Abstract
The present study examined SMAD4 expression in fine-needle aspiration cell blocks from patients with breast ductal carcinoma, in order to assess its viability as a prognostic marker. Using immunohistochemistry, the SMAD4 protein status of 86 breast ductal carcinoma fine-needle biopsies, from patients who underwent tumor resection at Beihua University Affiliated Hospital (Jilin, China) between 2002 and 2008, was characterized. The association between SMAD4 expression and clinicopathological parameters, as well as prognosis was assessed using the Mantel-Haenszel method and Cox proportional hazards regression. SMAD4 staining was observed in the cytoplasm and nucleus, and its expression was found to be decreased in ductal breast carcinoma as compared with adjacent normal breast epithelia. Patients with reduced SMAD4 expression levels tended to exhibit more poorly differentiated tumors, a higher risk of recurrence and shorter overall survival. These results demonstrated that the evaluation of SMAD4 protein status in fine-needle biopsy specimens of breast ductal carcinoma may provide additional prognostic information.
Keywords: ductal breast carcinoma, SMAD4, immunochemistry assay
Introduction
Worldwide, breast cancer is the most frequently diagnosed carcinoma amongst females, accounting for approximately one-third of all cancers in women (1). Although the mortality rate associated with breast cancer has decreased over the past 25 years, particularly in more economically developed countries, as a result of progress in earlier detection through mammography and the development of more efficacious adjuvant therapies (2–4), breast cancer currently remains the leading cause of cancer-associated mortality amongst females, worldwide (5). Based on Globocan 2008 (5), breast cancer accounted for 458,000 mortalities, and the age-standardized rate of breast cancer mortality was 13.7/100,000. Invasive ductal carcinoma is the most common subtype of invasive breast cancer, accounting for 80% of all diagnosed cases (6,7). This subtype is characterized by its histomorphological and genetic complexity, as well as its clinical diversity (6), all of which renders accurate histological grading difficult and thus perturbs the selection of appropriate treatment options.
Breast cancer is highly curable if diagnosed at an early stage and treated with the optimal strategies, thus it is clinically significant to have accurate prognostic markers that are able to aid guidance of the clinical management of the disease. Traditional prognostic factors, including axillary lymph node status, tumor size and histological grade, that currently guide the clinical management of breast cancer are not able to accurately predict treatment response. Technological advances over the last decade have provided insight into the molecular mechanisms associated with the development and progression of breast cancer, raising prospects that molecular markers may aid histological classification and the prediction of patient outcomes. A large number of molecular markers, including p53 (8,9) phosphatase and tensin homolog (10), BRCA (11,12), fragile histidine triad (13), human epidermal growth factor receptor-2 (14–16) and Ki-67 (17), have been investigated in breast cancer to determine their ability to predict prognosis; however, these studies have generated conflicting results (18,19). SMAD4, a tumor suppressor gene, has attracted significant attention as a potential molecular marker in the field of cancer research (20).
The SMAD4 gene encodes a protein that is involved in the transmission of chemical signals in the transforming growth factor-β (TGF-β) signal transduction pathway. The TGF-β signaling process is initiated when TGF-β binds to its transmembrane receptor, the TGF-β type II receptor (TβRII) on the cell surface, which recruits and activates TGF-β type I receptor (TβRI). In turn, activated TβRI phosphorylates SMAD family members 2 or 3, which heterodimerize with SMAD4. The SMAD4/SMAD protein complex then translocates from the cytoplasm into the nucleus where it transmits upstream signals by binding to specific DNA sequences to control the activity of particular genes and regulate cell proliferation, differentiation and extracellular matrix production (21). Abrogation of SMAD4 function may result in the breakdown of the TGF-β signaling pathway and loss of transcription of genes critical to cell cycle control (22), and cells may therefore evade TGF-β-mediated growth control and apoptosis (23).
SMAD4 has been found to play a confirmed yet complicated role in the regulation of the initiation, progression and prognostic outcome of breast cancer. Li et al (24) found that SMAD4 induced apoptosis in estrogen receptor-α (ERα)-positive breast cancer cells, and this was confirmed by a subsequent in vitro study which demonstrated that SMAD4 was essential for TGF-β-mediated inhibition of ERα estrogenic transcription activity (25). However, Deckers et al (26) reported that SMAD4/TGF-β-induced growth inhibition and apoptosis only occurred at early stages of breast cancer, and that in advanced disease, TGF-β induced the epithelial to mesenchymal transition (EMT) and metastasis of breast cancer cells to bone, effects which were critically dependent on SMAD4. The dichotomous function of TGF-β in breast cancer progression has been attributed to aberrant expression of SMAD4 or disruption of SMAD4 activity, which has been demonstrated to switch TGF-β from a repressor to an activator of ERα trans-activation (25). Furthermore, it has been reported that SMAD4/TGF-β-induced breast cancer cell invasion occurred via the upregulation of matrix metalloproteinase (MMP)-2 and -9 (27). Although there have been numerous studies investigating the role of SMAD4 in the tumorigenesis and progression of breast cancer (28–31), there is currently very limited information regarding the expression of SMAD4 in human breast cancer tissues and its potential prognostic significance.
In the present study, immunohistochemistry was used to examine the expression of SMAD4 in 86 ductal breast carcinoma tissues in comparison to corresponding adjacent normal tissue from the mammary glands. The expression profile was analyzed for correlations with established prognostic markers, as well as overall survival.
Materials and methods
Study population
Investigations were conducted on 86 patients with ductal breast carcinoma treated at Beihua University Affiliated Hospital (Jilin, China) between 2002 and 2008. The study group comprised 86 patients whose tumor material from fine-needle aspiration (FNA), adjacent normal breast epithelia tissue and clinicopathological data were available at the time the present study was being performed. All patients were surgically treated by mastectomy (partial or total) and axillary lymph node resection. No patient had received radiotherapy or chemotherapy prior to surgery. Sixty-two (72%) of the patients were treated with surgery and post-operative radiotherapy, while the remaining 24 (28%) were treated with surgery only. Indications for the requirement of post-operative adjuvant therapy included large, deeply-invasive tumors, close or positive surgical margins and lymphovascular invasion. All 86 primary ductal breast carcinoma specimens were from female patients. The median age of the group was 54 years (range, 28–79 years) and the median period of follow-up was 267 weeks (range, 112–423 weeks). The distribution of the tumors according to T and N stage classification of the 2010 American Joint Committee on Cancer staging criteria (32) is presented in Table I. Time to recurrence and overall survival were measured from the date of diagnosis.
Table I.
Distribution of 86 breast ductal carcinomas according to T and N stage.
| T stage | ||||||
|---|---|---|---|---|---|---|
| N stage | T1 | T2 | T3 | T4 | Total | |
| N0 | 9 | 16 | 11 | 2 | 38 | |
| N1 | 3 | 5 | 4 | 3 | 15 | |
| N2 | 3 | 13 | 7 | 3 | 26 | |
| N3 | 0 | 5 | 1 | 1 | 7 | |
| Total | 15 | 39 | 23 | 9 | 86 | |
The study was approved by the ethics committee of Beihua University, and informed consent was obtained from each patient once the purpose and nature of the study had been fully explained.
Immunohistochemical assay
The expression of SMAD4 was evaluated by immunohistochemical analysis of 5-µm sections of formalin-fixed, paraffin-embedded tumor tissues and associated normal breast epithelial tissues, which were mounted on superfrost slides (Thermo Fisher Scientific, Pittsburgh, PA, USA). Briefly, antigen retrieval was achieved by heating the slides at 95°C for 15 min in citrate buffer (10 mM sodium citrate buffer; pH 6.0; Shandong Hongshide Chemical Industry Co., Ltd., Linyi, China). Following extensive trials to optimize the protocol, the subsequent steps were selected and performed. Endogenous peroxidase was blocked using 3% hydrogen peroxide (Maixin, Fuzhou, China) in phosphate-buffered saline (PBS; Qingdao Jie Shi Kang Biotechnology Co., Ltd., Qingdao, China) for 30 min at room temperature. To eliminate nonspecific staining, the sections were then incubated with goat serum (Maixin) for 20 min at room temperature. Following this, the sections were incubated with rabbit monoclonal anti-SMAD4 antibody (ab40759; Abcam, Cambridge, UK) at 1:100 dilution at 4°C overnight. Following 3 washes in PBS, biotinylated anti-mouse, anti-rabbit and anti-goat immunoglobulins (Maixin) were applied for 15 min. Processing was then conducted in a humidified chamber at room temperature by the addition of streptavidin-peroxidase (Maixin) for 15 min, followed by 3,3′-diaminobenzidine (Maixin) and hydrogen peroxide for 5 min. The sections were counterstained with Mayer's hematoxylin (Guangzhou Xiuwei Commerce Co., Ltd., Guangzhou, China) Negative controls were obtained by omission of the primary antibody and substitution of the primary antibody with normal serum. The positive control comprised a section of a breast ductal cancer block (ab40759; Abcam) previously demonstrated to be positive for the marker, which was incorporated in each run.
Immunohistochemical results of semi-quantitative analysis
Stained specimens were viewed under a light microscope (CX31; Olympus Corporation, Tokyo, Japan) at objective magnifications of x200 and x400, and evaluated semi-quantitatively by at least 3 of the 4 independent experienced pathologists involved in the present study (Dr Nannan Liu, Dr Chunyan Yu, Dr Yanfen Shi and Dr Yuhe Liu) without prior knowledge of the clinicopathological parameters and clinical outcomes of the patients. The SMAD4 expression of ≥1000 tumor cells was evaluated for each section. The evaluation method was similar to that described by Taubert et al (33), based on the intensity of staining and proportion of stained cells, using normal breast tissue as a reference. The percentage of positive cells was thus scored as follows: 1, 1–10% positive cells; 2, 11–50%; 3, 51–75%; and 4, >75% positive cells. Staining intensity was scored as: 0, absent; 1, weak; 2, moderate; and 3, intense. The immunoreactive score (IRS) was calculated by multiplying the scores for the percentage of positive cells and the expression intensity (34), and were as follows: 0, no staining; 1–4, weak staining; 5–8, moderate staining; and 9–12, strong staining. An IRS of 1–12 (1≤IRS≤12) was considered to indicate an SMAD4-positive result. Results were validated by repeat staining once more on sequential sections from the same block. Variations in classification between pathologists occurred infrequently and were reconciled using a double-headed microscope.
Statistical analysis
Statistical analyses were conducted using SPSS software version 17.0 (SPSS Inc., Chicago, IL, USA). Associations between clinical outcomes (tumor recurrence and disease-specific survival) and SMAD4 expression of the tumors, clinicopathological variables [including clinical stage, pathological node status, tumor grade, age (as a continuous variable) and treatment (surgery plus adjuvant radiotherapy or surgery alone)] were analyzed using the Mantel-Haenszel method (35). All tests were 2-tailed and P<0.05 was considered to indicate a statistically significant difference. Cox proportional hazards regression (36) was then used to examine the independent effects of each significant variable.
Results
Recurrence and survival of patients
Among the 86 patients evaluated there were 22 (25.6%) recurrences, comprising 6 of the 24 patients treated with surgery alone and 16 of the 62 patients who received surgery and adjuvant radiotherapy and/or chemotherapy. Disease recurred in the locoregional area in 7 patients, and systemic recurrences arose in 15 patients. The total 5-year overall survival rate was 80% for the study group.
SMAD4 expression is decreased in breast ductal carcinoma
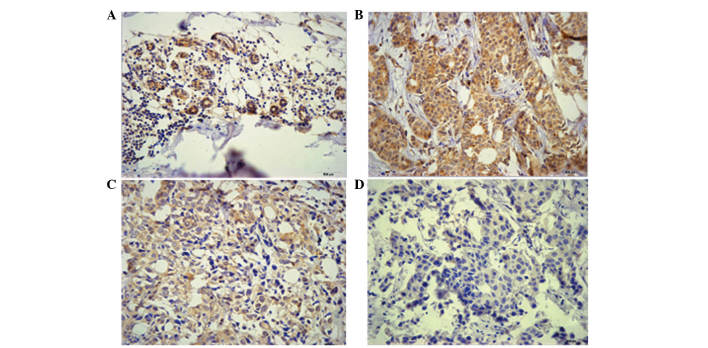
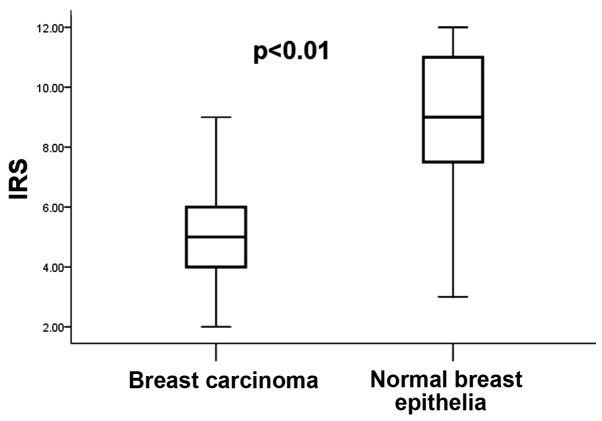
Immunostaining of SMAD4 was observed in the cytoplasm and nucleus of breast carcinoma cells, as well as normal breast epithelia (Fig. 1). As indicated in Fig. 1, the staining intensity varied between the tumor and normal epithelial tissues from various patients, however there was no significant difference in the intracellular localization of SMAD4 in the tumor tissues, compared with that of the normal breast epithelia. According to the IRS criteria, 12 cases (13.9%) of the tumor tissues were SMAD4-negative (IRS=0) and 74 cases (86.1%) stained positive (1≤IRS≤12). Of the SMAD4-positive cases, 37 (43%) were only weakly positive (1≤IRS≤4). By contrast, no negative staining was observed in the adjacent normal tissues, and 41 cases (48%) exhibited strong positive staining (9≤IRS≤12). In general, SMAD4-expression was significantly lower in breast carcinoma tissues (IRS, 4.03±2.12) than that in normal breast epithelia (IRS, 8.40±2.23; P<0.01; Fig. 2).
Figure 1.
Detection of SMAD4 in tumor tissues and surrounding normal breast epithelia by immunohistochemistry. Staining intensity varied between specimens. (A) Normal breast epithelia: Brown, normal breast lobules. Breast carcinoma: (B) Yellow, well differentiated; (C) pallide-flavens, moderately differentiated and (D) poor differentiated, colorless. Magnification, x400.
Figure 2.
Expression of SMAD4 as indicated by IRS in ductal carcinoma compared to the corresponding surrounding normal breast epithelia (n=86). The highest and lowest IRS values are shown (P<0.01, sign-test). IRS, immunoreactive score.
SMAD4 expression is negatively associated with histological tumor grade and poor prognosis
The expression of SMAD4 (negative: IRS=0 vs. positive: 1≤IRS≤12) was negatively associated with histological tumor grade. No significant correlation was detected between SMAD4 expression and other clinicopathological parameters evaluated [age, tumor size, World Health Organization (WHO) stage or nodal status] (Table II).
Table II.
Expression of SMAD4 in association with clinicopathological characteristics.
| SMAD4 expression | |||
|---|---|---|---|
| Variable | Negative | Positive | P-value |
| Age, years | 0.234 | ||
| ≤51 | 8 | 38 | |
| >51 | 4 | 37 | |
| Stage | 0.993 | ||
| I/II | 7 | 43 | |
| III/IV | 5 | 31 | |
| Histological grade | 0.008 | ||
| I | 0 | 12 | |
| II | 5 | 45 | |
| III | 7 | 17 | |
| Tumor size | 0.935 | ||
| ≤2 cm | 3 | 12 | |
| >2 cm | 9 | 62 | |
| Lymph node (n=48) | 0.563 | ||
| ≤10 | 4 | 37 | |
| >10 | 1 | 6 | |
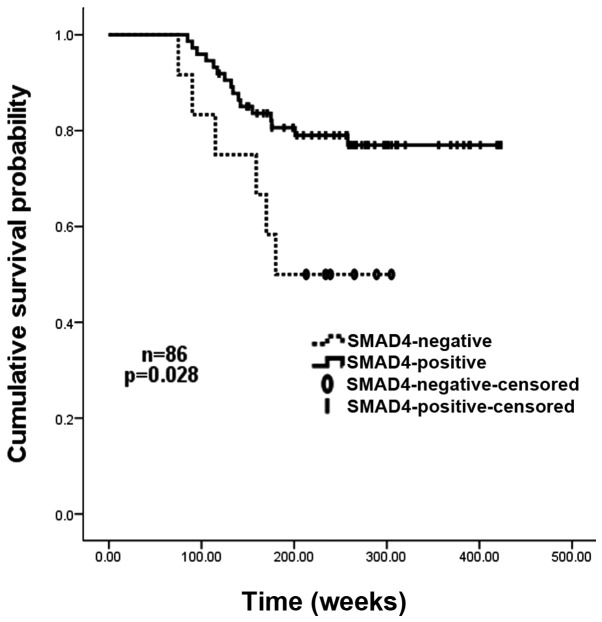
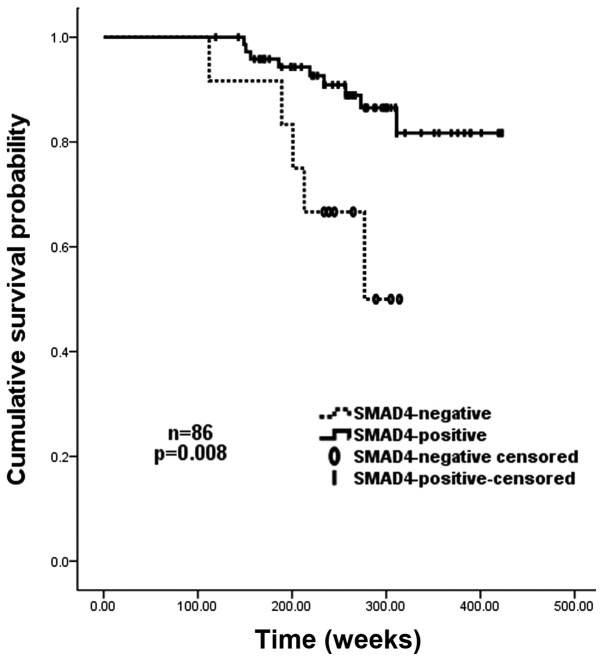
Univariate analyses revealed that negative SMAD4 expression (IRS=0), together with high histological grade (grade III vs. grades I and II), high lymph-node involvement (≥10 vs. <10), and large tumor size (≥5 cm vs. <5 cm) significantly predicted an increased risk of tumor recurrence and poor overall survival. Patients with negative SMAD4 expression had a 2-fold higher risk of recurrence than those with SMAD4-positive carcinomas (P=0.028). The Kaplan-Meier recurrence curve for SMAD4-positive and -negative cases is shown in Fig. 3. In addition, the overall survival of patients without SMAD4 expression was significantly shorter than that of those with tumors expressing SMAD4 (median survival, 212.5 vs. 298.5 weeks; P=0.011). The relative risk of mortality amongst patients with SMAD4-negative cancer was >3 times higher than that of those with SMAD4-positive tumors (P=0.008). The Kaplan-Meier survival curve for SMAD4-positive and negative cases is shown in Fig. 4. In the multivariate Cox model, SMAD4 expression remained a statistically significant marker of recurrence (P<0.05) once age, tumor size, WHO stage and nodal status had been taken into account, whereas histological grade remained significant when all variables except SMAD4 expression were included in the model (P<0.05). Cox regression analysis for survival indicated that the improved survival for those patients with SMAD4-positive cancers was independent of stage, pathological node status, age, and tumor size (P<0.05). Histological grade remained significant when SMAD4 expression was excluded from the model.
Figure 3.
Kaplan-Meier recurrence-free survival curves for 86 patients with breast carcinoma, grouped according to SMAD4 expression. SMAD4-negative patients demonstrated a significantly reduced recurrence-free survival rate compared with that of SMAD4-positive patients (P=0.028).
Figure 4.
Kaplan-Meier cancer-specific survival curves for 86 patients with breast carcinoma, grouped according to their SMAD4 expression status. SMAD4-negative patients had a significantly reduced overall survival rate compared with that of SMAD4-positive patients (P=0.008).
Discussion
In the current study, the expression of SMAD4 was investigated by immunohistochemical analysis, in 86 cases of human breast ductal carcinoma compared with that in the surrounding normal breast epithelia. The present data demonstrated that SMAD4 protein expression was decreased or even lost in tumor tissues, compared with that of normal breast epithelia. A decreasing trend in SMAD4 protein levels was detected from histological grade I to III, suggesting that SMAD4 may participate in the carcinogenesis and progression of breast ductal carcinoma. Notably, it was also demonstrated that among the 86 patients undergoing surgical resection of breast ductal carcinoma, the loss of SMAD4 expression was independently associated with poor prognosis. Patients with SMAD4-negative cancers exhibited a 2-fold greater risk of recurrence and a 3-fold higher risk of mortality than those with SMAD4-positive cancers. SMAD4 expression therefore provides additional prognostic information, even following adjustments for other clinicopathological parameters, including age, tumor size, stage and nodal status.
The present observation of decreased SMAD4 protein expression in breast ductal carcinoma was in agreement with the reports of Stuelten et al (37) and Ren et al (25), which also demonstrated that SMAD4 protein expression was markedly downregulated or lost in breast ductal carcinoma when compared with that in the normal breast epithelium. However, Ren et al (25) further observed that SMAD4 was highly expressed in the cytoplasm and nucleus of benign breast ductal epithelial cells, whereas it was only weakly expressed and largely restricted to the cytoplasm of cancer cells. Such a change in the intracellular localization of SMAD4 was not detected in the present study, which may be a result of sampling differences and the variability of antibody sensitivity. In addition, a small study conducted by Miller et al (38) reported reduced expression of the SMAD4 gene in a series of 26 invasive ductal carcinomas, where the protein expression of SMAD4 was also reduced accordingly. In contrast to the apparent loss of expression of SMAD4 protein associated with ductal breast carcinoma, Xie et al (39) detected SMAD4 protein expression in 98% of 367 cases of primary invasive ductal breast carcinoma, while only 9 cases failed to express SMAD4 protein in the study group using a tissue microarray analysis. Regardless of the discrepancies in the expression of SMAD4 protein amongst these studies, which may be attributed to the antibodies used, the distinct sample groups or the cut-offs used for the evaluation, it was suggested that the reduced expression of SMAD4 may occur in at least a subset of breast ductal carcinoma.
The mechanisms underlying the decrease in SMAD4 protein expression remain to be elucidated. SMAD4 (also known as MADH4/DPC4) homozygous deletion may represent one of the mechanisms of SMAD4 protein instability. This was evidenced by the results of a study by Zhong et al (40) who demonstrated that the frequency of SMAD4 homozygous deletion was ~6% in infiltrative ductal carcinoma, and that these samples were also negative for SMAD4 protein expression by immunohistochemistry. In addition, proteasome-mediated protein degradation may also have a role in SMAD4 protein inactivation. Jakob et al (41) identified a 9 bp in-frame deletion of SMAD4 (amino acids 49–51) in breast cancer cell line HCC1428 that did not affect the SMAD4 message but altered the levels of SMAD4 protein. There are likely additional mechanisms contributing to the loss of SMAD4 protein expression, for example epigenetic changes, and further investigations of the underlying cause are therefore required.
SMAD4 inactivation appears to result in a biologically more aggressive form of breast ductal carcinoma. The present data indicated that SMAD4 protein levels were significantly decreased from grade I to III, suggesting that SMAD4 inactivation is associated with the advanced disease state of breast cancer. Analogously, western blot analysis of breast cancer cells with increasing malignancy revealed that SMAD4 protein expression decreased concurrently with increasing malignancy of the tumor cell line, suggesting that decreased SMAD4 protein expression may accompany tumor progression from the early stages of cancer, as demonstrated in situ and in vivo (37). As the key intracellular mediator of transcriptional responses to TGF-β signaling, which is altered in various tumors (42), SMAD4 consistently transmits the TGF-β signal from the cell membrane to the nucleus. Inactivation of SMAD4 results in insensitivity to TGF-β growth inhibitive signaling (43), and is thus involved in tumorigenesis and progression (44,45). In addition, SMAD4 inactivation within an evolving neoplasm may indirectly influence the extracellular matrix to promote tumor growth. This was confirmed by a study, which found that TGF-β/SMAD induced breast cancer cell invasion via upregulation of MMP2 and 9 by TGF-β in a SMAD4-dependent manner (27).
The observation that SMAD4 expression is correlated with prognosis is also of interest. Despite one report, which demonstrated that loss of SMAD4 expression resulted in a trend for increased survival times in a subgroup of 197 cases of breast ductal carcinoma (N1 or ER-negative, respectively) (37), in the present series of 86 patients it was demonstrated that loss of SMAD4 expression was associated with increased risk of recurrence and shorter survival time. This finding corroborates the results of a study by Stuelten et al (37), which indicated that the survival curve of SMAD4-negative patients was marginally better than that of SMAD4-positive patients in terms of overall 5-year survival, although this difference was not statistically significant. These contradictory results may be due to the differences in sampling and clinical management of the disease.
In conclusion, the present study demonstrated that SMAD4 expression in ductal breast cancer was lower than that in normal adjacent breast epithelial tissue, and that SMAD4 expression levels were negatively correlated with histological grade of the tumor. In addition, decrease or loss of SMAD4 expression was correlated with higher risk of recurrence and shorter overall survival. Since in the majority of cases, the primary diagnosis of breast ductal carcinoma is made by pathological assessment of fine-needle aspiration, such samples of breast cancer are therefore critical in guiding clinical management of the disease.
Acknowledgements
The present study was supported by the initial Foundation for Doctor Research Project of Beihua University (no. 199500032) and the Science and Technology Bureau of Jilin (project no. 201536069).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. 2001;94:153–156. doi: 10.1002/ijc.1440. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Center MM, Desantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 3.Althuis MD, Dozier JM, Anderson WF, Devesa SS, Brinton LA. Global trends in breast cancer incidence and mortality 1973–1997. Int J Epidemiol. 2005;34:405–412. doi: 10.1093/ije/dyh414. [DOI] [PubMed] [Google Scholar]
- 4.Autier P, Boniol M, La Vecchia C, Vatten L, Gavin A, Héry C, Heanue M. Disparities in breast cancer mortality trends between 30 European countries: Retrospective trend analysis of WHO mortality database. BMJ. 2010;341:c3620. doi: 10.1136/bmj.c3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferlay J, Shin HR, Bray F, Forman D, Mathers CD, Parkin D. Estimates of worldwide burden of cancer in 2008: GLOBCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 6.Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol. 2011;223:307–317. doi: 10.1002/path.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakha EA, Putti TC, Abd El-Rehim DM, Paish C, Green AR, Powe DG, Lee AH, Robertson JF, Ellis IO. Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol. 2006;208:495–506. doi: 10.1002/path.1916. [DOI] [PubMed] [Google Scholar]
- 8.Linjawi A, Kontogiannea M, Halwani F, Edwardes M, Meterissian S. Prognostic significance of p53, bcl-2 and Bax expression in early breast cancer. J Am Coll Surg. 2004;198:83–90. doi: 10.1016/j.jamcollsurg.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Rohan TE, Li SQ, Hartwick R, Kandel RA. p53 Alterations and protein accumulation in benign breast tissue and breast cancer risk: A cohort study. Cancer Epidemiol Biomarkers Prev. 2006;15:1316–1323. doi: 10.1158/1055-9965.EPI-06-0195. [DOI] [PubMed] [Google Scholar]
- 10.Tsutsui S, Inoue H, Yasuda K, Suzuki K, Higashi H, Era S, Mori M. Reduced expression of PTEN protein and its prognostic implications in invasive ductal carcinoma of the breast. Oncology. 2005;68:398–404. doi: 10.1159/000086981. [DOI] [PubMed] [Google Scholar]
- 11.Wolff TA, Wilson JE. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility. Am Fam Physician. 2006;74:1759–1760. [PubMed] [Google Scholar]
- 12.Greggi S, Scala F, Laurelli G. Hereditery breast and ovarian cancer. Curr Probl Cancer. 2003;27:24–28. doi: 10.1067/mcn.2003.120003a. [DOI] [PubMed] [Google Scholar]
- 13.Gatalica Z, Lele SM, Rampy BA, Norris BA. The expression of Fhit protein is related inversely to disease progression in patients with breast carcinoma. Cancer. 2000;88:1378–1383. doi: 10.1002/(SICI)1097-0142(20000315)88:6<1378::AID-CNCR15>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 14.Allred DC, Clark GM, Tandon AK, Molina R, Tormey DC, Osborne CK, Gilchrist KW, Mansour EG, Abeloff M, Eudey L. Her-2/neu in node-negative breast cancer: Prognostic significance of overexpression influenced by the presence of in situ carcinoma. J Clin Oncol. 1992;10:599–605. doi: 10.1200/JCO.1992.10.4.599. [DOI] [PubMed] [Google Scholar]
- 15.Muss HB, Thor AD, Berry DA, Kute T, Liu ET, Koerner F, Cirrincione CT, Budman DR, Wood WC, Barcos M. C-erbB-2 expression and response to adjuvant therapy in women with node-positive early breast cancer. N Engl J Med. 1994;330:1260–1266. doi: 10.1056/NEJM199405053301802. [DOI] [PubMed] [Google Scholar]
- 16.Paik S, Bryant J, Park C, Fisher B, Tan-Chiu E, Hyams D, Fisher ER, Lippman ME, Wickerham DL, Wolmark N. ErbB-2 and response to doxorubicin in patients with axillary lymph node-positive, hormone receptor-negative breast cancer. J Natl Cancer Inst. 1998;90:1361–1370. doi: 10.1093/jnci/90.18.1361. [DOI] [PubMed] [Google Scholar]
- 17.Veronese SM, Gambacorta M, Gottardi O, Scanzi F, Ferrari M, Lampertico P. Proliferation index as a prognostic marker in breast cancer. Cancer. 1993;71:3926–3931. doi: 10.1002/1097-0142(19930615)71:12<3926::AID-CNCR2820711221>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Van Poznak C, Tan L, Panageas KS, Arroyo CD, Hudis C, Norton L, Seidman AD. Assessment of molecular markers of clinical sensitivity to single-agent taxane therapy for metastatic breast cancer. J Clin Oncol. 2002;20:2319–2326. doi: 10.1200/JCO.2002.08.125. [DOI] [PubMed] [Google Scholar]
- 19.De Placido S, De Laurentiis M, Carlomagno C, Gallo C, Perrone F, Pepe S, Ruggiero A, Marinelli A, Pagliarulo C, Panico L, et al. Twenty-year results of the Naples GUN randomized trial: Predictive factors of adjuvant tamoxifen efficacy in early breast cancer. Clin Cancer Res. 2003;9:1039–1046. [PubMed] [Google Scholar]
- 20.de Winter JP, Roelen BA, ten Dijke P, van der Burg B, van den Eijnden-van Raaij AJ. DPC4 (SMAD4) mediates transforming growth factor-beta1 (TGF-beta1) induced growth inhibition and transcriptional response in breast tumour cells. Oncogene. 1997;14:1891–1899. doi: 10.1038/sj.onc.1201017. [DOI] [PubMed] [Google Scholar]
- 21.Ali NA, McKay MJ, Molloy MP. Proteomics of Smad4 regulated transforming growth factor-beta signalling in colon cancer cells. Mol Biosyst. 2010;6:2332–2338. doi: 10.1039/c0mb00016g. [DOI] [PubMed] [Google Scholar]
- 22.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 23.Morén A, Itoh S, Moustakas A, Dijke P, Heldin CH. Functional consequences of tumorigenic missense mutations in the amino-terminal domain of Smad4. Oncogene. 2000;19:4396–4404. doi: 10.1038/sj.onc.1203798. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Wu L, Oelschlager DK, Wan M, Stockard CR, Grizzle WE, Wang N, Chen H, Sun Y, Cao X. Smad4 inhibits tumor growth by inducing apoptosis in estrogen receptor-positive breast cancer cells. J Biol Chem. 2005;280:27022–27028. doi: 10.1074/jbc.M505071200. [DOI] [PubMed] [Google Scholar]
- 25.Ren Y, Wu L, Frost AR, Grizzle W, Cao X, Wan M. Dual effects of TGF-beta on ERalpha-mediated estrogenic transcriptional activity in breast cancer. Mol Cancer. 2009;8:111. doi: 10.1186/1476-4598-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deckers M, van Dinther M, Buijs J, Que I, Löwik C, van der Pluijm G, ten Dijke P. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66:2202–2209. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- 27.Wiercinska E, Naber HP, Pardali E, van der Pluijm G, van Dam H, ten Dijke P. The TGF-β/Smad pathway induces breast cancer cell invasion through the up-regulation of matrix metalloproteinase 2 and 9 in a spheroid invasion model system. Breast Cancer Res Treat. 2011;128:657–666. doi: 10.1007/s10549-010-1147-x. [DOI] [PubMed] [Google Scholar]
- 28.Wang LH, Kim SH, Lee JH, Choi YL, Kim YC, Park TS, Hong YC, Wu CF, Shin YK. Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin Cancer Res. 2007;13:102–110. doi: 10.1158/1078-0432.CCR-06-1467. [DOI] [PubMed] [Google Scholar]
- 29.Kojima K, Vickers SM, Adsay NV, Jhala NC, Kim HG, Schoeb TR, Grizzle WE, Klug CA. Inactivation of Smad4 accelerates Kras (G12D)-mediated pancreatic neoplasia. Cancer Res. 2007;67:8121–8130. doi: 10.1158/0008-5472.CAN-06-4167. [DOI] [PubMed] [Google Scholar]
- 30.Kitamura T, Kometani K, Hashida H, Matsunaga A, Miyoshi H, Hosogi H, Aoki M, Oshima M, Hattori M, Takabayashi A, et al. SMAD4-deficient intestinal tumors recruit CCR1+ myeloid cells that promote invasion. Nat Genet. 2007;39:467–475. doi: 10.1038/ng1997. [DOI] [PubMed] [Google Scholar]
- 31.Yang L, Mao C, Teng Y, Li W, Zhang J, Cheng X, Li X, Han X, Xia Z, Deng H, et al. Targeted disruption of Smad4 in mouse epidermis results in failure of hair follicle cycling and formation of skin tumors. Cancer Res. 2005;65:8671–8678. doi: 10.1158/0008-5472.CAN-05-0800. [DOI] [PubMed] [Google Scholar]
- 32.Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. 7th. Springer; New York, NY: 2010. pp. 347–376. [Google Scholar]
- 33.Taubert H, Heidenreich C, Holzhausen HJ, Schulz A, Bache M, Kappler M, Eckert AW, Würl P, Melcher I, Hauptmann K, et al. Expression of survivin detected by immunohistochemistry in the cytoplasm and in the nucleus is associated with prognosis of leiomyosarcoma and synovial sarcoma patients. BMC Cancer. 2010;10:65. doi: 10.1186/1471-2407-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 35.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 36.Cox DR. Regression models and life tables. J Roy Statist Soc. 1972;34:187–220. [Google Scholar]
- 37.Stuelten CH, Buck MB, Dippon J, Roberts AB, Fritz P, Knabbe C. Smad4-expression is decreased in breast cancer tissues: A retrospective study. BMC Cancer. 2006;6:25. doi: 10.1186/1471-2407-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller DV, Leontovich AA, Lingle WL, Suman VJ, Mertens ML, Lillie J, Ingalls KA, Perez EA, Ingle JN, Couch FJ, et al. Utilizing nottingham prognostic index in microarray gene expression profiling of breast carcinomas. Mod Pathol. 2004;17:756–764. doi: 10.1038/modpathol.3800114. [DOI] [PubMed] [Google Scholar]
- 39.Xie W, Mertens JC, Reiss DJ, Rimm DL, Camp RL, Haffty BG, Reiss M. Alterations of Smad signaling in human breast carcinoma are associated with poor outcome: A tissue microarray study. Cancer Res. 2002;62:497–505. [PubMed] [Google Scholar]
- 40.Zhong D, Morikawa A, Guo L, Colpaert C, Xiong L, Nassar A, Chen C, Lamb N, Dong JT, Zhou W. Homozygous deletion of SMAD4 in breast cancer cell lines and invasive ductal carcinomas. Cancer Biol Ther. 2006;5:601–607. doi: 10.4161/cbt.5.6.2660. [DOI] [PubMed] [Google Scholar]
- 41.Jakob J, Nagase S, Gazdar A, Chien M, Morozova I, Russo JJ, Nandula SV, Murty VV, Li CM, Tycko B, et al. Two somatic biallelic lesions within and near SMAD4 in a human breast cancer cell line. Genes Chromosomes Cancer. 2005;42:372–383. doi: 10.1002/gcc.20142. [DOI] [PubMed] [Google Scholar]
- 42.Mesker WE, Liefers GJ, Junggeburt JM, van Pelt GW, Alberici P, Kuppen PJ, Miranda NF, van Leeuwen KA, Morreau H, Szuhai K, et al. Presence of a high amount of stroma and downregulation of SMAD4 predict for worse survival for stage I–II colon cancer patients. Cell Oncol. 2009;31:169–178. doi: 10.3233/CLO-2009-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fink SP, Swinler SE, Lutterbaugh JD, Massagué J, Thiagalingam S, Kinzler KW, Vogelstein B, Willson JK, Markowitz S. Transforming growth factor-beta-induced growth inhibition in a Smad4 mutant colon adenoma cell line. Cancer Res. 2001;61:256–260. [PubMed] [Google Scholar]
- 44.de Jong JS, van Diest PJ, van der Valk P, Baak JP. Expression of growth factors, growth inhibiting factors and their receptors in invasive breast cancer. I: An inventory in search of autocrine and paracrine loops. J Pathol. 1998;184:44–52. doi: 10.1002/(SICI)1096-9896(199801)184:1<44::AID-PATH984>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 45.Gobbi H, Dupont WD, Simpson JF, Plummer WD, Jr, Schuyler PA, Olson SJ, Arteaga CL, Page DL. Transforming growth factor-beta and breast cancer risk in women with mammary epithelial hyperplasia. J Natl Cancer Inst. 1999;91:2096–2101. doi: 10.1093/jnci/91.24.2096. [DOI] [PubMed] [Google Scholar]