Abstract
Asian soybean rust (ASR) caused by Phakopsora pachyrhizi is a devastating foliar disease affecting soybean production worldwide. Understanding nonhost resistance against ASR may provide an avenue to engineer soybean to confer durable resistance against ASR. We characterized a Medicago truncatula-ASR pathosystem to study molecular mechanisms of nonhost resistance. Although urediniospores formed appressoria and penetrated into epidermal cells of M. truncatula, P. pachyrhizi failed to sporulate. Transcriptomic analysis revealed the induction of phenylpropanoid, flavonoid and isoflavonoid metabolic pathway genes involved in the production of phytoalexin medicarpin in M. truncatula upon infection with P. pachyrhizi. Furthermore, genes involved in chlorophyll catabolism were induced during nonhost resistance. We further characterized one of the chlorophyll catabolism genes, Stay-green (SGR), and demonstrated that the M. truncatula sgr mutant and alfalfa SGR-RNAi lines showed hypersensitive-response-like enhanced cell death upon inoculation with P. pachyrhizi. Consistent with transcriptomic analysis, metabolomic analysis also revealed the accumulation of medicarpin and its intermediate metabolites. In vitro assay showed that medicarpin inhibited urediniospore germination and differentiation. In addition, several triterpenoid saponin glycosides accumulated in M. truncatula upon inoculation with P. pachyrhizi. In summary, using multi-omic approaches, we identified a correlation between phytoalexin production and M. truncatula defense responses against ASR.
Even though plants are immune to almost all pathogens in nature, enormous losses in crop production occur due to specific pathogen infestation in agricultural fields1. Plants have developed many strategies to defend themselves against pathogens. The first line of defense is the recognition of conserved molecules from pathogens, called pathogen-associated molecular patterns (PAMPs), by plant pattern-recognition receptors, resulting in PAMP-triggered immunity (PTI)2. The second line of defense that exists to defend plants from pathogens is effector-triggered immunity (ETI) or host-specific resistance based on the gene-for-gene interaction which is governed by individual plant resistance (R) genes and their corresponding avirulence (Avr) genes3. Breeding for disease resistance in crop plants has mainly relied on introducing R genes by crossing cultivated species with closely related wild species that are resistant to certain diseases. However, the resistant varieties produced by such an approach are not always durable. The pathogen can evolve to overcome the resistance by mutations in the Avr protein4. For example, wheat varieties that were resistant for over 50 years to wheat stem rust caused by Puccinia graminis f. sp. tritici now have compromised resistance against a new race of stem rust, Ug99, after it was first reported in Uganda in 19995. The Ug99 strain of P. graminis has become a potential threat to worldwide wheat cultivation6. In addition to wheat stem rust, other important diseases such as late blight of potato, banana black sigatoka disease, Asian soybean rust (ASR), rice blast and cassava streak virus are a threat to global food security1. Therefore, for the stability of worldwide food supply, there is a need for durable and sustainable resistance against important diseases7,8.
Pathogen outbreaks in important crop plants could have a significant negative impact on human life9. For example, the outbreak of ASR, caused by Phakopsora pachyrhizi, impacted the economy of South America in recent years10. It has been reported that the losses caused by ASR were ~2 billion US dollars in Brazil alone in 200310. Unlike wheat rust fungus that has high host specificity, the host range of ASR is broad11. The P. pachyrhizi fungus, belonging to Basidiomycetes, forms uredinia five to eight days after infection and then produces urediniospores by asexual reproduction12. Urediniospores can be dispersed by wind and germinate on the host plant to form appressoria. Unlike the cereal rust fungus that penetrates through stomata13, P. pachyrhizi directly penetrates into the epidermal cells of host plants. After penetration, P. pachyrhizi extends the infection hyphae and forms haustoria (feeding structures) in the mesophyll cells. Generally, haustoria formation is observed 24–48 hours after infection14. Six soybean accessions that are resistant to a particular P. pachyrhizi race have been identified, and the R genes (Rpp1–6) that confer resistance have also been identified15,16,17. However, none of the soybean accessions in the world show resistance to all races of P. pachyrhizi18. Therefore, timely application of fungicides is the only means to control ASR19.
Nonhost resistance, contrary to R-gene-mediated, host-specific resistance, is believed to be more robust and universal in plant defense systems20,21,22. In studies based on interactions between the barley powdery mildew pathogen and a nonhost plant, Arabidopsis, PEN (PENETRATION) genes of Arabidopsis involved in nonhost resistance have been identified23,24,25. PEN genes are essential for nonhost resistance, which prevents penetration by certain potentially pathogenic filamentous fungi26,27,28. These Arabidopsis PEN genes are reported to also be essential for nonhost resistance against ASR29,30. PEN-mediated resistance is broadly involved in nonhost pre-invasive defense against fungal pathogens24. Enhanced penetration was observed in the Arabidopsis pen3 mutant, suggesting pre-invasive defense in Arabidopsis as a component of nonhost resistance against ASR30. The Arabidopsis pen3 mutant did not completely compromise nonhost resistance against ASR, due to active apoplastic defense. Genetic studies using Arabidopsis to identify components of apoplastic defense against ASR will be challenging since the studies have to be done in the pen3 mutant background. Furthermore, based on analysis using the double mutants of salicylic acid (SA) or jasmonic acid (JA) signaling pathways in the pen3 mutant background, it has been suggested that both SA and JA signaling pathways are involved in nonhost resistance of Arabidopsis against ASR30. However, molecular mechanisms of nonhost post-invasive defense against fungal pathogens are still unclear.
In this study, we have characterized Medicago truncatula as a model nonhost pathosystem for ASR to investigate the molecular mechanisms of nonhost resistance. Based on transcriptome and metabolome analyses, we show the importance of phytoalexins such as isoflavonoids in M. truncatula to confer nonhost resistance against ASR. In addition, we demonstrate that the M. truncatula chlorophyll catabolism pathway genes were differently regulated during nonhost resistance to ASR and one of chlorophyll catabolism pathway mutant stay-green (sgr) has enhanced defense response to ASR.
Results
Medicago truncatula exhibits nonhost resistance to ASR
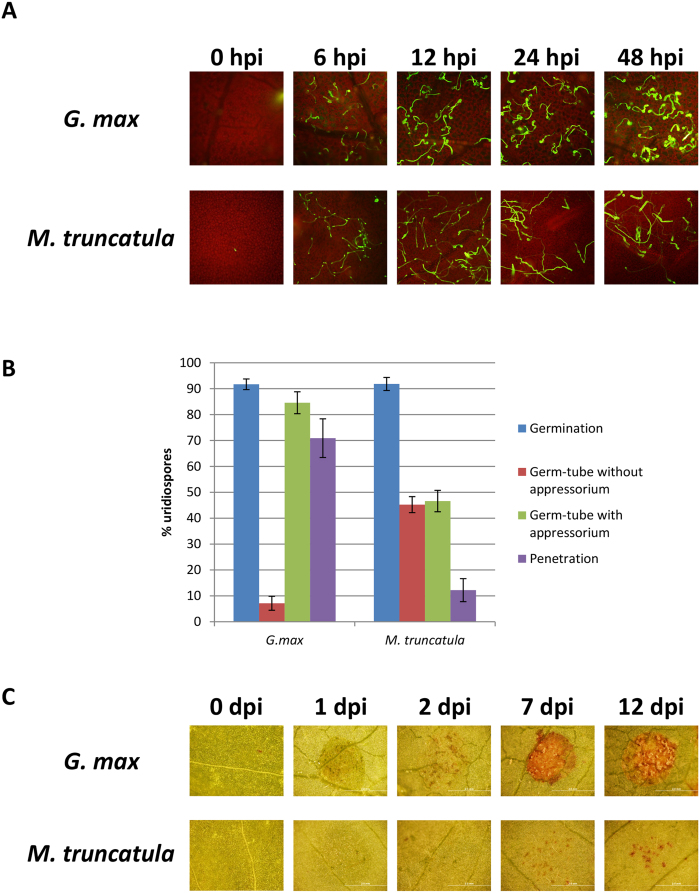
To investigate the molecular mechanisms of nonhost resistance, especially apoplastic defense, against ASR, we decided to use the model legume M. truncatula since P. pachyrhizi can penetrate M. truncatula. Furthermore, identification of genes in M. truncatula that confer nonhost resistance against ASR will provide an avenue to engineer soybean for durable resistance against ASR. To assess the feasibility of using M. truncatula as a model pathosystem to study nonhost resistance, leaves of both soybean (host) and M. truncatula (nonhost) were inoculated with urediniospores of ASR. In soybean, around 90% of urediniospores germinated, and 70% of them penetrated the epidermal tissue within 72 hours post inoculation (hpi). Meanwhile, although around 90% of urediniospores also germinated in M. truncatula, ~15% of them penetrated the epidermis (Fig. 1A,B), suggesting a weaker pre-invasive nonhost defense in M. truncatula against ASR when compared to Arabidopsis. In soybean, uredinia formation was observed within seven days post inoculation (dpi), and they produced urediniospores (Fig. 1C). However, M. truncatula did not produce urediniospores but produced localized cell death at the penetration site, suggesting an active apoplastic nonhost defense against ASR (Fig. 1C). Together, these results suggest that M. truncatula is a suitable model to study nonhost resistance against ASR.
Figure 1.
Disease and resistance phenotypes of soybean (Glycine max) and M. truncatula leaves inoculated with P. pachyrhizi. (A) Epifluorescence micrographs of P. pachyrhizi on abaxial leaf surfaces of soybean (G. max cv. Williams) and M. truncatula wild-type R108 at 6, 12, 24 and 48 hours post-inoculation (hpi). The 4-week-old soybean and M. truncatula plants were spray-inoculated with 1 × 105 spores/ml (0.001% Tween 20) of P. pachyrhizi using an artist air-brush. For fluorescence microscopy, fungal mycelia in the inoculated leaves were stained with 10 μg/mL WGA-Alexa Fluor 488 for 20 min at room temperature. After washing with PBS, the leaves were observed using an epifluorescence microscope. (B) Quantification of germination and pre-infection structures of P. pachyrhizi on abaxial leaf surfaces of soybean (G. max cv. Williams) and M. truncatula wild-type R108 at 72 hpi. Four-week-old soybean and M. truncatula plants were spot-inoculated with 10 μl of 1 × 104 spores/ml (0.001% Tween 20) of P. pachyrhizi. The formation of pre-infection structures was counted from 20 random fields on three independent leaves. The number of dead autofluorescing epidermal cells resulting from direct penetration of P. pachyrhizi were counted from 20 random fields per each inoculated site and are used to calculate the percentage of penetration. The experiments were repeated at least 3 times. (C) Disease symptom development of soybean (G. max cv. Williams) and M. truncatula abaxial leaf surfaces inoculated with P. pachyrhizi at 1, 2, 7 and 12 days post-inoculation (dpi). Four-week old soybean and M. truncatula plants were spot-inoculated on the adaxial side of the leaf with 10 μl of 1 × 104 spores/ml (0.001% Tween 20) of P. pachyrhizi. The inoculated leaves were maintained in a dew chamber for 24 hrs with 100% humidity maintained at 19 °C; 0-hrs-light/24-hrs-dark cycle. The leaves were then transferred to a growth chamber (22 °C/19 °C with 12 hrs-light/12 hrs-dark cycle) and incubated further to allow symptom development. Photographs were taken at time points indicated.
Transcriptome analysis identifies a correlation between phytoalexin production and M. truncatula defense responses against ASR
To further investigate the molecular mechanism of nonhost resistance of M. truncatula against ASR, we performed integrated transcriptome and metabolome analyses using samples derived from M. truncatula leaves inoculated with P. pachyrhizi. To identify host signaling pathways triggered by P. pachyrhizi infection, we carried out microarray analysis to monitor the expression profiles associated with nonhost resistance, including pre-invasive (12 hpi) and apoplastic defense (24 hpi), using Affymetrix GeneChip® Medicago Genome Array (Affymetrix). Using a Bonferroni corrected P value threshold of 1.1406e-006 and a threefold ratio cutoff, 2,704 and 3,386 genes were identified as induced or repressed, respectively, at 12 hpi when compared with the mock controls (Supplementary Figs S1 and S2; Supplementary Tables S1 and S2). Furthermore, at 24 hpi, 2,809 and 3,504 genes were identified as induced or repressed, respectively (Supplementary Figs S1 and S2; Supplementary Tables S3 and S4). ASR-regulated M. truncatula genes were classified into different functional groups using the new BIN structure developed by Goffard and Weiller31.
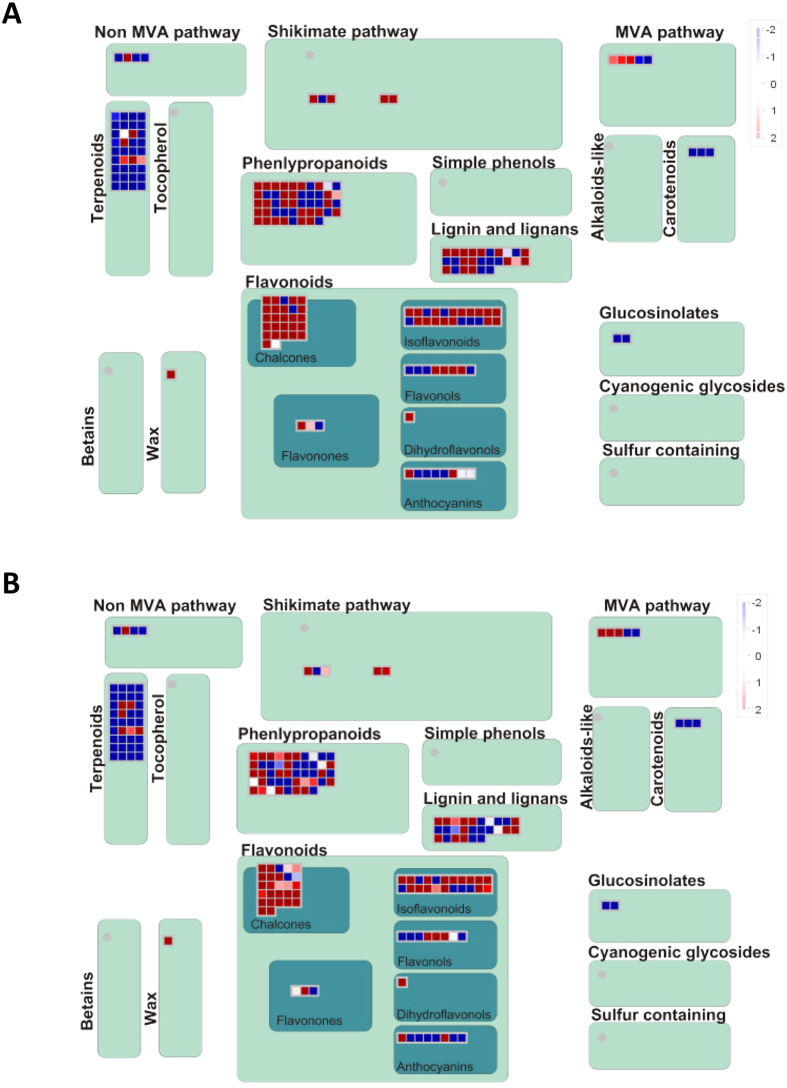
MAPMAN software was used to obtain an overview of genes belonging to various metabolic pathways in the nonhost resistance of M. truncatula to ASR (Fig. 2; Supplementary Figs S3 and S4). Using automated and manual annotation, genes represented in the M. truncatula microarray were classified into different functional groups and assigned BIN numbers. MAPMAN analysis revealed that there are no significant differences in primary metabolism pathways among the genes differentially regulated between 12 h and 24 h after inoculation with ASR (Fig. 2; Supplementary Figs S1, S2, S3 and S4). Analysis of gene expression profiles between P. pachyrhizi-infected and mock-inoculated M. truncatula leaves revealed that 149 and 160 genes (around 5%) belonging to secondary metabolism were induced at 12 or 24 hpi, respectively. (Supplementary Figs S1, S2 and S3). In addition, the genes involved in photosynthesis, including light reactions and photorespiration, were significantly suppressed during nonhost interactions (Supplementary Figs S1, S2 and S3).
Figure 2.
MAPMAN illustration of M. truncatula Affymetrix data showing the genes involved in secondary metabolism during M. truncatula-P. pachyrhizi interactions at 12 (A) and 24 (B) hours post-inoculation (hpi). Four-week old M. truncatula plants were spray-inoculated with 1 × 105 spores/ml (0.001% Tween 20) of P. pachyrhizi; total RNA was isolated from leaves inoculated with P. pachyrhizi at 12 and 24 hpi, or mock treatment. Three biological replicates were used for each inoculation. Each biological replicate consisted of a pool of eight plants. Red indicates upregulation, and blue indicates downregulation.
Several genes related to phytohormones including salicylic acid (SA), jasmonic acid (JA), ethylene (ET), auxin, cytokinin and gibberellin were differently regulated during nonhost interactions (Supplementary Fig. S4; Supplementary Tables S5, S6, S7 and S8). Several SA-regulated Pathogenesis-related (PR) genes were also induced during nonhost resistance (Supplementary Tables S5 and S7). Further, several genes involved in ethylene biosynthesis were induced during nonhost interactions (Supplementary Tables S5 and S7). These genes included 1-Aminocyclopropane-1-carboxylate synthase (ACC synthase) and 1-Aminocyclopropane-1-carboxylate oxidase (ACC oxidase). P. pachyrhizi also induced the expression of genes encoding ET-responsive transcription factors (AP2/ERF family members) (Supplementary Fig. S4; Supplementary Tables S6 and S7). In addition, several genes involved in JA biosynthesis were induced during nonhost interactions. These genes included Lipoxygenases (LOX), Allene oxide synthase (AOS) and 12-Oxophytodienoic acid 10,11-reductase (OPR) (Supplementary Fig. S1). However, the expression of several other members of JA-related genes including LOX, Allene oxide cyclase (AOC) and OPR were downregulated during nonhost resistance (Supplementary Fig. S2; Supplementary Tables S6 and S8). These results indicate a correlation between up-regulation of SA-, ET- and JA-mediated signaling pathways and nonhost resistance of M. truncatula against ASR.
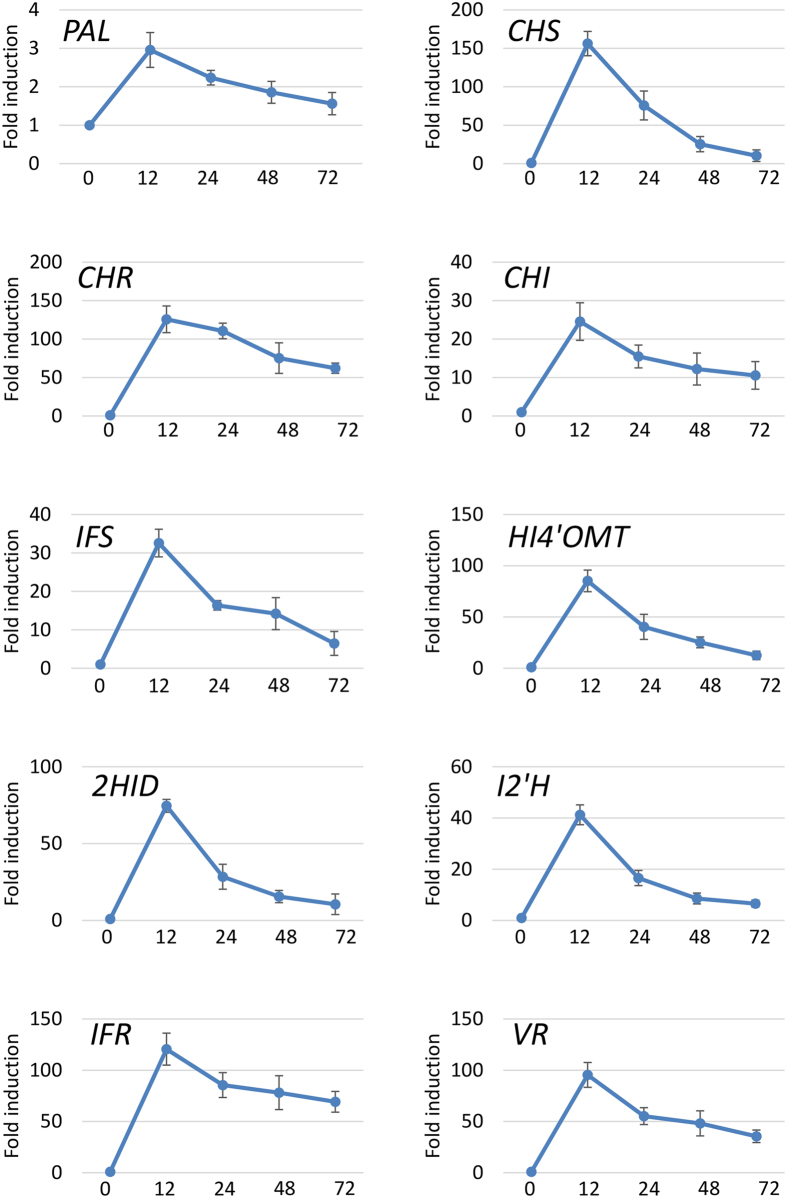
As shown in Fig. 2 and Supplementary Figs S1 and S2, microarray analysis revealed a significant induction of transcripts derived from genes encoding proteins associated with phenylpropanoid, flavonoid and isoflavonoid metabolic pathways in M. truncatula leaves at 12 and 24 hpi with P. pachyrhizi (Fig. 2). Interestingly, the genes involved in the phytoalexin production pathway were induced and the genes involved in the terpenoid production pathway were suppressed in response to P. pachyrhizi inoculation (Fig. 2). Furthermore, real-time quantitative RT-PCR (RT-qPCR) analysis confirmed the induction of genes involved in the phytoalexin medicarpin metabolic pathway, including Phenylalanine ammonia-lyase (PAL), Chalcone synthase (CHS), Chalcone reductase (CHR), Chalcone isomerase (CHI), Isoflavone synthase (IFS), Isoflavone 4′-O-methyltransferase (HI4′OMT), 2-Hydroxyisoflavanone dehydratase (2HID), Isoflavone 2′-hydroxylase (I2′H), Isoflavone reductase (IFR) and Vestitone reductase (VR) (Fig. 3). The expression profiles of these genes in response to P. pachyrhizi inoculation were almost identical. The transcripts of these genes were significantly induced within 12 hpi and slightly decreased until 72 hpi with P. pachyrhizi. These results indicate a correlation between phytoalexin medicarpin pathway and nonhost resistance against ASR.
Figure 3.
Expression profiles of genes involved in phenylpropanoid and flavonoid pathways in M. truncatula inoculated with P. pachyrhizi. M. truncatula leaves were inoculated with P. pachyrhizi at 12, 24, 48 and 72 hours post-inoculation (hpi) or mock control (0 time). Expression profiles are presented for genes encoding Phenylalanine ammonia-lyase (PAL), Chalcone synthase (CHS), Chalcone reductase (CHR), Chalcone isomerase (CHI), Isoflavone synthase (IFS), Isoflavone 4′-O-methyltransferase (HI4′OMT), 2-Hydroxyisoflavanone dehydratase (2HID), Isoflavone 2′- hydroxylase (I2′H), Isoflavone reductase (IFR) and Vestitone reductase (VR). The expression of genes was evaluated by RT-qPCR with gene-specific primer sets (Supplementary Table S10). The values represent the fold induction compared to mock control. Three biological replicates were used for each inoculation. The Ubiquitin gene was used as an internal control. Bars represent the means ± standard deviation (SD).
The chlorophyll catabolism pathway is upregulated by P. pachyrhizi infection
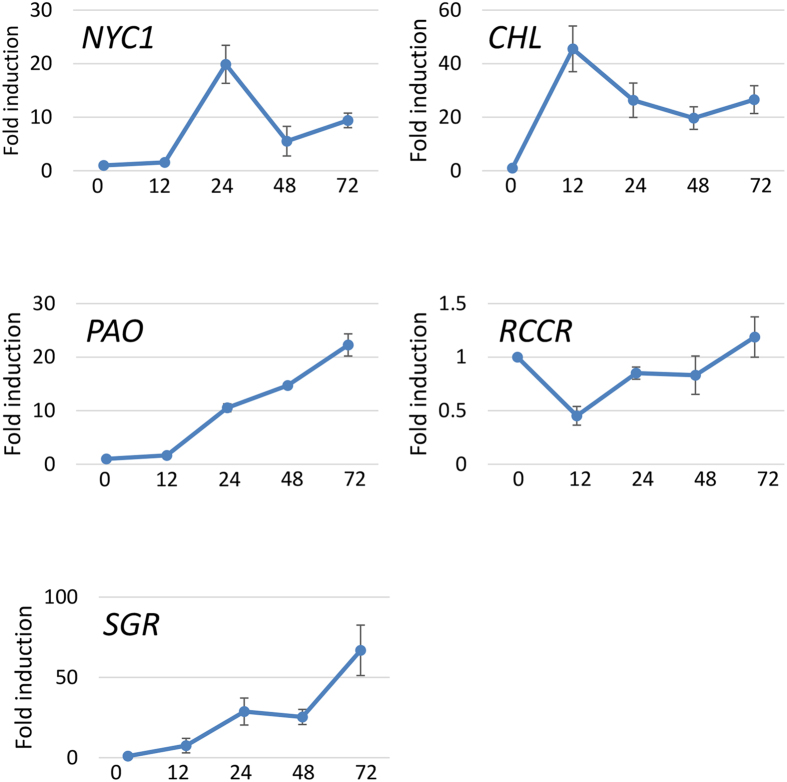
Transcriptome analysis revealed downregulation of a large number of genes involved in photosynthesis in response to P. pachyrhizi inoculation (Supplementary Figs S2 and S3). At the same time, many genes with exception of Red chlorophyll catabolite reductase (RCCR) that encode enzymes involved in the chlorophyll catabolism pathway, including Chlorophyll b reductase (NYC1), Chlorophyllase (CHL), Pheophorbide a oxygenase (PAO) and Stay-green (SGR), were upregulated in response to P. pachyrhizi inoculation (Supplementary Table. S9). Previous studies have demonstrated an important link between light and disease resistance, based on the mutants involved in the chlorophyll degradation pathway in Arabidopsis32,33. To further investigate the molecular mechanism of the chlorophyll catabolism pathway in nonhost resistance of M. truncatula against ASR, expression analysis of NYC1, CHL, PAO, RCCR and SGR was performed by RT-qPCR. Consistent with the microarray results (Supplementary Table S9), the expression of NYC1, CHL, PAO and SGR was significantly induced by ASR (Fig. 4). Interestingly, the expression of RCCR was suppressed 12 hours after inoculation with ASR (Fig. 4; Supplementary Table S9). Among these genes, NYC1, CHL, PAO and RCCR are known to encode enzymes involved in chlorophyll degradation34. Although SGR is believed to regulate the enzymes involved in chlorophyll degradation and involved in disease symptom development35,36,37, the precise function of SGR in plant defense is still unknown. Therefore, we decided to further characterize the role of SGR in nonhost resistance against ASR.
Figure 4.
Expression profiles of genes involved in the chlorophyll catabolism pathway in M. truncatula inoculated with P. pachyrhizi. M. truncatula leaves were inoculated with P. pachyrhizi at 12, 24, 48 and 72 hours post-inoculation (hpi) or mock control (0 time). Expression profiles are presented for genes encoding Chlorophyll b reductase (NYC1), Chlorophyllase (CHL), Pheophorbide A oxygenase (PAO), Red chlorophyll catabolite reductase (RCCR) and Stay-green (SGR). The expression of genes was evaluated by RT-qPCR with gene-specific primer sets (Supplementary Table S10). The values represent the fold induction compared to mock control. Three biological replicates were used for each inoculation. The Ubiquitin gene was used as an internal control. Bars represent the means ± standard deviation (SD).
M. truncatula sgr mutant shows enhanced defense responses to ASR
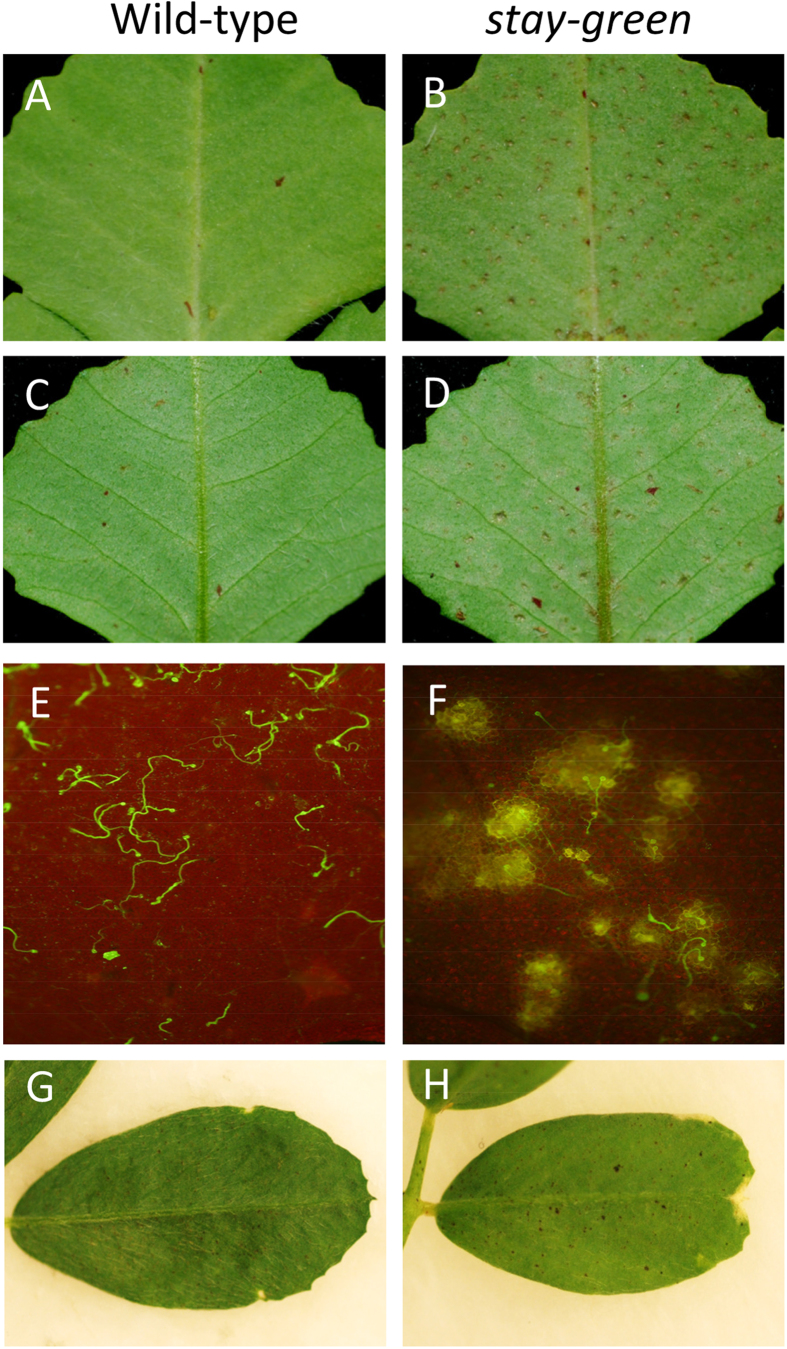
To investigate the functional role of SGR in nonhost resistance of M. truncatula against ASR, we inoculated the previously characterized sgr mutant38 and its corresponding wild type (R108) with P. pachyrhizi and observed for symptom development. Previous study has demonstrated that dark-induced senescence was delayed in the M. truncatula sgr mutant38. As shown in Fig. 5 and as expected, no visible symptoms were observed on both adaxial (Fig. 5A) and abaxial (Fig. 5C) leaf surfaces of wild-type R108 at 48 hpi with P. pachyrhizi (Fig. 5E). Wild-type M. truncatula produces HR-like cell death at the penetration site only seven days after inoculation (Fig. 1C). Interestingly, necrotic symptoms that look like hypersensitive response (HR) cell death were observed in the sgr mutant 48 hpi with P. pachyrhizi (Fig. 5B,D). Microscopic observation also showed autofluorescence around the P. pachyrhizi penetration site in the sgr mutant (Fig. 5F).
Figure 5.
Response of M. truncatula wild-type, stay-green (sgr) mutant, alfalfa wild-type and alfalfa SGR-RNAi line to P. pachyrhizi. (A–D) Response of M. truncatula wild-type R108 (A,C) and sgr mutant (B,D) to P. pachyrhizi at 48 hours post-inoculation (hpi) on adaxial (A,B) or abaxial (C,D) leaf surfaces. Four-week old M. truncatula wild-type R108 and sgr mutant were spray-inoculated with 1 × 105 spores/ml (0.001% Tween 20) of P. pachyrhizi using an artist air-brush. (E,F) Epifluorescence micrographs of WGA-Alexa Fluor 488-stained pre-infection structures of P. pachyrhizi on abaxial leaf surfaces of M. truncatula wild-type R108 (E) or sgr mutant (F) at 48 hpi. Fungal mycelia in the inoculated leaves were stained with 10 μg/mL WGA-Alexa Fluor 488 for 20 min at room temperature. After washing with PBS, the leaves were observed using an epifluorescence microscope. (G,H) Response of alfalfa wild-type Regan SY4D (G) or alfalfa SGR-RNAi line (H) to P. pachyrhizi at 48 hours post-inoculation (hpi) on abaxial leaf surfaces.
To further confirm the HR cell death phenotype of sgr, we utilized another nonhost plant, alfalfa (Medicago sativa) which is a close relative of M. tuncatula. RNAi lines of alfalfa that downregulates SGR has been previously developed and characterized for the sgr phenotype38. Wild-type alfalfa (Regan SY4D) and SGR-RNAi lines were inoculated with P. pachyrhizi. As expected, no visible symptoms were observed on abaxial leaf surfaces of wild-type alfalfa at 48 hpi with P. pachyrhizi (Fig. 5G). Consistent with the phenotype of M. truncatula sgr mutant, necrotic symptoms were observed in the SGR-RNAi lines 48 hpi with P. pachyrhizi (Fig. 5H). Together, these results indicate that sgr mutant and SGR-RNAi lines have enhanced defense responses against ASR infection.
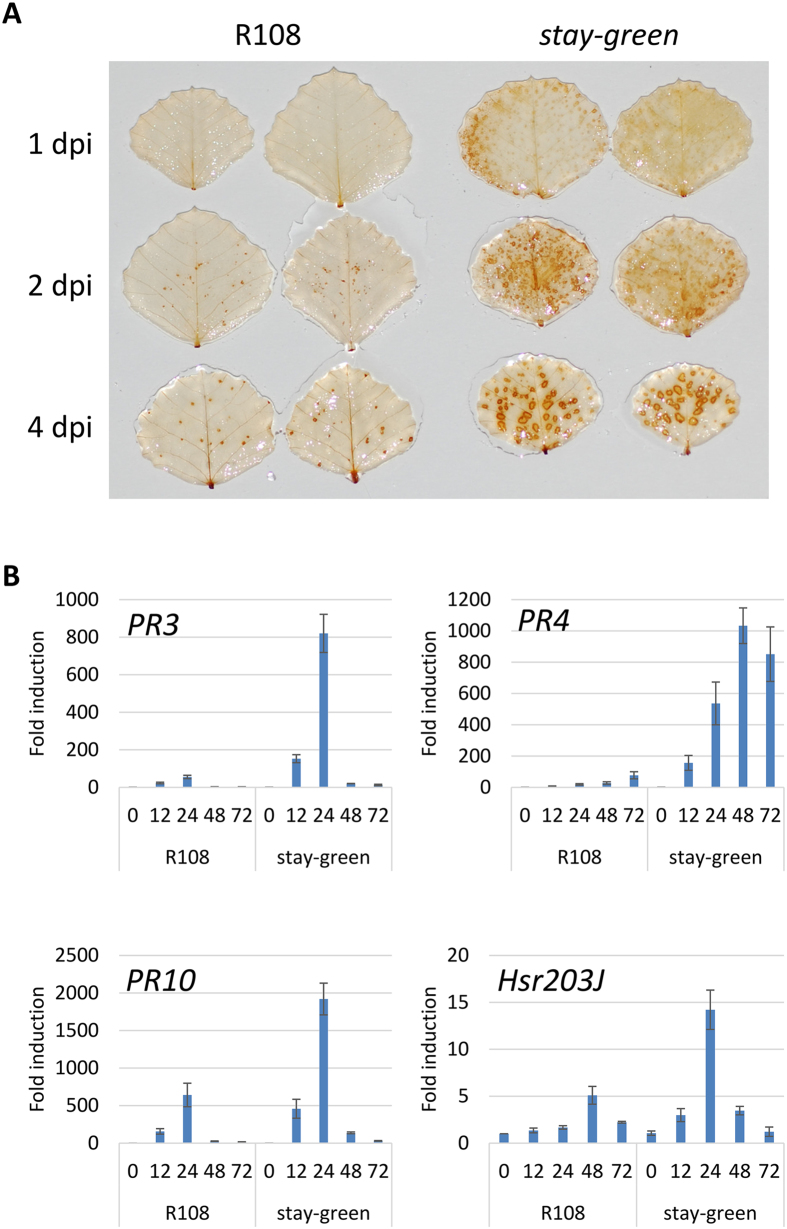
To further study the defense responses of the sgr mutant against P. pachyrhizi, we checked the accumulation of reactive oxygen species (ROS) and the expression of defense-related genes in the sgr mutant and its wild-type R108. To visualize the accumulation of ROS, especially hydrogen peroxide, we stained the inoculated leaves with DAB (3,3′-Diaminobenzidine). As shown in Fig. 6A, a slight accumulation of hydrogen peroxide was observed as brown-colored spots around the penetration sites at 2 dpi in wild type (Fig. 6A). Strikingly, a significantly greater accumulation of hydrogen peroxide was observed within 1 dpi in the sgr mutant when compared to wild type, and it gradually increased up to 4 dpi suggesting that the sgr mutant accumulates more ROS compared to wild type in response to P. pachyrhizi inoculation (Fig. 6A). Although the expression of PR genes, including PR3, PR4 and PR10, was induced in response to P. pachyrhizi in both wild type and the sgr mutant, the induction levels of these genes were significantly higher in the sgr mutant when compared to the wild type (Fig. 6B). The expression of M. truncatula Hsr203J, an orthologous gene for tobacco Hsr203J whose expression is rapidly induced in HR cell death, was highly (~14-fold) induced 24 hrs after inoculation with P. pachyrhizi in the sgr mutant (Fig. 6B). In contrast, the Hsr203J induction was delayed in the wild type by 24 hrs when compared to the sgr mutant and was induced only ~five-fold after inoculation with P. pachyrhizi (Fig. 6B). Together, these results suggest that the sgr mutant has enhanced defense responses against ASR.
Figure 6.
The production of reactive oxygen species and gene expression profiles of M. truncatula wild-type R108 and the stay-green (sgr) mutant to P. pachyrhizi. (A) 3,3-diaminobenzidine (DAB) staining for detection of hydrogen peroxide. Four-week-old M. truncatula wild-type R108 and sgr mutant were spray-inoculated with 1 × 105 spores/ml (0.001% Tween 20) of P. pachyrhizi, then incubated with DAB for 1, 2 and 4 days post-inoculation (dpi). (B) Expression profiles of genes involved in defense responses in M. truncatula inoculated with P. pachyrhizi. M. truncatula leaves spray-inoculated with P. pachyrhizi were sampled at 12, 24, 48 and 72 hours post-inoculation (hpi) or mock control (0 time). Expression profiles are presented for genes encoding Pathogenesis-related protein 3 (PR3), Pathogenesis-related protein 4 (PR4), pathogenesis-related protein 10 (PR10) and Hsr203J. The expression of genes was evaluated by RT-qPCR with gene-specific primer sets (Supplementary Table S10). The values represent the fold induction compared to mock control. Three biological replicates were used for each inoculation. The Ubiquitin gene was used as an internal control. Bars represent the means ± standard deviation (SD).
SGR plays an important role for chlorosis symptom development
Previous studies have shown that Arabidopsis STAYGREEN has an important role for disease symptom development, especially chlorosis, for both bacterial and fungal pathogens including Pseudomonas syringae pv. tomato DC3000 and Alternaria brassicicola34. To investigate the molecular basis of STAYGREEN in disease development in legumes, we inoculated M. truncatula sgr mutant and alfalfa SGR-RNAi lines38 with an anthracnose pathogen, Colletotrichum trifolii. Consistent with the previous report in Arabidopsis34, chlorosis symptom development in M. truncatula sgr mutant and alfalfa SGR-RNAi lines after inoculation with C. trifolii was delayed compared with wild type (Supplementary Fig. S5). These results indicate that M. truncatula SGR also has a role for disease development, especially chlorosis symptoms.
Metabolome analysis identifies a correlation between phytoalexins and M. truncatula defense responses against ASR
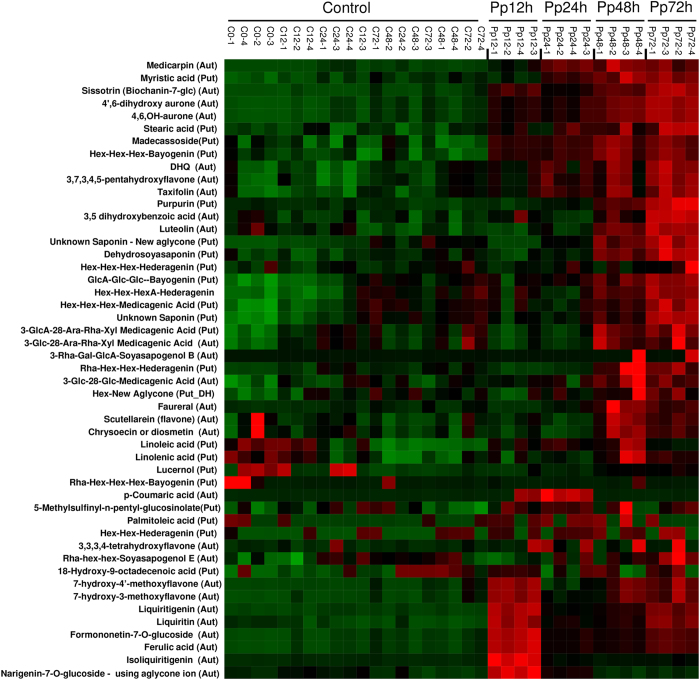
Transcriptome analysis revealed a significant induction of genes involved in phytoalexin medicarpin pathways in M. truncatula at 12 and 24 hpi with P. pachyrhizi (Figs 2 and 3; Supplementary Fig. S3). To investigate whether M. truncatula accumulated the phytoalexin medicarpin and intermediate metabolites such as isoflavones and flavones during nonhost resistance response against ASR, metabolome analysis was carried out using total metabolites derived from P. pachyrhizi-inoculated M. truncatula leaves at 12, 24, 48 and 72 hpi, and mock-inoculated controls. Consistent with the transcriptome analysis, a significant accumulation of intermediate metabolites such as isoflavones and flavones, including narigenin-7-O-glucoside, formononetin-7-O-glucoside, liquiritin, liquiritigenin, 7-hydroxy-3-methoxyflavone and 7-hydroxy-4′-methoxyflavone was observed 12 hpi with P. pachyrhizi when compared to the control (Fig. 7). Interestingly, their induction levels decreased 12 hpi, whereas the accumulation of medicarpin was observed at 24, 48 and 72 hpi (Fig. 7). Taken together with the transcriptome data, these results suggest a correlation between isoflavonoid metabolic pathways including the medicarpin pathway and nonhost resistance against ASR.
Figure 7.
Heat map illustration of metabolites during M. truncatula-P. pachyrhizi interactions at 12, 24, 48 and 72 hours post-inoculation (hpi). Four-week-old soybean and M. truncatula plants were spray-inoculated with 1 × 105 spores/ml (0.001% Tween 20) of P. pachyrhizi. Total metabolites derived from P. pachyrhizi-inoculated leaves at 12, 24, 48 and 72 hpi, and water-treated controls were isolated in 80% methanol. The metabolites were quantified by UPLC system fitted with a hybrid quadrupole time-of-flight (QTOF) Premier mass spectrometer. Four biological replicates were used for each inoculation. Red indicates upregulation, and green indicates downregulation. Put and Aut mean compound confirmation as putative and authentic, respectively.
In addition to phytoalexins, metabolome analysis identified a significant accumulation of triterpenoid saponins, such as madecassoside, Hex-Hex-Hex-bayogenin, dehydrosoyasaponin, Hex-Hex-Hex-hederagenin, GlcA-Glc-Glc-bayogenin, Hex-Hex-HexA-hederagenin, Hex-Hex-Hex-medicagenic acid, 3-GlcA-28-Ara-Rha-Xyl medicagenic acid, 3-Glc-28-Ara-Rha-Xyl medicagenic acid, 3-Rha-Gal-GlcA-soyasapogenol B, Rha-Hex-Hex-hederagenin and 3-Glc-28-Glc-medicagenic acid, mainly at 72 hpi with P. pachyrhizi (Fig. 7). We understand very little about the role of saponins in nonhost resistance.
M. truncatula phytoalexin medicarpin and its intermediate formononetin-7-O-glucoside suppress the pre-infection structure formation of P. pachyrhizi
Transcriptome and metabolome analyses revealed a significant induction of phytoalexin medicarpin pathway during nonhost resistance of M. truncatula against P. pachyrhizi (Figs 2,3 and 7; Supplementary Fig. S3). To further confirm the importance for phytoalexin during nonhost resistance, we studied germination and differentiation of P. pachyrhizi urediniospores on a hydrophobic surface in the presence of phytoalexin medicarpin or its intermediate compound formononetin-7-O-glucoside (ononin). Interestingly, the germination of urediniospores was slightly inhibited in the presence of either medicarpin or ononin when compared to mock control (Fig. 8). In addition, the formation of pre-penetration structures including germ-tubes and appressoria were significantly suppressed by treatment with either medicarpin or ononin (Fig. 8). Inhibition was more prominent at higher concentration. These results clearly indicate that phytoalexin medicarpin plays an important role in nonhost resistance of M. truncatula against P. pachyrhizi.
Figure 8.
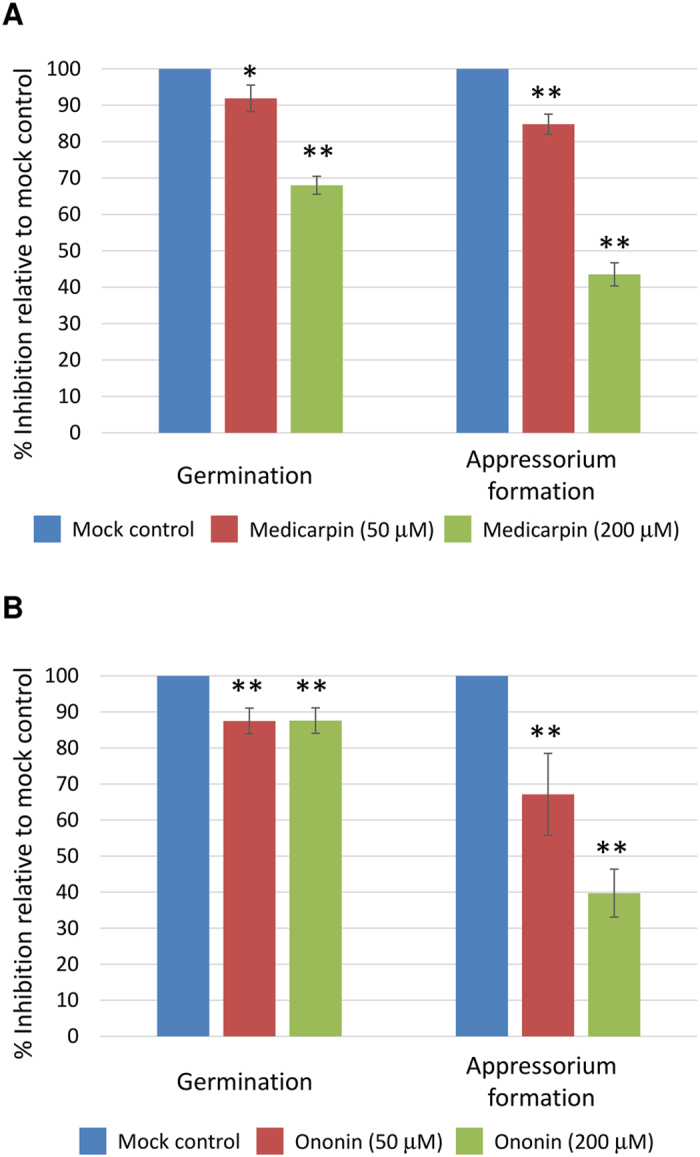
Effect of phytoalexin medicarpin and its intermediate formononetin-7-O-glucoside (ononin) on differentiation of urediniospores of P. pachyrhizi. P. pachyrhizi urediniospores were incubated with DMSO (mock control) or medicarpin or ononin to observe pre-infection structure formation. The percentage of germination and appressorium formation were counted 18 hours after treatment. Bars represent the means ± standard deviation (SD) of eight replications from two independent experiments for each data point. Asterisks indicate significant difference evaluated using paired Student’s t test (*P < 0.05, **P < 0.01).
Discussion
In this study, we characterized a model pathosystem between M. truncatula and ASR to investigate the molecular mechanism of nonhost resistance (Fig. 1). We identified the key genes or pathways for nonhost disease resistance of M. truncatula against ASR and demonstrated that a large number of genes involved in phenylpropanoid and (iso) flavonoid biosynthetic pathways that produce phytoalexin medicarpin were upregulated after inoculation with P. pachyrhizi (Figs 2 and 3; Supplementary Fig. S3). Furthermore, a significant accumulation of the metabolite medicarpin was detected 72 hours after inoculation with P. pachyrhizi (Fig. 7). Consistent with the results from transcriptome and metabolome analyses during nonhost resistance, phytoalexin medicarpin significantly inhibited the pre-infection structure formation of P. pachyrhizi (Fig. 8). These results clearly suggest a correlation between phenylpropanoid and (iso) flavonoid pathways leading to medicarpin production and nonhost resistance in M. truncatula against fungal pathogens such as P. pachyrhizi. Interestingly, similar results were observed in soybean transcriptome analysis for Rpp2-mediated disease resistance against ASR wherein the expression of phenylpropanoid pathway genes, such as PAL, was induced during infection39. It was reported that PAL1-silenced soybean plants compromised Rpp2-mediated disease resistance against ASR40. In addition, the expression of genes involved in the phenylpropanoid pathway was also reported to be induced in Rpp3-mediated disease resistance in soybean41. It is known that the phenylpropanoid pathway has an important role in providing precursors not only for the production of antimicrobial metabolites, such as phytoalexins, but also for lignin biosynthesis42. Together with our present study, these results suggest that both R-gene-mediated resistance and nonhost resistance utilize the phenylpropanoid pathway to produce antimicrobial metabolites.
Metabolome analysis identified a significant accumulation of flavones and isoflavones, precursors for the production of antimicrobial metabolites, after inoculation with P. pachyrhizi (Fig. 7). The accumulation of these metabolites correlated with the expression profiles of genes involved in phenylpropanoid and flavonoid pathways (Figs 2 and 3). We also detected a significant accumulation of triterpenoid saponins 72 hpi with P. pachyrhizi (Fig. 7). However, unlike phenylpropanoid and flavonoid pathways, no significant induction of genes involved in triterpenoid saponins biosynthesis pathways was observed in transcriptome analysis (Fig. 2). This is probably due to the early time points (12 and 24 hpi) used for gene expression analyses. It is known that triterpenoid saponins function as important antimicrobial compounds and have beneficial effects for human health43. However, the biosynthetic pathway of saponins is still poorly understood44. Recent studies have shown that despite the accumulation of medicagenic acid, a major component in M. truncatula saponins in leaves, the transcripts of CYP716A12 that encode the key enzyme for saponin biosynthesis were highly expressed in roots45. Furthermore, M. truncatula UGT73F3, which encodes glycosyltransferases in triterpene saponin biosynthesis, was also mainly expressed in roots44. It has been demonstrated that mutants corresponding to these genes showed severe growth inhibition indicating that triterpene saponins may have an important role in plant developmental processes. In addition, Papadopoulou et al.46 have shown that saponin-deficient (sad) mutants of oat species Avena strigose were compromised in their resistance to a variety of fungal pathogens. Triterpene 12,13β-epoxy-16β-hydroxy-β-amyrin is also shown to be an important antifungal compound47 (Geisler et al., 2013). Medicagenic acid saponins are shown to have antifungal activity to Sclerotium rolfsii, Rhizoctonia solani, Trichoderma viride, Aspergillus niger and Fusarium oxysporum48. Antifungal activity of saponin glycosides, including hederagenin 3-O-β-D-glucopyranoside and medicagenic acid 3-O-β-D-glucopyranoside, is demonstrated49. Thus, it is likely that triterpene saponins have dual functions involved in defense responses and in plant developmental processes. Further precise investigation of the expression profiles is needed to understand the function of triterpene saponins in nonhost resistance of M. truncatula.
In recent years, the importance of chloroplasts in plant immunity has been reported50. There are numerous reports showing that pathogens target host chloroplast to cause disease51,52,53. Furthermore, there is an important link between light and disease resistance. The intermediate metabolites derived from chlorophyll degradation are known to generate ROS which function as signaling molecules to cause cell death, that play a role in disease resistance32,54,55. Consistent with these reports, the present study demonstrated that the expression of genes involved in chlorophyll catabolism, including NYC1, CHL, PAO and SGR, was significantly induced by inoculation with P. pachyrhizi (Fig. 4; Supplementary Table S9). On the other hand, the expression of genes related to photosynthetic pathways was suppressed during P. pachyrhizi infection (Supplementary Figs S2 and S3). These results suggest that optimization of chlorophyll catabolism could be essential for protecting plants against the toxicity of ROS, especially when the photosynthetic activity is suppressed by stresses such as pathogen infection. Interestingly, similar results were observed in a previous study of Arabidopsis SGR. It was reported that the expression of AtSGR was induced after inoculation with an avirulent bacterial pathogen, Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) carrying AvrRpm155, suggesting that SGR may play a role in plant defense. Furthermore, transcriptome analysis for soybean demonstrated that the expression of genes involved in chlorophyll catabolism including SGR and CHL was induced in the compatible interactions with P. pachyrhizi41. However, the induction of SGR and CHL was ~2.5-fold in contrary to ~40-fold induction of SGR and CHL during nonhost resistance in M. truncatula (Fig. 4). Consistent with the fact that pathogen effectors modulate host defense machinery, we speculate that P. pachyrhizi utilizes effectors to interfere with the expression of chlorophyll catabolism pathway genes, especially SGR, to suppress plant defense.
A M. truncatula Tnt1 insertion collection56,57 was previously utilized to identify a sgr mutant line38. We used the M. truncatula sgr mutant to characterize the function of SGR in plant defense response. A previous study of the Arabidopsis no chlorosis1 (noc1/sgr1) mutant with hemibiotrophic bacterial pathogen Pst DC3000 and necrotrophic fungal pathogen Alternaria brassicicola has shown that SGR plays a role in chlorosis symptom development34. It was also reported that the expression of AtSGR1 was induced during Pst DC3000 or A. brassicicola infection34. Interestingly, our study demonstrated that the necrotic cell death associated with high levels of ROS accumulation was observed in the M. truncatula sgr mutant inoculated with P. pachyrhizi suggesting a negative role of SGR in HR-like cell death (Fig. 6). In contrast, earlier studies reported a positive role for SGR in cell death phenomenon. Overexpression of AtSGR in Arabidopsis caused spontaneous, HR-like necrotic cell death, and AtSGR RNAi lines showed a delay in HR upon inoculation with an avirulent pathogen55. A negative role of MtSGR in HR-like cell death is intriguing. In addition, it has been reported that SGR is essential for recruiting the enzymes involved in chlorophyll catabolism during senescence by binding to light-harvesting complex II37. Therefore, it is likely that SGR may function as a key regulator, not only for leaf senescence and disease symptom development, but also for plant immunity through feedback regulation in certain plant species. P. pachyrhizi could utilize its effectors to interfere with a plant’s chlorophyll catabolism pathway for pathogenicity by inducing SGR to increase chlorosis in infected tissue. In addition, the induction of SGR in M. truncatula inhibits cell death related to HR, and this helps with the virulence of the pathogen. However, the suppression of HR alone by SGR is perhaps not sufficient for P. pachyrhizi to cause disease in M. truncatula (Fig. 1). The exact role of SGR in HR-like cell death needs further investigation using overexpression lines for SGR.
Methods
Plant growth conditions and pathogen inoculation assay
Seeds of M. truncatula ecotype R108 and sgr mutant (NF2089) were scarified for 8 min using concentrated sulfuric acid, washed thrice with distilled water and germinated on moist filter papers. Two days after germination in darkness at 24 °C, 12 seedlings were transferred to soil (one seedling per cell in 6 × 12 celled trays). Following three weeks’ incubation in the greenhouse, the plants were transferred to growth chambers located in a USDA-APHIS approved BSL2 + facility to conduct ASR inoculation assays.
Pathogen inoculation and cytological studies were conducted as described previously58. An isolate of the ASR pathogen P. pachyrhizi, from Illinois, was maintained on the susceptible soybean cultivar (Glycine max cv. Williams) in a growth chamber at 22 °C/19 °C with 12-hrs-light/12-hrs-dark cycle (100–150 μmol m–2 s–1). Fresh urediniospores were collected using a gelatin capsule spore collector designed by the CDL, St. Paul, MN, and suspended in distilled water with 0.001% Tween 20. The 4-week-old M. truncatula plants were spray-inoculated with 1 × 105 spores/ml (0.001% Tween 20) using an artist air-brush (Paasche Airbrush Co. Chicago, IL, USA) set at 2 PSI with a portable air-pump (Gast Mfd Co. Benton Harbor, MI, USA) for uniform spore deposition. The inoculated plants were maintained in a dew chamber for 24 hrs with 100% humidity maintained at 19 °C; 0-hrs-light/24-hrs-dark cycle. The plants were then transferred to a growth chamber (22 °C/19 °C with 12 hrs-light/12 hrs-dark cycle) and incubated further to allow symptom development.
To quantify the formation of pre-penetration structures, approximately 100 spores of P. pachyrhizi in 10-μL aliquots were placed on the abaxial surface of the detached leaves of 4-week-old soybean (Glycine max cv. Williams) or M. truncatula wild-type R108 and incubated in dark overnight and then transferred to a growth chamber (22 °C/19 °C with 12-h-light/12-h-dark cycle). At 24 hours after inoculation, the leaves were stained with 10 μg/mL WGA-Alexa Fluor 488 (WGA-Alexa Fluor® 488; Invitrogen, Corp., Carlsbad, CA, USA) supplemented with 0.05% Tween 20 to visualize the fungal germ-tubes and appressoria. The subsequent developments were followed 72 hours after inoculation, and the germ-tubes forming differentiated appressoria were counted as appressoria. The differentiated germ-tubes without appressoria that grew on the surface were also counted from 20 random fields on three independent leaves. The number of dead autofluorescing epidermal cells resulting from direct penetration of P. pachyrhizi were counted 72 hours after inoculation from 20 random fields per each inoculated site and are used to calculate the percentage of penetration.
Alfalfa anthracnose pathogen, C. trifolii race 1 was maintained on potato dextrose agar media. Two-weeks-old cultures of conidia were harvested, washed in water, and re-suspended in sterile distilled water. Leaves from 4-week-old wild-type R108 and sgr mutant were harvested and spot- inoculated with 10 μL of suspension 1 × 106 spores/mL in 0.005% Tween 20. The disease symptom development was recorded 4 days after inoculation.
Light microscopy
Initial interactions of P. pachyrhizi with M. truncatula were recorded by direct observation of inoculated leaves using an Olympus stereo (SZX19, Olympus Corporation Co. Ltd., Tokyo, Japan) or compound microscopes (BX 41, Olympus Corporation Co. Ltd., Tokyo, Japan) equipped with fluorescence attachment. For fluorescence microscopy, fungal mycelia were stained with wheat germ agglutinin (WGA) coupled to green fluorescent dye Alexa Fluor 488 as described previously59. Inoculated leaves were stained with 10 μg/mL WGA-Alexa Fluor 488 by a brief vacuum infiltration in phosphate-buffered saline (PBS) followed by a 20 min incubation at room temperature. For microscopic observations, after washing with PBS, whole or sections of the leaf were placed on a glass slide and mounted using a cover glass with Dow Corning® (Midland, MI, USA) high vacuum grease for microscopy. Fluorescence microscopy to document the infection process was done using Olympus epifluorescence microscope.
Detection of hydrogen peroxide
The generation of hydrogen peroxide was detected using 3,3′-diaminobenzidine (DAB) staining as described previously53. Inoculated leaves were placed in 1 mg/ml DAB-HCl (pH 3.8). After incubation for 6 hours at room temperature, chlorophyll was removed with 95% ethanol.
Transcriptome analysis
The inoculated leaves were gently ground by mortar and pestle with liquid nitrogen. The powdered samples were split into two aliquots for transcriptome and metabolome analyses. Total RNA was purified from inoculated leaves using TRIzol reagent (Invitrogen). For microarray analysis, the total RNA was purified using the RNAeasy MinElute Cleanup Kit (Qiagen, Valencia, CA, USA), and 500 ng of total RNA was used for hybridization with the Affymetrix Medicago GeneChip. Three biological replicates were used for each inoculation. Each biological replicate consisted of a pool of eight plants. Experimental and statistical procedures were as described previously59. Briefly, data normalization between chips was conducted using RMA (Robust Multi-chip Average60). Presence/absence call for each probe set was obtained using dCHIP61. Gene selections based on associative t-test62 (Dozmorv and Centola, 2003) were made using Matlab (MathWorks, Natick, MA, USA). A selection threshold of 3 for transcript ratios (where applicable) and a Bonferroni correction P value threshold of 1.1406e-006 were used. Differentially expressed genes between mock and ASR inoculation were selected, and functional annotation of genes was performed as described previously58. Microarray data are available in the ArrayExpress database ( www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-2911.
Real-time quantitative RT-PCR analyses
Real-time quantitative RT-PCR (RT-qPCR) was performed as described59 using the gene-specific primers designed based on the target sequence (Supplemental Table S10). Total RNA was treated with Turbo DNase (Ambion) to eliminate genomic DNA, and 5 μg of DNase-treated RNA was reverse-transcribed using Superscript III reverse transcriptase (Invitrogen) with oligo d(T)20 primers. The cDNA (1:10) was then used for RT-qPCR. The RT-qPCR was performed using gene-specific primer sets (Supplementary Table S10) and Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) in an optical 384-well plate with an ABI Prism 7900 HT sequence detection system (Applied Biosystems). Melt-curve analysis was performed to monitor primer-dimer formation and to check amplification of gene-specific products. The average threshold cycle (CT) values calculated from triplicate biological samples were used to determine the fold expression relative to the controls. Primers specific for Ubiquitin were used to normalize small differences in template amounts.
Metabolome analysis
The total metabolites were extracted from 10 mg of dried samples in 80% methanol containing 18 ng/μl of umbelliferone (Sigma-Aldrich, St. Louis, MO, USA) as an internal standard. Samples were agitated for 2 hours at room temperature followed by centrifugation at 2900 × g for 30 minutes. The supernatant was transferred to a 2 mL autosampler vial. Five microliters of the solution were injected on to a Waters Acquity UPLC system fitted with a hybrid quadrupole time-of-flight (QTOF) Premier mass spectrometer (MS; Waters). A reverse-phase, 1.7-μm UPLC BEH C18, 2.1 × 150 mm column (Waters) was used for separations. The mobile phase consisted of eluent A (0.05% [v/v] formic acid/water) and eluent B (acetonitrile), and separations were achieved using a linear gradient of 95 to 30% A over 30 min, 30 to 5% A over 3 min, and 5 to 95% A over 3 min. The flow rate was 0.56 mL/min, and the column temperature was maintained at 60 °C. Masses of the eluted compounds were detected in the negative ESI mode from 50 to 2,000 mass-to-charge ratio. The QTOF Premier mass spectrometer was operated under the following instrument parameters: desolvation temperature of 375 °C; desolvation nitrogen gas flow of 850 liters/h; capillary voltage of 2.9 kV; cone voltage of 48 eV; and collision energy of 10 eV. The MS system was calibrated using sodium formate, and raffinose was used as the lockmass. Metabolites were identified based on accurate masses and retention times relative to authentic standards. Data Bridge (Mass Lynx version 4.1) was used to convert the raw data files to NetCDF. Relative abundances were calculated using MET-IDEA, and the peak areas were normalized by dividing each peak area by the value of the internal standard peak area.
In vitro urediniospore germination and differentiation assay
Two hundred microliters of P. pachyrhizi urediniospores (1 × 106 spores/mL) were mixed with DMSO (mock control) or 5 mM medicarpin or 5 mM ononin at a final concentration of 50 μM or 200 μM of medicarpin or ononin. These urediniospores were incubated in 6-well cell plastic culture plates (BD Falcon) for 18 h in dark. After incubation, the number of P. pachyrhizi spores that germinated and germ tubes with and without differentiated appressoria were counted from eight random fields. Average of eight observations was used to calculate the percentage of spore germination and germ tubes with and without an appressorium. These experiments were repeated more than two times.
Additional Information
How to cite this article: Ishiga, Y. et al. Transcriptomic and metabolomic analyses identify a role for chlorophyll catabolism and phytoalexin during Medicago nonhost resistance against Asian soybean rust. Sci. Rep. 5, 13061; doi: 10.1038/srep13061 (2015).
Supplementary Material
Acknowledgments
We thank Dr. Zeng-Yu Wang (Noble Foundation) for seeds of M. truncatula sgr mutant and alfalfa SGR-RNAi lines, Dr. Kazuhiro Toyoda for medicarpin, Jackie Kelley for editing the manuscript and Stacy Allen for generating the microarray data. Financial source: This work was supported by The Samuel Roberts Noble Foundation and in part by BASF Plant Sciences, Germany and Program to Disseminate Tenure Tracking System, MEXT, Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions Y.I., S.R.U. and U.G. designed and performed experiments; Y.I., S.R.U. and K.S.M. wrote the manuscript; Y.T. performed the transcriptome analysis; D.H. performed the metabolome analysis.
References
- Pennisi E. Armed and dangerous. Science. 327, 804–805 (2010). [DOI] [PubMed] [Google Scholar]
- Zipfel C. Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol. 20, 10–16 (2008). [DOI] [PubMed] [Google Scholar]
- Ellis J., Dodds P. & Pryor T. Structure, function and evolution of plant disease resistance genes. Curr Opin Plant Biol. 3, 278–284 (2000). [DOI] [PubMed] [Google Scholar]
- Ayliffe M., Singh R. & Lagudah E. Durable resistance to wheat stem rust needed. Curr Opin Plant Biol. 11, 187–192 (2008). [DOI] [PubMed] [Google Scholar]
- Singh R. P. et al. The emergence of Ug99 races of the stem rust fungus is a threat to world wheat production. Annu Rev Phytopathol. 49, 465–481 (2011). [DOI] [PubMed] [Google Scholar]
- Stokstad E. Plant pathology. Deadly wheat fungus threatens world’s breadbaskets. Science. 315, 1786–1787 (2007). [DOI] [PubMed] [Google Scholar]
- Kou Y. & Wang S. Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol. 13, 181–185 (2010). [DOI] [PubMed] [Google Scholar]
- St Clair D. A. Quantitative disease resistance and quantitative resistance Loci in breeding. Annu Rev Phytopathol. 48, 247–268 (2010). [DOI] [PubMed] [Google Scholar]
- Stukenbrock E. H. & McDonald B. A. The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol. 46, 75–100 (2008). [DOI] [PubMed] [Google Scholar]
- Yorinori J. T. et al. Epidemics of soybean rust (Phakopsora pachyrhizi) in Brazil and Paraguay from 2001 to 2003. Plant Disease. 89, 675–677 (2005). [DOI] [PubMed] [Google Scholar]
- Slaminko T. L., Miles M. R., Frederick R. D., Bonde M. R. & Hartman G. L. New legume hosts of Phakopsora pachyrhizi based on greenhouse evaluations. Plant Disease. 92, 767–771 (2008). [DOI] [PubMed] [Google Scholar]
- Goellner K. et al. Phakopsora pachyrhizi, the causal agent of Asian ASR. Mol Plant Pathol. 11, 169–177 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read N. D., Kellock L. J., Collins T. J. & Gundlach A. M. Role of topography sensing for infection-structure differentiation in cereal rust fungi. Planta. 202, 163–170 (1997). [Google Scholar]
- Koch E., Ebrahimnesbat F. & Hoppe H. H. Light and electron-microscopic studies on the development of ASR (Phakopsora pachyrhizi Syd) in susceptible soybean leaves. Phytopathology. 106, 302–320 (1983). [Google Scholar]
- Garcia A. et al. Molecular mapping of ASR (Phakopsora pachyrhizi) resistance genes: discovery of a novel locus and alleles. Theor Appl Genet. 117, 545–553 (2008). [DOI] [PubMed] [Google Scholar]
- Hartwig E. E. & Bromfield K. R. Relationships among three genes conferring specific resistance to rust in soybeans. Crop Science. 23, 237–239 (1983). [Google Scholar]
- Bromfield K. R. & Hartwig E. E. Resistance to ASR and mode of inheritance. Crop Science. 20, 254–255 (1980). [Google Scholar]
- Yamaoka Y. Recent outbreak of rust diseases and the importance of basic biological research for controlling rusts. J Gen Plant Pathol. 80, 375–388 (2014). [Google Scholar]
- Ivancovich A. ASR in Argentina. Plant Disease. 89, 667–668 (2008). [DOI] [PubMed] [Google Scholar]
- Heath M. C. Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol. 3, 315–319 (2000). [DOI] [PubMed] [Google Scholar]
- Lipka U., Fuchs R., Kuhns C., Petutschnig E. & Lipka V. Live and let die—Arabidopsis nonhost resistance to powdery mildews. Eur J Cell Biol. 89, 194–199 (2010). [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M. & Mysore K. S. Nonhost resistance against bacterial pathogens: retrospectives and prospects. Annu Rev Phytopathol. 51, 407–427 (2013). [DOI] [PubMed] [Google Scholar]
- Collins N. C. et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 425, 973–977 (2003). [DOI] [PubMed] [Google Scholar]
- Lipka V. et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 310, 1180–1183 (2005). [DOI] [PubMed] [Google Scholar]
- Stein M. et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 18, 731–746 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egusa M., Miwa T., Kaminaka H., Takano Y. & Kodama M. Nonhost resistance of Arabidopsis thaliana against Alternaria alternata involves both pre- and postinvasive defenses but is collapsed by AAL-toxin in the absence of LOH2. Phytopathology. 103, 733–740 (2013). [DOI] [PubMed] [Google Scholar]
- Nakao M., Nakamura R., Kita K., Inukai R. & Ishikawa A. Non-host resistance to penetration and hyphal growth of Magnaporthe oryzae in Arabidopsis. Sci Rep. 1, 171 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada C. et al. Nonhost resistance in Arabidopsis-Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol Plant Microbe Interact. 19, 270–279 (2006). [DOI] [PubMed] [Google Scholar]
- Langenbach C., Campe R., Schaffrath U., Goellner K. & Conrath U. UDP-glucosyltransferase UGT84A2/BRT1 is required for Arabidopsis nonhost resistance to the Asian ASR pathogen Phakopsora pachyrhizi. New Phytol. 198, 536–545 (2013). [DOI] [PubMed] [Google Scholar]
- Loehrer M., Langenbach C., Goellner K., Conrath U. & Schaffrath U. Characterization of nonhost resistance of Arabidopsis to the Asian soybean rust. Mol Plant Microbe Interact. 21, 1421–1430 (2008). [DOI] [PubMed] [Google Scholar]
- Goffard N. & Weiller G. Extending MapMan: application to legume genome arrays. Bioinformatics. 22, 2958–2959 (2006). [DOI] [PubMed] [Google Scholar]
- Mach J. M., Castillo A. R., Hoogstraten R. & Greenberg J. T. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA. 98, 771–776 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Tanner G., Anders I., Roca M. & Hörtensteiner S. Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci USA. 100, 15259–15264 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. Chlorophyll degradation during senescence. Annu Rev Plant Biol. 57, 55–77 (2006). [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 14, 155–162 (2009). [DOI] [PubMed] [Google Scholar]
- Mecey C. et al. A critical role of STAYGREEN/Mendel’s I locus in controlling disease symptom development during Pseudomonas syringae pv tomato infection of Arabidopsis. Plant Physiol. 157, 1965–1974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y. et al. STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell. 24, 507–518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C. et al. From model to crop: functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol. 157, 1483–1496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel M. et al. Distinct biphasic mRNA changes in response to Asian soybean rust infection. Mol Plant Microbe Interact. 20, 887–899 (2007). [DOI] [PubMed] [Google Scholar]
- Pandey A. K. et al. Functional analysis of the Asian soybean rust resistance pathway mediated by Rpp2. Mol Plant Microbe Interact. 24, 194–206 (2011). [DOI] [PubMed] [Google Scholar]
- Schneider K. T. et al. Biphasic gene expression changes elicited by Phakopsora pachyrhizi in soybean correlate with fungal penetration and haustoria formation. Plant Physiol. 157, 355–371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M. A. et al. Genome-wide analysis of phenylpropanoid defence pathways. Mol Plant Pathol. 11, 829–846 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin J. M., Kuzina V., Andersen S. B. & Bak S. Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry. 72, 435–457 (2011). [DOI] [PubMed] [Google Scholar]
- Naoumkina M. A. et al. Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell. 22, 850–866 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli M. et al. Medicago truncatula CYP716A12 is a multifunctional oxidase involved in the biosynthesis of hemolytic saponins. Plant Cell. 23, 3070–3081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulou K., Melton R. E., Leggett M., Daniels M. J. & Osbourn A. E. Compromised disease resistance in saponin-deficient plants. Proc Natl Acad Sci USA. 96, 12923–12928 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler K. et al. Biochemical analysis of a multifunctional cytochrome P450 (CYP51) enzyme required for synthesis of antimicrobial triterpenes in plants. Proc Natl Acad Sci USA. 110, E3360–3367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehavi U. et al. Synthesis and antifungal activity of medicagenic acid saponins on plant pathogens: modification of the saccharide moiety and the 23 alpha substitution. Carbohydr Res. 244, 161–169 (1993). [DOI] [PubMed] [Google Scholar]
- Saniewska A., Jarecka A., Bialy Z. & Jurzysta M. Antifungal activity of saponins originated from Medicago hybrida against some ornamental plant pathogens. Acta Agrobot. 59, 51–58 (2006). [Google Scholar]
- Nomura H. et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat Commun. 3, 926 (2012). [DOI] [PubMed] [Google Scholar]
- Ciuffetti L. M., Manning V. A., Pandelova I., Betts M. F. & Martinez J. P. Host-selective toxins, Ptr ToxA and Ptr ToxB, as necrotrophic effectors in the Pyrenophora tritici-repentis-wheat interaction. New Phytol. 187, 911–919 (2010). [DOI] [PubMed] [Google Scholar]
- Ishiga Y., Ishiga T., Wangdi T., Mysore K. S. & Uppalapati S. R. NTRC and chloroplast-generated reactive oxygen species regulate Pseudomonas syringae pv. tomato disease development in tomato and Arabidopsis. Mol Plant Microbe Interact. 25, 294–306 (2012). [DOI] [PubMed] [Google Scholar]
- Ishiga Y. et al. The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol. 181, 147–160 (2009). [DOI] [PubMed] [Google Scholar]
- Hirashima M., Tanaka R. & Tanaka A. Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol. 50, 719–729 (2009). [DOI] [PubMed] [Google Scholar]
- Mur L. A. et al. Accumulation of chlorophyll catabolites photosensitizes the hypersensitive response elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 188, 161–174 (2010). [DOI] [PubMed] [Google Scholar]
- Tadege M. et al. Large scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347 (2008). [DOI] [PubMed] [Google Scholar]
- Tadege M., Wang T. L., Wen J., Ratet P. & Mysore K. S. Mutagenesis and Beyond! Tools for understanding legume biology. Plant Physiol. 151, 978–984 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati S. R. et al. Loss of abaxial leaf epicuticular wax in Medicago truncatula irg1/palm1 mutants results in reduced spore differentiation of anthracnose and nonhost rust pathogens. Plant Cell. 24, 353–370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati S. R. et al. Global gene expression profiling during Medicago truncatula-Phymatotrichopsis omnivora interaction reveals a role for jasmonic acid, ethylene, and the flavonoid pathway in disease development. Mol Plant Microbe Interact. 22, 7–17 (2009). [DOI] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B. & Speed T. P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4, 249–264 (2003). [DOI] [PubMed] [Google Scholar]
- Li C. & Wong W. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 98, 31–36 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov I. & Centola M. An associative analysis of gene expression array data. Bioinformatics. 19, 204–211 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.