CF-DU1 Homo sapiens cDNA clone UI-CF-DU1-ado-e-06-0-UI 3′, mRNA sequence |
| A_23_P8981 | 12.29509 | Up | STAR | O | Homo sapiens steroidogenic acute regulator (STAR), nuclear gene encoding mitochondrial protein, transcript variant 1, mRNA |
| A_23_P321846 | 15.988988 | Up | KCNS1 | O | Homo sapiens potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 (KCNS1), mRNA |
| A_24_P54900 | 12.008987 | Up | LNX1 | O | Homo sapiens ligand of numb-protein X 1 (LNX1), mRNA |
| A_24_P219474 | 14.443749 | Up | MGAT5B | O | Homo sapiens mannosyl (alpha-1,6-)- glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B (MGAT5B), transcript variant 1, mRNA |
| A_23_P144847 | 10.753042 | Up | CDH12 | O | Homo sapiens cadherin 12, type 2 (N-cadherin 2) (CDH12), mRNA |
| A_23_P318616 | 11.135667 | Up | LRTM2 | O | Homo sapiens leucine-rich repeats and transmembrane domains 2 (LRTM2), mRNA |
| A_32_P142818 | 10.828055 | Up | DLX1 | O | Homo sapiens distal-less homeobox 1 (DLX1), transcript variant 1, mRNA |
| A_23_P140858 | 11.060884 | Up | O | Homo sapiens ataxin 2-binding protein 1 (A2BP1), transcript variant 4, mRNA | |
| A_23_P2543 | 15.522975 | Up | CUX2 | O | Homo sapiens cut-like 2 (Drosophila) (CUTL2), mRNA |
| A_23_P337642 | 12.069429 | Up | ATP2B3 | O | Homo sapiens ATPase, Ca++ transporting, plasma membrane 3 (ATP2B3), transcript variant 1, mRNA |
| A_24_P380311 | 26.18467 | Up | CAMK2A | N | Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha (CAMK2A), transcript variant 1, mRNA |
| A_23_P13822 | 10.42801 | Up | STYK1 | O | Homo sapiens serine/threonine/tyrosine kinase 1 (STYK1), mRNA |
| A_23_P65918 | 11.070756 | Up | ITPKA | O | Homo sapiens inositol 1,4,5-trisphosphate 3-kinase A (ITPKA), mRNA |
| A_23_P157027 | 11.949467 | Up | O | Homo sapiens hypothetical protein LOC 285878, mRNA (cDNA clone IMAGE: 5299807) | |
| A_23_P22723 | 12.559601 | Up | ATP2B3 | O | Homo sapiens ATPase, Ca++ transporting, plasma membrane 3 (ATP2B3), transcript variant 1, mRNA |
| A_23_P132175 | 11.3289 | Up | RTN4R | O | Homo sapiens reticulon 4 receptor (RTN4R), mRNA |
| A_23_P79968 | 10.7763815 | Up | PCSK2 | O | Homo sapiens proprotein convertase subtilisin/kexin type 2 (PCSK2), mRNA |
| A_23_P105803 | 11.011639 | Up | FGF9 | A | Homo sapiens fibroblast growth factor 9 (glia-activating factor) (FGF9), mRNA |
| A_23_P53137 | 11.880983 | Down | HBG1 | O | Homo sapiens hemoglobin, gamma A (HBG1), mRNA |
| A_32_P385587 | 15.667379 | Down | ALAS2 | O | Homo sapiens aminolevulinate, delta-, synthase 2 (sideroblastic/hypochromic anemia) (ALAS2), nuclear gene encoding mitochondrial protein, transcript variant 1, mRNA |
| A_23_P121596 | 19.073265 | Down | PPBP | I | Homo sapiens pro-platelet basic protein (chemokine [C-X-C motif] ligand 7) (PPBP), mRNA |
| A_32_P168342 | 10.181584 | Down | C6orf25 | O | G6b protein precursor [Source: Uniprot/SWISSPROT; Acc: O95866] |
| A_23_P87346 | 14.329434 | Down | HBD | O | Homo sapiens hemoglobin, delta (HBD), mRNA |
| A_24_P79403 | 13.201708 | Down | PF4 | O | Homo sapiens platelet factor 4 (chemokine [C-X-C motif] ligand 4) (PF4), mRNA |
A: angiogenesis, D: death, I: inflammation, N: neuron, O: others.
Table 5.
Clinical presentation and gene expression (embolization)
| ProbeName | Fold change | Regulation | Common name | Category | Description |
|---|---|---|---|---|---|
| A_23_P62857 | 12.33438 | Down | A_23_P62857 | O | PLA2G2A |
| A_23_P73526 | 13.889064 | Down | CITED1 | O | Homo sapiens Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 (CITED1), mRNA |
| A_23_P121064 | 32.46276 | Down | PTX3 | I | Homo sapiens pentraxin-related gene, rapidly induced by IL-1 beta (PTX3), mRNA |
| A_32_P107372 | 15.64205 | Down | GBP1 | I | Homo sapiens guanylate binding protein 1, interferon-inducible, 67kDa (GBP1), mRNA |
| A_23_P78037 | 15.200374 | Down | CCL7 | I | Homo sapiens chemokine (C-C motif) ligand 7 (CCL7), mRNA |
| A_23_P161698 | 16.020575 | Down | MMP3 | D | Homo sapiens matrix metallopeptidase 3 (stromelysin 1, progelatinase) (MMP3), mRNA |
| A_32_P377880 | 13.725988 | Down | GDNF | N | Glial cell line-derived neurotrophic factor precursor (Astrocyte- derived trophic factor 1) (ATF-1) |
| A_32_P5417 | 17.92484 | Down | CA946373 | O | CA946373 ni04a06.x1 Human lacrimal gland: ni Homo sapiens cDNA clone ni04a06 5′, mRNA sequence |
| A_23_P62890 | 16.525377 | Down | GBP1 | O | Homo sapiens guanylate binding protein 1, interferon-inducible, 67kDa (GBP1), mRNA |
| A_23_P52067 | 12.040656 | Down | GRHL3 | O | Homo sapiens grainyhead-like 3 (Drosophila) (GRHL3), transcript variant 2, mRNA |
| A_24_P932887 | 45.99536 | Down | SPOCD1 | O | Homo sapiens cDNA FLJ39908 fis, clone SPLEN2017620 |
| A_23_P63254 | 12.1611395 | Down | SFN | O | Homo sapiens stratifin (SFN), mRNA |
| A_24_P335092 | 50.85907 | Down | SAA1 | O | Homo sapiens serum amyloid A1 (SAA1), transcript variant 1, mRNA |
| A_23_P336554 | 11.759418 | Down | IL1RAP | I | Homo sapiens interleukin 1 receptor accessory protein (IL1RAP), transcript variant 2, mRNA |
| A_23_P431388 | 24.716013 | Down | SPOCD1 | O | Homo sapiens SPOC domain containing 1 (SPOCD1), mRNA |
| A_32_P15544 | 7.8432913 | Down | PRIMA1 | O | Homo sapiens proline rich membrane anchor 1 (PRIMA1), mRNA |
| A_24_P923854 | 15.754891 | Down | AF113674 | O | Homo sapiens clone FLB1727 PRO0398 mRNA, complete cds |
| A_23_P104073 | 11.630592 | Down | S100A3 | N | Homo sapiens S100 calcium binding protein A3 (S100A3), mRNA |
| A_32_P116488 | 11.215696 | Down | THC2677011 | O | Unknown |
| A_24_P379521 | 28.073503 | Down | BM702245 | O | UI-E-CQ1-aey-h-03-0-UI.r1 UI-E-CQ1 Homo sapiens cDNA clone UI-E-CQ1-aey-h-03-0-UI 5′, mRNA sequence |
| A_23_P306203 | 20.476131 | Down | SAA2 | O | Homo sapiens serum amyloid A2 (SAA2), mRNA |
A: angiogenesis, D: death, I: inflammation, N: neuron, O: others.
Table 6.
Clinical presentation and gene expression (high-flow)
| ProbeName | Fold change | Regulation | Common name | Category | Description |
|---|---|---|---|---|---|
| A_24_P933319 | 13.79898 | Down | RAB3B | O | Ras-related protein Rab-3B [Source: Uniprot/SWISSPROT; Acc: P20337] |
| A_23_P415541 | 26.580494 | Down | GPR26 | O | Homo sapiens G protein-coupled receptor 26 (GPR26), mRNA |
| A_32_P51005 | 12.428625 | Down | AL834342 | O | Homo sapiens mRNA; cDNA DKFZp761P2314 (from clone DKFZp761P2314) |
| A_32_P66804 | 12.633398 | Down | PTPRN2 | O | Homo sapiens protein tyrosine phosphatase, receptor type, N polypeptide 2 (PTPRN2), transcript variant 1, mRNA |
| A_23_P256470 | 56.132885 | Down | NPY | N | Homo sapiens neuropeptide Y (NPY), mRNA |
| A_32_P183367 | 17.20502 | Down | BRUNOL4 | O | PREDICTED: Homo sapiens bruno-like 4, RNA binding protein (Drosophila) (BRUNOL4), mRNA |
| A_32_P152195 | 11.834058 | Down | STAC2 | O | Homo sapiens SH3 and cysteine rich domain 2 (STAC2), mRNA |
| A_24_P307964 | 21.591301 | Down | SOHLH1 | O | Homo sapiens spermatogenesis and oogenesis specific basic helix-loop-helix 1 (SOHLH1), mRNA |
| A_32_P323 | 13.151337 | Down | BC037323 | O | Homo sapiens cDNA clone IMAGE: 5261489 |
| A_32_P3476 | 16.78356 | Down | RPRML | O | Homo sapiens reprimo-like (RPRML), mRNA |
| A_24_P393571 | 15.406029 | Down | GDA | O | Homo sapiens guanine deaminase (GDA), mRNA |
| A_23_P392654 | 13.023042 | Down | SPHKAP | O | Homo sapiens SPHK1 (sphingosine kinase type 1) interacting protein (SKIP), mRNA |
| A_24_P479551 | 14.176087 | Down | UBE2QL1 | O | Homo sapiens mRNA, clone: TH049G03 |
| A_24_P944714 | 11.678925 | Down | ENST00000381655 | O | Probable phospholipid-transporting ATPase IB (EC 3.6.3.1) (ATPase class I type 8A member 2) (ML-1) |
| A_32_P197156 | 27.140347 | Down | BI758260 | O | 603029911F1 NIH_MGC_114 Homo sapiens cDNA clone IMAGE:5200131 5′, mRNA sequence [BI758260] |
| A_23_P162010 | 15.891171 | Down | CCKBR | O | Homo sapiens cholecystokinin B receptor (CCKBR), mRNA |
| A_23_P10025 | 11.819316 | Down | NELL2 | O | Homo sapiens NEL-like 2 (chicken) (NELL2), mRNA |
| A_23_P36795 | 14.929889 | Down | SYT1 | N | Homo sapiens synaptotagmin I (SYT1), mRNA |
| A_23_P67569 | 21.238386 | Down | O | Homo sapiens plasticity-related gene 2 (PRG2), mRNA | |
| A_32_P25295 | 28.055656 | Down | NEUROD2 | N | Homo sapiens neurogenic differentiation 2 (NEUROD2), mRNA |
| A_24_P940006 | 11.538975 | Down | EFNB3 | A | Homo sapiens ephrin-B3 (EFNB3), mRNA |
| A_32_P84369 | 12.537511 | Down | FAM153B | O | Homo sapiens hypothetical protein LOC202134 (LOC202134), mRNA |
| A_23_P429601 | 25.979113 | Down | GALNTL5 | O | Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5 (GALNTL5), mRNA |
| A_23_P215283 | 11.123473 | Down | TAC1 | O | Homo sapiens tachykinin, precursor 1 (TAC1), transcript variant beta, mRNA |
| A_23_P50928 | 14.404265 | Down | C1QL2 | O | Homo sapiens complement component 1, q subcomponent-like 2 (C1QL2), mRNA |
| A_23_P60775 | 10.825105 | Down | BRUNOL5 | O | Homo sapiens bruno-like 5, RNA binding protein (Drosophila) (BRUNOL5), mRNA |
| A_23_P357207 | 13.065118 | Down | MRAP2 | O | Homo sapiens chromosome 6 open reading frame 117 (C6orf117), mRNA |
| A_23_P204885 | 11.051615 | Down | PCDH20 | O | Homo sapiens protocadherin 20 (PCDH20), mRNA |
| A_24_P548966 | 15.763181 | Down | RAB3B | O | Ras-related protein Rab-3B [Source:Uniprot/SWISSPROT;Acc:P20337] |
| A_23_P92730 | 19.459343 | Down | HSPB3 | D | Homo sapiens heat shock 27kDa protein 3 (HSPB3), mRNA |
| A_23_P252817 | 20.574722 | Down | SST | O | Homo sapiens somatostatin (SST), mRNA |
| A_23_P428485 | 10.119454 | Down | SPHKAP | O | Homo sapiens SPHK1 (sphingosine kinase type 1) interacting protein (SKIP), mRNA |
| A_32_P45844 | 13.05868 | Down | BX110856 | O | BX110856 Soares adult brain N2b4HB55Y Homo sapiens cDNA clone IMAG-p998M09331, mRNA sequence |
| A_23_P368794 | 22.000626 | Down | TCERG1L | O | Homo sapiens transcription elongation regulator 1-like (TCERG1L), mRNA |
| A_24_P266131 | 15.002172 | Down | FSTL4 | O | Homo sapiens follistatin-like 4 (FSTL4), mRNA |
| A_23_P100022 | 16.488022 | Down | SV2B | O | Homo sapiens synaptic vesicle glycoprotein 2B (SV2B), mRNA |
| A_23_P324706 | 11.529245 | Down | FAM153A | O | Homo sapiens mRNA for KIAA0752 protein, partial cds |
| A_23_P51019 | 13.003195 | Down | SCN2A | O | Homo sapiens sodium channel, voltage-gated, type II, alpha subunit (SCN2A), transcript variant 1, mRNA |
| A_23_P408195 | 28.795895 | Down | TMEM155 | O | Homo sapiens transmembrane protein 155 (TMEM155), mRNA |
| A_24_P38290 | 10.143327 | Down | TAC1 | O | Homo sapiens tachykinin, precursor 1 (TAC1), transcript variant beta, mRNA |
A: angiogenesis, D: death, I: inflammation, N: neuron, O: others.
Discussion
AVMs seem to have unique and relatively homogeneous molecular abnormalities that can be detected at the mRNA and protein levels. Most studies have focused on the abnormal expression of vascular endothelial growth factor and its receptors3,8,12–15) or angiogenesis or cell death-related factors and receptors. Moreover, we reported that the death receptor pathway and the NF-kappaB pathway were upregulated in AVMs.9,11) These results indicate that dynamic vascular remodeling and neuronal death occur in and around the nidus of AVMs.16–20) The majority of these studies, however, have focused on only one or a few genes or protein products. Here, using microarray analysis, we were able to dissect numerous molecular pathways that interact with or counteract each other within the same samples. Our findings were, in general, consistent with previously published findings, especially for genes showing a statistically significant difference between AVMs and controls.3,8,12,15,21)
One previous study reported an increase in IL6 protein levels in AVM tissue. In addition, the GG genotype of the IL6 174G > C promoter polymorphism was associated with the clinical presentation of intracranial hemorrhage in AVMs.8,13) As for MMP9, Hashimoto et al.22) reported that AVM samples had higher levels of total MMP9, active MMP9, pro-MMP9, TIMP1, and TIMP3 than controls. In contrast, TIMP4 levels were higher in the control brain than in the AVM specimens. In addition, MMP9 was reported to be localized to the endothelial cell/peri-endothelial cell layer and infiltrating neutrophils of AVMs. Regarding IL1, we found that IL1R2 was elevated in our AVM samples. Fontanella et al.16) suggested that functional polymorphisms within the IL1 complex gene are associated with AVMs and influence the clinical characteristics of the disease, supporting a role for proinflammatory cytokines in disease etiopathogenesis.23) IL1β promoter polymorphisms were reported to be associated with AVM susceptibility and an increased risk of intracranial hemorrhage in the AVM clinical course.16,23) These results suggest that the inflammatory pathways, including the IL1β cytokine, play an important role in intracranial hemorrhage. In previous studies, elevated IL6 was strongly associated with IL8 and MMP12, which were both elevated at the gene level in this study.8,13) We and others have reported brain infiltration of various types of inflammatory cells in and around the nidus of AVMs.10,24) We identified several chemokine genes to be elevated in AVMs; chemokines may be released by these infiltrating cells.10,24) Previously we also showed reduced neuronal density around the nidus,11) which may be related to our observed alterations in neuron-related genes. Our gene microarray data may help us to establish further hypotheses for testing. For example, microarray data showed inflammation-related genes including IL-8 and IL-6. These observations may lead us to anti-inflammatory treatment against AVMs. To establish this hypothesis, further study is necessary to confirm that inflammation increase the risk of AVM rupture. In addition, MMP family including MMP9, MMP12, and MMP3 changed. This observation may lead us to perform other further analysis using MMP inhibitors.
In this study, we analyze gene expression focusing on the neuron-, death-, angiogenesis-, and inflammation-related genes. Because we and others indicated the role of these pathways in cerebral AVMs.5,9–14,16,21,23,24) Decreased expression of neuron-related genes indicate the loss of neurons in and around the nidus. Increased expressions of death-related genes indicate cellular death of neurons, infiltrating and vascular cells. In addition, increased expression of angiogenesis and inflammation-related genes may show upregulation of these events.
We also analyzed associations between gene expression and the clinical presentation or treatment of AVMs (the presence or absence of hemorrhage, deep-draining veins, embolization, and high-flow), focusing on the neuron-, death-, angiogenesis-, and inflammation-related genes. A deep-draining system may cause venous congestion, which can lead to neuronal loss. However, our data did not indicate neuronal loss because FGF9, which can induce angiogenesis, was upregulated. We focused on inflammation-related genes in relation to preoperative embolization, and demonstrated downregulation of several genes in embolized samples. This may indicate that these changes are not related to preoperative embolization but instead to the operative process itself (two of these samples had intraoperative hemorrhage). In the high-flow samples, neuron-related genes, including neuropeptide Y (NPY), synaptotagmin 1 (SYT1), neurogenic differentiation 2 (NeuroD), and ephrin B3 (EFNB3) were downregulated. This may indicate that neurons and neuronal networks were injured in high-flow AVMs, and may correlate with our previous finding of neuronal loss in the perinidal area.11)
One of the limitations of this study, and of most previous studies, is the small sample size, which can lead to false-negative or false-positive results. Clinical samples may show significant variation in the levels of a specific gene or its product, which may reflect different stages and severity of the disease or simply interindividual variation. One more limitation of the study, during surgical process gene expression may be affected with local ischemia, inflammation, mechanical compression and coagulation. Microarray analysis on a large number of clinical specimens with a well-characterized clinical background is necessary to validate our findings. In addition, it should be noted that the correlation between gene expression and that of its protein product is extremely variable. Transcription efficiency, post-transcriptional modification, and protein metabolism can all independently affect gene and protein levels.
In conclusion, we examined gene expression in AVMs by microarray analysis. Using our data, we are able to generate and test new hypotheses to explore AVM pathophysiology. Microarray analysis is a useful technique to study clinical specimens from patients with brain vascular malformations.
References
- 1). Buis DR, Van Den Berg R, Lagerwaard FJ, Vandertop WP: Brain arteriovenous malformations: from diagnosis to treatment. J Neurosurg Sci 55: 39– 56, 2011. [PubMed] [Google Scholar]
- 2). Cockroft KM, Jayaraman MV, Amin-Hanjani S, Derdeyn CP, McDougall CG, Wilson JA: A perfect storm: how a randomized trial of unruptured brain arteriovenous malformations' (ARUBA's) trial design challenges notions of external validity. Stroke 43: 1979– 1981, 2012. [DOI] [PubMed] [Google Scholar]
- 3). Friedlander RM: Clinical practice. Arteriovenous malformations of the brain. N Engl J Med 356: 2704– 2712, 2007. [DOI] [PubMed] [Google Scholar]
- 4). van Beijnum J, van der Worp HB, Buis DR, Al-Shahi Salman R, Kappelle LJ, Rinkel GJ, van der Sprenkel JW, Vandertop WP, Algra A, Klijn CJ: Treatment of brain arteriovenous malformations: a systematic review and meta-analysis. JAMA 306: 2011– 2019, 2011. [DOI] [PubMed] [Google Scholar]
- 5). Ozduman K, Ozkan A, Yildirim O, Pamir MN, Gunel M, Kilic T: Temporal expression of angiogenesis-related genes in developing neonatal rodent retina: a novel in vivo model to study cerebral vascular development. Neurosurgery 66: 538– 543; discussion 543, 2010. [DOI] [PubMed] [Google Scholar]
- 6). Sasahara A, Kasuya H, Akagawa H, Ujiie H, Kubo O, Sasaki T, Onda H, Sakamoto Y, Krischek B, Hori T, Inoue I: Increased expression of ephrin A1 in brain arteriovenous malformation: DNA microarray analysis. Neurosurg Rev 30: 299– 305; discussion 305, 2007. [DOI] [PubMed] [Google Scholar]
- 7). Takagi Y, Kikuta K, Moriwaki T, Aoki T, Nozaki K, Hashimoto N, Miyamoto S: Expression of thioredoxin-1 and hypoxia inducible factor-1α in cerebral arteriovenous malformations: possible role of redox regulatory factor in neoangiogenic property. Surg Neurol Int 2: 61, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8). Yao JS, Zhai W, Fan Y, Lawton MT, Barbaro NM, Young WL, Yang GY: Interleukin-6 upregulates expression of KDR and stimulates proliferation of human cerebrovascular smooth muscle cells. J Cereb Blood Flow Metab 27: 510– 520, 2007. [DOI] [PubMed] [Google Scholar]
- 9). Aziz MM, Takagi Y, Hashimoto N, Miyamoto S: Activation of nuclear factor κB in cerebral arteriovenous malformations. Neurosurgery 67: 1669– 79; discussion 1679–80, 2010. [DOI] [PubMed] [Google Scholar]
- 10). Aziz MM, Takagi Y, Hashimoto N, Miyamoto S: Expression and activation of STAT family proteins in cerebral arteriovenous malformations. World Neurosurg 78: 487– 497, 2012. [DOI] [PubMed] [Google Scholar]
- 11). Takagi Y, Kikuta K, Nozaki K, Fujimoto M, Hayashi J, Hashimoto N: Neuronal expression of Fas-associated death domain protein and caspase-8 in the perinidal parenchyma of cerebral arteriovenous malformations. J Neurosurg 106: 275– 282, 2007. [DOI] [PubMed] [Google Scholar]
- 12). Chen G, Zheng M, Shu H, Zhan S, Wang H, Zhou D, Zeng S, Tang K, Feng L: Macrophage migration inhibitory factor reduces apoptosis in cerebral arteriovenous malformations. Neurosci Lett 508: 84– 88, 2012. [DOI] [PubMed] [Google Scholar]
- 13). Chen Y, Pawlikowska L, Yao JS, Shen F, Zhai W, Achrol AS, Lawton MT, Kwok PY, Yang GY, Young WL: Interleukin-6 involvement in brain arteriovenous malformations. Ann Neurol 59: 72– 80, 2006. [DOI] [PubMed] [Google Scholar]
- 14). Gao P, Chen Y, Lawton MT, Barbaro NM, Yang GY, Su H, Ling F, Young WL: Evidence of endothelial progenitor cells in the human brain and spinal cord arteriovenous malformations. Neurosurgery 67: 1029– 1035, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15). Takagi Y, Kikuta K, Nozaki K, Hashimoto N: Early regrowth of juvenile cerebral arteriovenous malformations: report of 3 cases and immunohistochemical analysis. World Neurosurg 73: 100– 107, 2010. [DOI] [PubMed] [Google Scholar]
- 16). Fontanella M, Rubino E, Crobeddu E, Gallone S, Gentile S, Garbossa D, Ducati A, Pinessi L, Rainero I: Brain arteriovenous malformations are associated with interleukin-1 cluster gene polymorphisms. Neurosurgery 70: 12– 17, 2012. [DOI] [PubMed] [Google Scholar]
- 17). Hashimoto T, Lawton MT, Wen G, Yang GY, Chaly T, Jr, Stewart CL, Dressman HK, Barbaro NM, Marchuk DA, Young WL: Gene microarray analysis of human brain arteriovenous malformations. Neurosurgery 54: 410– 423; discussion 423–425, 2004. [DOI] [PubMed] [Google Scholar]
- 18). Inoue S, Liu W, Inoue K, Mineharu Y, Takenaka K, Yamakawa H, Abe M, Jafar JJ, Herzig R, Koizumi A: Combination of linkage and association studies for brain arteriovenous malformation. Stroke 38: 1368– 1370, 2007. [DOI] [PubMed] [Google Scholar]
- 19). Oikawa M, Kuniba H, Kondoh T, Kinoshita A, Nagayasu T, Niikawa N, Yoshiura K: Familial brain arteriovenous malformation maps to 5p13-q14, 15q11-q13 or 18p11: linkage analysis with clipped fingernail DNA on high-density SNP array. Eur J Med Genet 53: 244– 249, 2010. [DOI] [PubMed] [Google Scholar]
- 20). Shenkar R, Elliott JP, Diener K, Gault J, Hu LJ, Cohrs RJ, Phang T, Hunter L, Breeze RE, Awad IA: Differential gene expression in human cerebrovascular malformations. Neurosurgery 52: 465– 477; discussion 477–478, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21). Starke RM, Komotar RJ, Hwang BY, Hahn DK, Otten ML, Hickman ZL, Garrett MC, Sisti MB, Lavine SD, Meyers PM, Solomon RA, Connolly ES, Jr: Systemic expression of matrix metalloproteinase-9 in patients with cerebral arteriovenous malformations. Neurosurgery 66: 343– 348; discussion 348, 2010. [DOI] [PubMed] [Google Scholar]
- 22). Hashimoto T, Wen G , Lawton MT , Boudreau NJ , Bollen AW , Yang GY , Barbaro NM , Higashida RT , Dowd CF , Halbach VV , Young WL , University of California, San Francisco BAVM Study Group : Abnormal expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in brain arteriovenous malformations. Stroke 34: 925– 931, 2003. [DOI] [PubMed] [Google Scholar]
- 23). Kim H, Hysi PG, Pawlikowska L, Poon A, Burchard EG, Zaroff JG, Sidney S, Ko NU, Achrol AS, Lawton MT, McCulloch CE, Kwok PY, Young WL: Common variants in interleukin-1-Beta gene are associated with intracranial hemorrhage and susceptibility to brain arteriovenous malformation. Cerebrovasc Dis 27: 176– 182, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24). Chen Y, Zhu W, Bollen AW, Lawton MT, Barbaro NM, Dowd CF, Hashimoto T, Yang GY, Young WL: Evidence of inflammatory cell involvement in brain arteriovenous malformations. Neurosurgery 62: 1340– 1349; discussion 1349–1350, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]