Abstract
The mechanisms underlying drug resistance in colorectal cancer (CRC) treatment remain to be fully elucidated. Therefore, the present study aimed to investigate the underlying mechanism resistance to a widely used anticancer drug, 5-Fluorouracil (5-FU). Nuclear factor-erythroid 2-related factor 2 (Nrf2) is an important transcription factor involved in cellular protection. In the present study, it was hypothesized that the epigenetic modification of Nrf2 may be a potential target for 5-FU resistance in CRC treatment. Protein and messenger RNA levels of Nrf2, heme oxygenase-1 (HO-1), DNA methylases and DNA methyltransferases were determined and DNA methylation analysis for the Nrf2 promoter was performed in a human CRC control (SNU-C5) and resistant (SNU-C5R) cell line. The results demonstrated that Nrf2 expression levels, nuclear translocation and promoter binding were significantly increased in SNU-C5R cells compared with SNU-C5 cells. Elevated levels of activated Nrf2 in SNU-C5R cells resulted in the increased protein expression and activity of HO-1. In addition, increased production of reactive oxygen species (ROS) and upregulation of ten-eleven translocation (TET)1 were observed in SNU-C5R cells compared with SNU-C5 cells. Furthermore, methylation analysis revealed Nrf2 promoter cytosine-phosphate-guanine island hypomethylation in 5-FU-treated cells. In conclusion, the results indicated that 5-FU-induced ROS production resulted in the upregulation of TET1 expression and function. In addition, these results indicated that TET-dependent demethylation of the Nrf2 promoter upregulated Nrf2 and HO-1 expression, which induced cellular protection mechanisms, ultimately leading to drug resistance.
Keywords: colorectal cancer, SNU-C5 cells, 5-fluorouracil, cytosine-phosphate-guanine islands, epigenetics
Introduction
Chemotherapy is widely used for the treatment of cancer; however, acquired drug resistance is considered to be a substantial obstacle for effective chemotherapy (1). Drug resistance is a phenomenon that occurs due to a combination of factors and may involve individual differences in patients as well as genetic and epigenetic variations in tumors (2,3). Numerous previous studies have demonstrated that non-mutational microRNA (miRNA)-mediated gene expression has a crucial role in acquired drug resistance (4–6).
The classic antimetabolite, 5-Fluorouracil (5-FU) is widely used in colon cancer therapy and results in cytotoxic effects that induce cell death through affecting nucleoside metabolism (7). However, acquired 5-FU resistance is a severe hindrance for the clinical treatment of cancer (7,8). Although several studies have attempted to elucidate the molecular events that result in 5-FU resistance, limited evidence has been provided for the role of epigenetic modification (9–11). Therefore, it is important to reveal the distinct mechanisms involved in 5-FU resistance.
The multidrug-resistant phenotype observed in adriamycin-resistant breast cancer cells was found to be accompanied by epigenetic modification and overexpression of DNA methyltransferase (DNMT) genes (12). This therefore implicated the involvement of DNMTs in DNA hypermethylation, which has been reported to be involved in the onset of anticancer drug resistance in cancer patients. However, DNA demethylases as well as ten-eleven translocation enzymes (TETs), TET1, TET2 and TET3, are able to reverse this methylation process through the conversion of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine, 5-formylcytosine (5-fC) and 5-carboxylcytosine, eventually resulting in the production of cytosine (13,14). These modified bases act as intermediates for the DNA demethylation process as well as enhance the epigenetic variation of genomic DNA.
Nuclear factor erythroid-2-related factor 2 (Nrf2) is a transcription factor that has been suggested to be associated with cancer development and progression, including in lung (15), breast (16) and colorectal cancer (17). Nrf2 enables the adaptation of normal cells to oxidants and electrophiles that are generated by harmful exogenous agents, in order to reactive oxygen species and their secondary metabolites (18). Keap1 is a negative regulator of Nrf2 and its main function is to serve as an adaptor for cullin3/ring box1 (Cul3/Rbx1) E3 ubiquitin ligase complex (19,20) Under physiological conditions, Nrf2 is principally repressed by Keap1, which functions as an intracellular redox sensor, targeting Nrf2 for proteasomal degradation (21). Once a cell is exposed to oxidative stress, Keap1 releases Nrf2, which translocates to the nucleus and activates antioxidant response elements and xenobiotics element genes (including NQO1). This results in the protein expression of growth factors and receptors, drug efflux pumps, drug-metabolizing enzymes, heat shock proteins and various transcription factors (22,23).
Two previous studies have investigated Keap1/Nrf2 in colorectal cancer (CRC) cells (24,25). Activation of the Keap1/Nrf2 signaling pathway mediates protective responses to mitigate nitric oxide (NO)-induced damage and may contribute to the resistance of CRC cells to NO-induced cytotoxicity (23). Arlt et al reported that Nrf2 activity was elevated in colon cancer, resulting in overexpression of the proteasome subunit proteins and thus increased proteasome activity (25).
The present study aimed to explore the mechanisms underlying anticancer drug resistance in CRC cells through investigating epigenetic modification in the promoter DNA of the nuclear factor-erythroid 2-related factor 2 (Nrf2) gene.
Materials and methods
Cell culture
The human SNU-C5 CRC cell line and the 5-FU-resistant SNU-C5R cell line were purchased from the cell bank of the Chinese Academy of Science (Shanghai, China). Cells were cultured in RPMI-1640 medium (Invitrogen Life Technologies, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) at 37°C in a 5% CO2 incubator. A total of 1×104 SNU-C5R cells/well were then subcultured at 37°C in 140 mM 5-FU twice per week for >6 months in order to establish stable drug-resistant cell lines (26).
Cell proliferation assay
Cell viability was assessed using a colorimetric assay with the tetrazolium salt, MTT. Transfection with short interference (si)RNA was performed using the JetSI Transfection Reagent for siRNA (Polyplus-Transfection, Illkirch, France) at 50 nM, according to the manufacturer's instructions. After 24 h of transfection with siRNA, the cells were exposed to different concentrations of 5-FU (0, 10 or 100 µM; Sigma-Aldrich, St. Louis, MO, USA) for 72 h at room temperature. MTT (0.5 mg/ml; Sigma-Aldrich) was then added to each well and incubated for 3 h at room temperature. Plates were centrifuged at 4,500 × g for 5 min at room temperature, the medium was then removed and 100 µl acidic isopropanol (40 mM) was added to solubilize the crystals. The absorbance was measured at 570 mm using a microplate reader (Victor 3; Perkin Elmer, Turku, Finland).
Reactive oxygen species (ROS) detection
Cells (3×105/well) were seeded onto six-well plates and administered 25 mM dichlorodihydrofluorescein diacetate (DCF-DA; 30 µl). Next, 2,7-dichlorofluorescein (DCF) fluorescence levels were then detected using a flow cytometer (BD FACSCanto™; BD Bioscience, Franklin Lakes, NJ, USA). CellQuest software (version 6.0; BD Biosciences) was used to analyze the flow cytometry results. In order to evaluate the production of intracellular ROS, image analysis was performed through seeding cells (2×105/well) onto a coverslip-loaded six-well plate. Cells were then stained with 1 µM H2-DCFH-DA (Invitrogen Life Technologies) in phosphate-buffered saline (PBS) for 30 min at room temperature. Cells were then washed twice with PBS and visualized using an Eclipse TE2000-U fluorescent microscope (Nikon Corp., Tokyo, Japan) using a green filter (450–490 nm).
Protein blot analysis
Radioimmunoprecipitation assay buffer (Cell Signaling Technology, Inc., Danvers, MA, USA) was used to lyse cells. A total of 10–20 µg/µl soluble proteins were separated using 12% SDS-PAGE and then blotted onto polyvinylidene fluoride membranes. Membranes were then blocked using 5% non-fat dry milk for 1 h at room temperature prior to incubation overnight at 4°C with the following primary antibodies for: Heme oxygenase-1 (HO-1; cat. no. sc-10789), Nrf2 (cat. no. sc-722;), TET1 (cat. no. HPA019032), TET2 (cat. no. sc-136926), TET3 (cat. no. sc-139186), DNMT3B (cat. no. sc-130740) and β-actin (cat. no. sc-130657), which were all rabbit polyclonal IgG antibodies, used at a dilution of 1:200 and purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), with the exception of TET1 that was obtained from Sigma-Aldrich. In addition, DNMT1 (cat. no. ab19905; rabbit polyclonal IgG), phospho-Nrf2 (cat. no. ab76026; rabbit monoclonal IgG), TATA box binding protein (TBP; cat. no. ab52701; mouse polyclonal IgG) and DNMT3A (cat. no. ab23565; mouse polyclonal IgG) were purchased from Abcam (Cambridge, MA, USA) and used at a dilution of 1:200. Membranes were washed with PBS and then incubated with secondary antibodies (goat anti-rabbit polyclonal IgG; cat. no. SAB3700843; dilution, 1:500; Sigma-Aldrich) for 1 h at room temperature. Membranes were incubated with luminol reagent (Thermo Fisher Scientific Inc., Rockford, IL, USA) and the bands were visualized using a Bio-Rad XRS system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from SNU-C5 and SNU-C5R cells using Quick-RNA™MicroPrep solution (Zymo Research Corp., Irvine, CA, USA). iScript™ Reverse Transcription Supermix for real-time PCR (Bio-Rad Laboratories, Inc.) was used to reverse transcribe column purified total RNA, which was analyzed using a MiniOpticon™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) using SsoFast™ EvaGreen® Supermix (Bio-Rad Laboratories, Inc.) as previously described (27). ProbeFinder software (Roche, Basel, Switzerland) was used to design the primers, which were then commercially synthesized by Shanghai Generay Biotech Co. (Shanghai, China; http://www.generay.com.cn/). The primers used were as follows: Nrf2 forward, 5′-GAGAGCCCAGTCTTCATTGC-3′, and reverse, 5′-TTGGCTTCTGGACTTGGAAC-3′; GAPDH forward, 5′-AACGTGTCAGTGGTGGACCTG-3′, and reverse, 5′-AGTGGGTGTCGCTGTTGAAGT-3′. The PCR conditions were as follows: denaturation program (95°C for 10 min), followed by an amplification and quantification program repeated 40 times (95°C for 15 sec, 60°C for 10 sec, 72°C for 60 sec). Agarose gel electrophoresis (2%; Sigma-Aldrich) was then used to visualize the PCR products of the primers, which were verified by DNA sequencing that was performed by Shanghai Generay Biotech Co. All reactions were performed in triplicate and three independent experiments were run. Serial dilutions of a reference sample were used to construct standard curves for determining the individual PCR amplification efficiencies; this was included in each quantitative run in order to correct for variations in product amplification. Standard curves were used to obtain relative copy numbers, which were normalized to the values obtained for Gapdh, the internal control. CFX manager 3.1 software (Bio-Rad Laboratories, Inc.) was employed for data acquisition, analysis and determining PCR efficiencies.
Bisulfite genomic DNA sequencing
The genomic DNA of SNU-C5 and SNU-C5R cells was subjected to bisulfite conversion using an EZ DNA Methylation-Direct™ kit (Zymo Research Corp.). Amplification of the bisulfite-modified DNA was then performed through bisulfite sequencing PCR (Bio-Rad T100; Bio-Rad Laboratories, Inc.) using Platinum® PCR SuperMix High Fidelity (Invitrogen Life Technologies) with human Nrf2 promoter-specific primers: Nrf2 forward, 5′-TGAGATATTTTGCACATCCGATA-3′ and reverse, 5′-ACTCTCAGGGTTCCTTTACACG-3′. Subsequently, the PCR amplification products were purified through gel extraction with Zymoclean™ Gel DNA recovery kit (Zymo Research Corp.). A TOPO TA Cloning® kit (Invitrogen Life Technologies) was then used to clone the purified products into pCR®4-TOPO vectors. The recombinant plasmids were transformed into One Shot® TOP10 chemically competent E. coli (Invitrogen Life Technologies) using the calcium chloride transformation method (28). The plasmid DNA of ~10 independent clones of each amplicon was isolated using a PureLink™Quick PlasmidMiniprep kit (Invitrogen Life Technologies) and then sequenced in order to determine the cytosine-phosphate-guanine methylation status. Clones with an insert with N 99.5% bisulfite conversion, i.e. non-methylated cytosine residues to thymine, were included in the present study and the remaining were excluded. Bisulfite Sequencing DNA Methylation Analysis software (http://biochem.jacobs-university.de/BDPC/BISMA/) was then used to analyze the sequenced data of each clone for DNA methylation in the Nrf2 promoter, using default filtering threshold settings.
Statistical analysis
All statistical analyses were performed with the SPSS 20.0 software (IBM SPSS, Armonk, NY, USA). Quantitative data are expressed as the mean ± standard deviation. The statistically significant differences between patient and control groups were tested using t-test and ANOVA test. P<0.05 was considered to indicate a statistically significant difference.
Results
Chemosensitivity of SNU-C5R cells to 5-FU
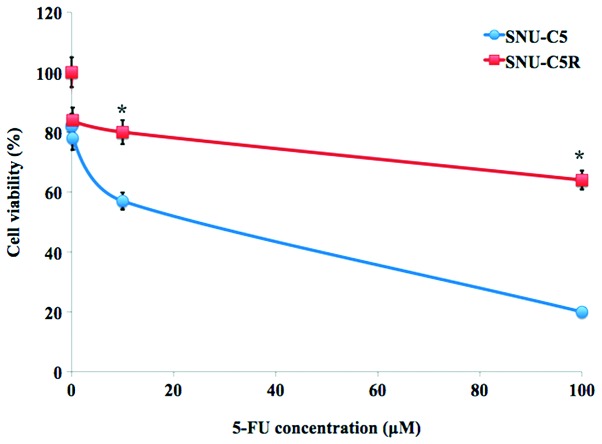
The relative chemosensitivity of the SNU-C5R 5-FU-resistant cell line was confirmed using an MTT cell viability assay. As shown in Fig. 1, 5-FU treatment for 72 h resulted in a dose-dependent suppression of cell growth in SNU-C5 cells, which was significantly decreased compared with that of the SNI-C5R cells (Fig. 1). The 5-FU concentrations causing a 50% growth inhibition as compared with control cells (IC50) were calculated by a modified Kärbers method (29). The IC50 value for 5-FU in SNU-C5R cells was 118.7±4.9 µM, whereas the corresponding value for their parental SNU-C5 cells was 23.2±3.4 µM (P<0.05).
Figure 1.
Chemosensitivity of 5-FU-resistant human SNU-C5R colorectal cancer cells. SNU-C5 and SNU-C5R cells were exposed to the indicated concentration of 5-FU for 72 h. An MTT assay was then used to determine the cell viability of the two cell lines. Values are presented as the mean ± standard error of the mean for three independent experiments. *P<0.05 vs. SNU-C5 cells. 5-FU, 5-fluorouracil.
Intracellular ROS levels in SNU-C5 and SNU-C5R cells
Flow cytometric analysis data revealed that ROS levels were significantly elevated in SNU-C5R cells [fluorescence intensity (FI), 340] compared with SNU-C5 cells (FI, 130; P<0.05) (Fig. 2A). Fluorescent microscopic imaging confirmed these results, as it demonstrated that the ROS green FI was markedly enhanced in SNU-C5R cells compared with SNU-C5 cells (Fig. 2B). This therefore suggested that SNU-C5R cells were exposed to an increased level of oxidative stress conditions compared with SNU-C5 cells.
Figure 2.
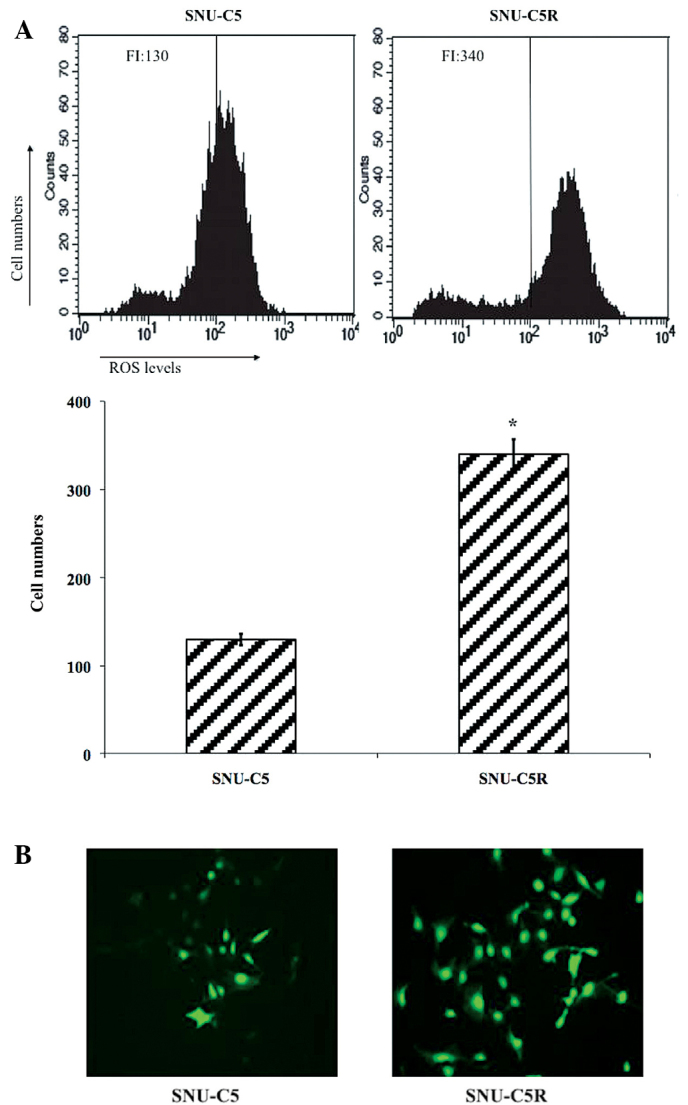
Intracellular ROS levels in SNU-C5 and 5-fluorouracil-resistant SNU-C5R cells. (A) ROS levels were detected by flow cytometric analysis and (B) fluorescent microscopic imaging was performed following H2-dichlorodihydrofluorescein diacetate staining. Values are presented as the mean ± standard error of the mean for three independent experiments. *P<0.05 vs. SNU-C5 cells. ROS, reactive oxygen species.
Antioxidant and DNA methylation-associated protein expression in SNU-C5 and SNU-C5R cells
HO-1 and Nrf2 protein levels were observed to be increased in SNU-C5R cells compared with SNU-C5 cells (Fig. 3A). In addition, the expression levels of the epigenetic modification-associated proteins, in terms of those for DNA methylation, was investigated by assessing the protein levels of DNMTs DNMT1, DNMT3A and DNMT3B as well as the DNA demethylases, including TET1, TET2 and TET3. The results revealed that DNMT expression was not significantly different between SNU-C5 and SNU-C5R cells, whereas TET expression was elevated in SNU-CR cells compared with SNU-C5 cells (Fig. 3B).
Figure 3.
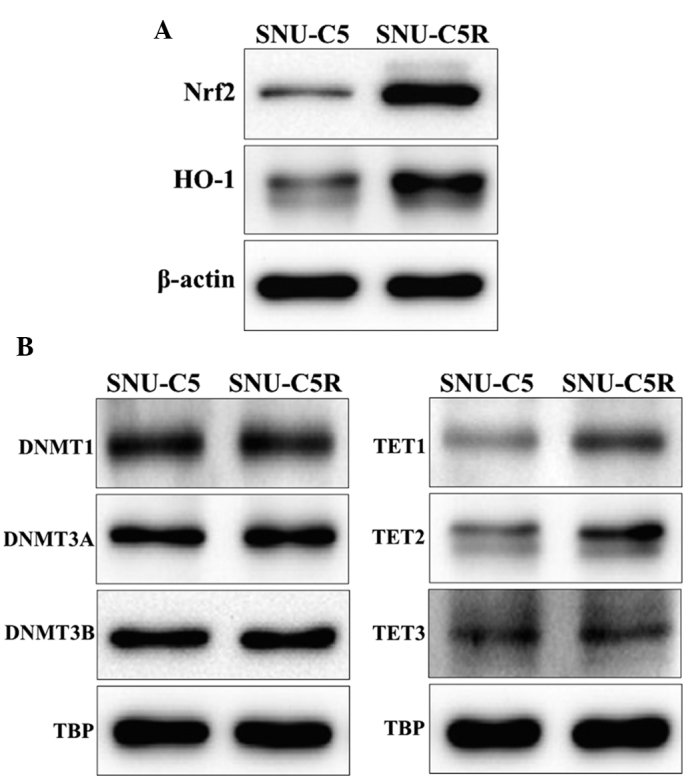
Antioxidant and DNA methylation-associated protein levels in SNU-C5 and 5-fluorouracil-resistant SNU-C5R cells. (A) Protein blots of antioxidants Nrf2 and HO-1. (B) Protein blots of DNA methylases DNMT1, DNMT3A and DNMT3B as well as DNA demethylases TET1, TET2 and TET3. β-actin and TBP were used as loading controls. Nrf2, nuclear factor-erythroid 2-related factor 2; HO-1, heme oxygenase-1; DNMT. DNA methyltransferase; TET, ten-eleven translocation enzyme; TBP, TATA box binding protein.
Epigenetic analysis and gene expression of Nrf2 in SNU-C5 and SNU-C5R cells
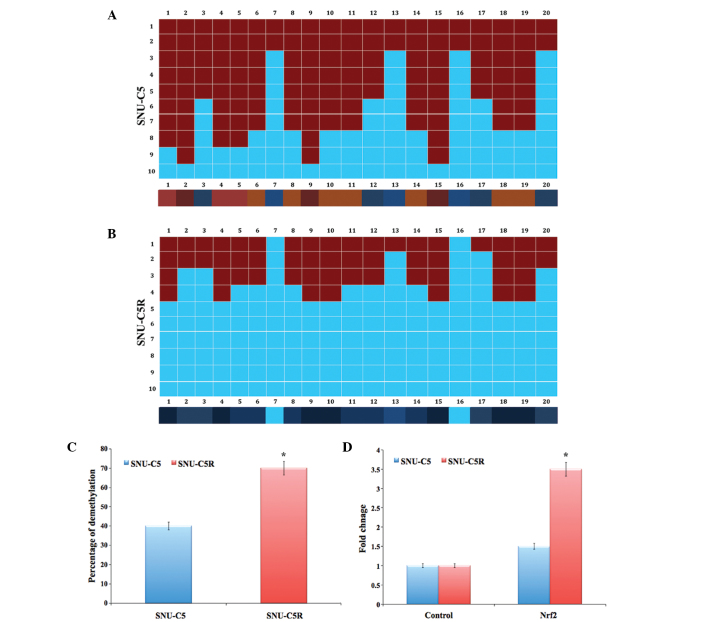
The bisulphate DNA sequencing of the Nrf2 promoter region revealed significant demethylation in SNU-C5R cells compared with that of SNU-C5 cells (P<0.05) (Fig. 4A and B). The overall percentage of Nrf2 promoter demethylation was ~40% in SNU-C5 cells and ~70% in SNU-C5R cells (P<0.05) (Fig. 4C). In addition, messenger (m)RNA levels of Nrf2 in SNU-C5R cells exhibited a 3.5-fold increase, which was significantly increased compared with the 1.5-fold increase observed in SNU-C5 cells (P<0.05); increases were compared to that of the control house keeping gene (Fig. 4D).
Figure 4.
Status of Nrf2 promoter DNA methylation and Nrf2 genes in SNU-C5 and 5-fluorouracil-resistant SNU-C5R cells. Bisulphite genomic DNA sequencing of SNU-C5 cells showing (A) average methylated CpG dinucleotides and (B) highly demethylated CpG dinucleotides in the region between −423 and −126 of Nrf2 promoter. A total of 11 individual clones of the bisulfite-converted DNA sequences were analyzed for DNA methylation, containing 20 CpG dinucleotides, of the Nrf2 promoter by Bisulfite Sequencing DNA Methylation Analysis software using default filtering threshold settings. Columns, CpG dinucleotide sites; red squares, methylated CpG dinucleotides; blue squares, unmethylated CpG dinucleotides. Color gradient bar indicates that red regions contain more methylated CpG dinucleotides and blue regions contain more unmethylated CpG dinucleotides. (C) Quantification of the percentage of Nrf2 demethylation. (D) Reverse transcription quantitative polymerase chain reaction analyses of SNU-C5R cells showing the increased expressions of Nrf2 messenger RNAs. Values are presented as the mean ± standard error of the mean for three independent experiments. *P<0.05 vs. SNU-C5 cells. Nrf2, nuclear factor-erythroid 2-related factor 2; CpG, cytosine-phosphate-guanine.
Discussion
Despite the occurrence of drug resistance, 5-FU was used as the standard treatment for CRC patients for several years (30,31). Novel therapeutic strategies for CRC are required; thus, elucidating the underlying mechanisms of 5-FU resistance is essential for the generation of novel molecular targeted therapies for overcoming drug resistance (7,11,31). There has been increasing evidence to suggest the role of epigenetic alterations in drug resistance in numerous types of cancer, such as CRC (32–35); this therefore indicates that epigenetic modification may occur following the administration of 5-FU. In the present study, the epigenetic status of Nrf2, a master transcription factor for major protective genes, was compared between the normal CRC SNU-C5 cell line and the 5-FU-resistant SNU-C5R cell line. Nrf2 is involved in the regulation of numerous genes, such as HO-1, which has a role in protecting cells against the chemical and oxidative stresses that are activated in CRC cells (25). In addition, 5-FU induces the activation of Nrf2, the mechanism of which was reported to occur via enhanced ROS production following drug treatment in the human HT-29 colon cancer cells (36). The results of the present study were consistent with these previous studies, as significant increases were observed in ROS production as well as Nrf2 and HO-1 expression in SNU-C5R cells compared with SNU-C5 cells. The upregulated expression and activity of HO-1 and Nrf2 in SNU-C5R cells indicated that Nrf2 protein translocation from the cytosol to the nucleus was enhanced and that Nrf2 interacted with the AU-rich element sequence in the HO-1 promoter region, therefore inducing cellular protection.
Previous studies have demonstrated that ROS promote epigenetic modifications, which therefore alter the genome and have a crucial role in carcinogenesis (37). In particular, ROS production was reported to be associated with the modification of DNA methylation patterns (38). In the present study, increased ROS production was observed in SNU-C5R cells compared with the SNU-C5 cells. Subsequently, the expression levels of DNA methylation-associated proteins, including DNMT1, DNMT3A and DNMT3B, as well as DNA demethylases, including TET1, TET2 and TET3, were investigated. The results clearly demonstrated that there was no change in the levels of DNMTs, whereas TET protein levels were markedly increased in SNU-C5R cells compared with SNU-C5 cells. TET1 is the main enzyme that catalyzes 5-mC oxidation to 5-fC; the remaining TET proteins, TET2 and TET3, were reported to compensate for reduced levels of TET1 (39,40). In the present study, bisulphate DNA sequence analysis revealed a high level of demethylation in the 5-FU-resistant cells, SNU-C5R. In concurrence with these results, Nrf2 mRNA levels were found to have increased ~3.5-fold in the drug-resistant cells.
In conclusion, the results of the present study suggested that 5-FU was responsible for inducing excess ROS production, resulting in epigenetic alterations. Increased levels of TET proteins further confirmed the epigenetic alterations; however, no difference was observed in DNMT protein levels, which suggested that they were not involved in the methylation process. Therefore, demethylation was confirmed. Furthermore, promoter DNA demethylation of the Nrf2 gene was confirmed by elevated levels of Nrf2 mRNA in drug-resistant SNU-C5R cells. Therefore, epigenetic modification in Nrf2 may provide a potential novel therapeutic target for counteracting the resistance of CRC cells against 5-FU treatment.
References
- 1.Kang H, Kim C, Lee H, Kim W, Lee EK. Post-transcriptional controls by ribonucleoprotein complexes in the acquisition of drug resistance. Int J Mol Sci. 2013;14:17204–17220. doi: 10.3390/ijms140817204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raguz S, Yagüe E. Resistance to chemotherapy: New treatments and novel insights into an old problem. Br J Cancer. 2008;99:387–391. doi: 10.1038/sj.bjc.6604510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan DS, Gerlinger M, Teh BT, Swanton C. Anti-cancer drug resistance: Understanding the mechanisms through the use of integrative genomics and functional RNA interference. Eur J Cancer. 2010;46:2166–2177. doi: 10.1016/j.ejca.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Mishra PJ. The miRNA-drug resistance connection: A new era of personalized medicine using noncoding RNA begins. Pharmacogenomics. 2012;13:1321–1324. doi: 10.2217/pgs.12.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra PJ, Bertino JR. MicroRNA polymorphisms: The future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, Xu X, Zhu Y, Li S, Zheng X, Xie L. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36:62–68. doi: 10.1007/s10059-013-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang N, Yin Y, Xu SJ, Chen WS. 5-Fluorouracil: Mechanisms of resistance and reversal strategies. Molecules. 2008;13:1551–1569. doi: 10.3390/molecules13081551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters GJ, Backus HH, Freemantle S, van Triest B, Codacci-Pisanelli G, et al. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta. 2002;1587:194–205. doi: 10.1016/S0925-4439(02)00082-0. [DOI] [PubMed] [Google Scholar]
- 9.Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, Longley DB, Caldas C, Johnston PG. Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res. 2006;66:2765–2777. doi: 10.1158/0008-5472.CAN-05-2693. [DOI] [PubMed] [Google Scholar]
- 10.Karasawa H, Miura K, Fujibuchi W, Ishida K, Kaneko N, et al. Down-regulation of cIAP2 enhances 5-FU sensitivity through the apoptotic pathway in human colon cancer cells. Cancer Sci. 2009;100:903–913. doi: 10.1111/j.1349-7006.2009.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurokawa K, Tanahashi T, Iima T, Yamamoto Y, Akaike Y, Nishida K, Masuda K, Kuwano Y, Murakami Y, Fukushima M, Rokutan K. Role of miR-19b and its target mRNAs in 5-fluorouracil resistance in colon cancer cells. J Gastroenterol. 2012;47:883–895. doi: 10.1007/s00535-012-0547-6. [DOI] [PubMed] [Google Scholar]
- 12.Segura-Pacheco B, Perez-Cardenas E, Taja-Chayeb L, Chavez-Blanco A, Revilla-Vazquez A, Benitez-Bribiesca L, Duenas-González A. Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine. J Transl Med. 2006;4:32. doi: 10.1186/1479-5876-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mc to 5hmc conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PloS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nioi P, Nguyen T. A mutation of Keap1 found in breast cancer impairs its ability to repress Nrf2 activity. Biochem Biophys Res Commun. 2007;362:816–821. doi: 10.1016/j.bbrc.2007.08.051. [DOI] [PubMed] [Google Scholar]
- 17.Hanada N, Takahata T, Zhou Q, et al. Methylation of the KEAP1 gene promoter region in human colorectal cancer. BMC Cancer. 2012;12:66. doi: 10.1186/1471-2407-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 24.Li CQ, Kim MY, Godoy LC, Thiantanawat A, Trudel LJ, Wogan GN. Nitric oxide activation of Keap1/Nrf2 signaling in human colon carcinoma cells. Proc Natl Acad Sci USA. 2009;106:14547–14551. doi: 10.1073/pnas.0907539106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arlt A, Bauer I, Schafmayer C, et al. Increased proteasome subunit protein expression and proteasome activity in colon cancer relate to an enhanced activation of nuclear factor E2-related factor 2 (Nrf2) Oncogene. 2009;28:3983–3996. doi: 10.1038/onc.2009.264. [DOI] [PubMed] [Google Scholar]
- 26.Jung GR, Kim KJ, Choi CH, Lee TB, Han SI, Han HK, Lim SC. Effect of betulinic acid on anticancer drug-resistant colon cancer cells. Basic Clin Pharmacol Toxicol. 2007;101:277–285. doi: 10.1111/j.1742-7843.2007.00115.x. [DOI] [PubMed] [Google Scholar]
- 27.Bourhy P, Bremont S, Zinini F, Giry C, Picardeau M. Comparison of real-time PCR assays for detection of pathogenic Leptospira spp. in blood and identification of variations in target sequences. J Clin Microbiol. 2011;49:2154–2160. doi: 10.1128/JCM.02452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dagert M, Ehrlich SD. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979;6:23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- 29.Liu GF. Median effective dose. In: Jiang ZJ, editor. Medical Statistics. 1st. People's Medical Publishing House; Beijing: 1997. pp. 136–153. [Google Scholar]
- 30.Was H, Dulak J, Jozkowicz A. Heme oxygenase-1 in tumor biology and therapy. Curr Drug Targets. 2010;11:1551–1570. doi: 10.2174/1389450111009011551. [DOI] [PubMed] [Google Scholar]
- 31.Becker JC, Fukui H, Imai Y, Sekikawa A, Kimura T, Yamagishi H, Yoshitake N, Pohle T, Domschke W, Fujimori T. Colonic expression of heme oxygenase-1 is associated with a better long-term survival in patients with colorectal cancer. Scand J Gastroenterol. 2007;42:852–858. doi: 10.1080/00365520701192383. [DOI] [PubMed] [Google Scholar]
- 32.Arnold CN, Goel A, Boland CR. Role of hMLH1 promoter hypermethylation in drug resistance to 5-fluorouracil in colorectal cancer cell lines. Int J Cancer. 2003;106:66–73. doi: 10.1002/ijc.11176. [DOI] [PubMed] [Google Scholar]
- 33.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 34.Dworkin AM, Huang TH, Toland AE. Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis and treatment. Semin Cancer Biol. 2009;19:165–171. doi: 10.1016/j.semcancer.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balch C, Huang TH, Brown R, Nephew KP. The epigenetics of ovarian cancer drug resistance and resensitization. Am J Obstet Gynecol. 2004;191:1552–1572. doi: 10.1016/j.ajog.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 36.Akhdar H, Loyer P, Rauch C, Corlu A, Guillouzo A, Morel F. Involvement of Nrf2 activation in resistance to 5-fluorouracil in human colon cancer HT-29 cells. Eur J Cancer. 2009;45:2219–2227. doi: 10.1016/j.ejca.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 37.Ziech D, Franco R, Pappa A, Panayiotidis MI. Reactive oxygen species (ROS) - induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res. 2011;711:167–173. doi: 10.1016/j.mrfmmm.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Donkena KV, Young CY, Tindall DJ. Oxidative stress and DNA methylation in prostate cancer. Obstet Gynecol Int. 2010;2010:302051. doi: 10.1155/2010/302051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudenko A, Dawlaty MM, Seo J, Cheng AW, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]