Abstract
Nearly half of bladder cancer patients experience recurrences. Reliable predictors of this recurrent phenotype are needed to guide surveillance and treatment.
Objective
To identify genetic variants that modify bladder cancer prognosis focusing on genes involved in major biological carcinogenesis processes (apoptosis, proliferation, DNA repair, hormone regulation, immune surveillance, and cellular metabolism).
Subjects and methods
We analyzed variant genotypes hypothesized to modify these processes in 563 urothelial-cell carcinoma cases enrolled in a population-based study of incident bladder cancer conducted in New Hampshire, U.S.A.
After diagnosis, cases were followed over time to ascertain recurrence and survival status, making this one of the first population-based studies with detailed prognosis data.
Cox proportional hazards regression was used to assess the relationship between SNPs and prognosis endpoints.
Results
Aldehyde dehydrogenase 2 (ALDH2) variants had shorter time to first bladder cancer recurrence (adjusted non-invasive HR 1.90 95%CI 1.29-2.78).
We observed longer survival among bladder cancer cases with non-invasive tumors associated with DNA repair XRCC4 heterozygous genotype compared with wildtype (adjusted HR 0.53 95%CI 0.38-0.74).
Time to recurrence was shorter for patients who had a variant allele in vascular cellular adhesion molecule 1 (VCAM1) and were treated with immunotherapy (P interaction <0.001).
Conclusion
Our analysis suggests candidate prognostic SNPs that could guide personalized bladder cancer surveillance and treatment.
Keywords: bladder cancer, polymorphism, prognosis, recurrence, survival, DNA repair, immune
Introduction
In the Western world, bladder cancer is the fourth most common cancer in men, and eighth most common in women [1]. Bladder cancer generally carries a favorable prognosis, but rates of recurrent disease are high, with over 40% having a recurrence in some form [2]. Bladder cancer is among the most expensive malignancies to treat due to the need for frequent, painful screening procedures to identify recurrences [3, 4]. Predictors of recurrent bladder cancer include tumor multiplicity, tumor size, T category, presence of carcinoma in situ, tumor grade, and patient gender [5]. Refining our ability to identify those patients at high-risk of recurrent disease for more frequent screening and aggressive treatment could help to improve quality of life for all bladder cancer patients [4].
Genetic variations in prognosis and response to treatment have been observed for many cancers [6]. A number of different mechanisms have adapted to respond to specific types of exogenous damage or aberrant cellular behavior to prevent or repair damage to macromolecules, and to remove damaged cells. We hypothesized that bladder cancer prognosis may be affected by genetic variation in each of these major carcinogenesis mechanisms. We tested whether genetic variation in each of these major processes dysregulated in carcinogenesis that could modify bladder cancer prognosis: apoptosis, DNA repair, hormone, immune surveillance, metabolism, proliferation, neural, telomere, and transport. Our study is the first population-based bladder cancer study in the U.S. with long-term case follow-up for recurrence. We performed analyses of hypothesized prognosis-related SNPs by carcinogenesis process, and identified interactions and sets of genes.
Subjects and Methods
Study group
We identified all cases of bladder cancer among New Hampshire residents, ages 25 to 74 years, in two sequential phases based on cancer diagnosis date (Phase I 1 July 1994–30 June 1998, Phase II 1 July 1998–30 June 2001) from the New Hampshire State Cancer Registry. Detailed methods have been described previously [16, 17]. Briefly, we interviewed a total of 857 bladder cancer cases, which was 85% of the cases confirmed to be eligible for the study. Slides and blocks were re-reviewed by the study pathologist to assess tumor histology, stage and grade using both the 1973 and 2004 WHO classifications of bladder tumors. The stage assigned by the pathologist was used for tumors <stage 2A, while Cancer Registry data on stage was used for higher stage. After re-review, 21 were non-cancer and 7 undetermined, leaving n=829 tumors which were either deemed cancerous by histopathology re-review (~90%), or the original diagnosis if the pathology materials were unavailable. The histologic composition of these tumors was 817 urothelial-cell (transitional-cell) carcinoma (660 papillary urothelial cell carcinoma, 133 urothelial cell carcinoma, 4 papillary carcinoma, 20 carcinoma NOS), as well as 2 spindle cell carcinoma, 3 small cell carcinoma, 5 squamous cell carcinoma, 2 adenocarcinoma. We omitted n=9 participants with primary diagnosis dates >2 years prior to the diagnosis date assumed in the interview. Most (>95%) of the subjects in this study are of self-reported Caucasian origin; thus restricting our genetic analyses to n=786 Caucasians did not appreciably alter the results. Analyses were further restricted to cases with known tumor stage (omitting n=3), leaving a total of n=783 participants in the analysis.
Personal interview
Informed consent was obtained from each participant and all procedures and study materials were approved by the Committee for the Protection of Human Subjects at Dartmouth College (approved study registration number: 11697). Consenting participants underwent a detailed in-person interview, usually at their home.
Disease status and the main objectives of the study were not disclosed to the interviewers. To ensure consistent quality of the study interviewer, interviews were tape recorded with the consent of the participants and routinely monitored by the interviewer supervisor. Subjects were asked to provide a blood sample (buccal sample was requested in the case of a refusal). Samples were maintained at 4°C and sent via courier to the study laboratory at Dartmouth within 24 hours for processing and analysis.
Genotyping
DNA was isolated using Qiagen genomic DNA extraction kits (QIAGEN Inc., Valencia, CA) from peripheral circulating blood lymphocyte specimens harvested at the time of interview. Genotyping was performed on all DNA samples of sufficient concentration (representing n=563 of the urothelial cell carcinoma cases in the analytic dataset), using the GoldenGate Assay system (Illumina, Inc., San Diego, CA). Genotyped SNPs were mostly those included on the Illumina Cancer Panel (1,421 SNPs in approximately 400 hypothesized cancer-related genes) (Supplementary Table I). SNPs were selected within coding, intronic and flanking regions hypothesized to be potentially functional of the genes of interest, including a median of 3 SNPs per gene. Results reported were restricted to SNPs with a minimum minor allele frequency of 5% and genotype call data available on at least 90% of our samples (n=1,367 SNPs). Samples repeated on multiple GoldenGate plates yielded the same call for 99.9% of SNPs and 99.5% of samples submitted were successfully genotyped.
The main goal of the statistical analysis was to assess the relationship between genetic variation in nine major carcinogenesis processes and bladder cancer susceptibility and prognosis. These nine processes are: Apoptosis, DNA Repair, Immune, Hormone, Metabolism, Neural, Proliferation, Telomere, and Transport or Signaling. We grouped SNPs that were involved in each process according to the Database for Annotation, Visualization, and Integrated Discovery (DAVID) Gene Ontology (GO) search engine [18]. The number of SNPs analyzed by functional group was: (metabolism n=340, proliferation n=304, immune n=243, DNA repair n=224, transport / signaling n=170, apoptosis n=135, hormone n=68, telomere n=27, neural n=11) (see full SNP list in Supplemental Table I).
Follow-up
Information on bladder cancer recurrences was obtained from medical records provided by the treating hospital(s) (both in and outpatient records, including any pathology reports) covering the follow-up period. Records were reviewed by an experienced, certified tumor registrar to abstract the data on bladder tumors occurring subsequent to the incident tumor. Hospital registry data were used if the medical record could not be obtained. The first recurrent tumor was defined as any tumor identified a disease-free remission period, more than 90 days after the date of initial primary bladder tumor diagnosis. These recurrent tumors include those of the same level of invasiveness, as well as those which have progressed to higher stage/grade. Persistent primary tumors that did not have a remission period are excluded from the analysis of recurrence (n=7). Time to recurrence was calculated as the time between the initial diagnosis date and the date of the first recurrence event. For progression analysis, the diagnosis of a tumor with a greater stage or grade than the initial primary bladder tumor was considered the event. If no events were reported, the patient was last seen documented in the medical record was used for censoring. Status (alive or dead) was determined as of January 13, 2011 using the Social Security and the National Death Indices (NDI). Survival time was calculated from the date of initial diagnosis to date of death. Data on the initial course of treatment were obtained from the State Cancer Registry and were verified by medical record review (immunotherapy 13.4%, chemotherapy 1.7%, radiotherapy 0.4%, transurethral resection 88.5%, cystectomy 2.1%). Analyses of genetic variation in relation to prognosis were restricted to the urothelial-cell carcinomas with genotype data available (n=563).
Statistical analysis
Bladder cancer prognosis associated with the individual SNPs was analyzed separately within each of the five biologic pathways of interest. Median times to first recurrence or death were calculated using the Kaplan-Meier method. Time to the first bladder tumor recurrence and survival analyses were performed using Cox-proportional hazards regression analysis in SAS version 9.3. Analyses were performed overall, and then separately for non-invasive tumors (stage 0, I), and invasive tumors (stage II, III, IV). The standard prognostic model included adjustment for age at diagnosis, gender, smoking (never, former, continuing), as well as tumor size (<3cm, ≥3cm), and multiplicity (single, >1) low grade, high grade, presence of Cis, and treatment (immunotherapy, chemotherapy, radiotherapy, transurethral resection, cystectomy). P values represent two-sided statistical tests. To correct for multiple comparisons within each group of SNPs, we also calculated P-values adjusted for the False Discovery Rate (FDR) for the total number of SNPs within a functional group. We report SNPs with FDR adjusted P-values <0.25. Statistical significance was considered at P<0.001 and we restricted our report to SNPs that also meet this cutoff. Kaplan-meier plots were examined and reported results are restricted to those with hazards that are proportional. To avoid reporting unstable estimates, we set the minimum cell size to n=20 for the main effects reported. Hardy-Weinberg equilibrium testing was performed by comparing the observed genotype distribution in the participants to that expected for a population in equilibrium (freq A = p; freq a = q; p + q = 1, freq AA = p2, freq Aa = 2pq, freq aa= q2) using a Chi-square test. We performed an a priori assessment of power for these analyses using a range of conservative Bonferroni corrected P-values (0.05/number of SNPs interrogated per pathway), which resulted in alphas ranging from 0.005 – 0.0001. With a variant SNP prevalence of 33%, we estimate 80% power to detect a minimum HR of 1.6 with n=520 patients with alpha=0.005, while we could detect a minimum HR of 1.85 with n=540 for an alpha=0.0001.
We constructed receiver operating characteristic curves and calculated the area under the curve (AUC) to evaluate the specificity and sensitivity of predicting 5-year recurrence and 10-year survival. Patients not experiencing the endpoint within the time interval were omitted from the model. Interactions between SNPs in relation to the time-to-event endpoints recurrence and survival were detected using our Survival - Multifactor Dimensionality Reduction (Surv-MDR) algorithms run in R 2.6.1 [19]. This method modifies the nonparametric multifactor dimensionality reduction (MDR) constructive induction algorithm to use the log-rank test. We ran MDR separately on each functional group of SNPs and selected the best SNP-SNP MDR model as the one with the lowest average prediction error. An error rate of 50% is expected under the null hypothesis. Statistical significance is determined using permutation testing. Here, the case-control labels are randomized 1000 times and the entire MDR model fitting procedure repeated on each randomized dataset to determine the expected distribution of testing accuracies under the null hypothesis. We used 10-fold cross-validation and 1000-fold permutation testing to reduce the chances of making a type I error due to multiple testing [20, 21].
To evaluate the multiplicative vs. additive nature of the interactions between the SNP allele combinations predicted by Surv-MDR, we included interaction terms in a Cox-proportional hazards model with adjustment for age at diagnosis, gender, smoking (never, former, current), as well as tumor size (<3cm, ≥3cm), and multiplicity (single, >1), low grade, high grade, presence of carcinoma in situ (Cis), and treatment (immunotherapy, chemotherapy, radiotherapy, transurethral resection, cystectomy). As the number of invasive cases was small in this population-based study, we analyzed of recurrence and survival interactions within the non-invasive cases only.
The Gene Set Analysis (GSA) method was performed in R version 2.7.2 to assess the significance of the each of the functional groups in relation to the time-to-event endpoints. GSA ranks the SNPs by their correlation with the endpoint (positive for decreased risk, negative for increased risk). An enrichment score is calculated as a running-sum statistic that increases when the next SNP down the list is in the same functional group, but decreases when it is not. Thus, enrichment scores with the maximum deviation from zero are achieved when multiple SNPs of the same functional group are ranked close together at the very top or bottom of the ranked correlation list. The peak enrichment score is the highest positive score, indicating that the variant form of the SNP is protective, or the lowest negative score, indicating that the variant form of the SNP is a risk factor. The False Discovery Rate (FDR) is then calculated using 1000 permutations of the endpoint to estimate the probability that the enrichment score represents a false positive finding. The enrichment score for GSA is a “maxmean” statistic, computed by averaging the positive parts of each Z-score in a given pathway, as well as the negative parts. We choose the Z-score that is larger in absolute value. GSA uses permutation testing to determine the P-value. Using these methods, we first analyzed the SNPs broken into nine major carcinogenesis pathways: apoptosis, DNA repair, hormone, immune surveillance, metabolism, proliferation, neural, telomere, and transport. We also applied a highly detailed assessment of SNPs grouped into 600 Gene Ontology (GO) biological processes using the Pathway Studio ResNet 8 database (Ariadne Genomics).
Results
Then genotyped population had similar characteristics as the overall case group (Table 1). As our study is population-based and included both community and academic facilities throughout the state, the majority (84%) of our cases were non-muscle invasive at initial diagnosis. Medical data were reviewed on 94% of the cases, with a median duration of follow-up of 5.4 years and 75% of the cases were followed >1.6 years. Half of the cases experienced at least one recurrence during the follow-up period and 40% were deceased (Table 1). Hardy Weinberg equilibrium P-values of the prognosis-associated SNPs in the bladder cancer case population were (ALDH2_08 rs2238151 0.32, IGF1_24 rs5742714 0.04, XRCC4_07 rs2662238 0.28, DRD4_15 rs4987059 0.35, RB1CC1_50 rs35402311 0.76, IL15RA_04 rs2228059 0.87, MBL2 rs2099902 0.06, TNKS_03 rs34206126 0.15, APC_09 rs2229992 0.76, VCAM1_05 rs3176879 0.05, IL1B_01 rs16944 0.70, IFNG_07 rs1861494 0.61).
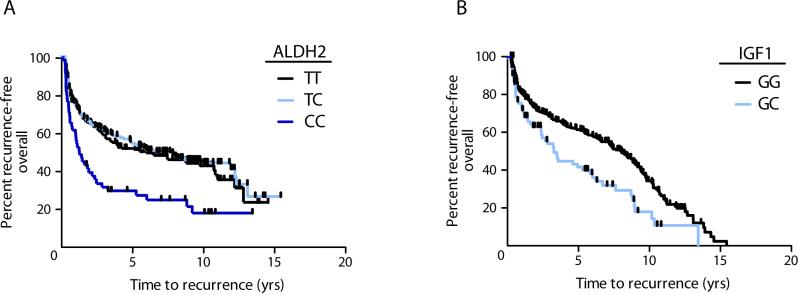
Within each of the nine major carcinogenesis processes, relationships between SNPs and bladder cancer recurrence or survival were assessed using Cox regression with adjustment for age, gender, smoking, stage, grade, treatment. Assuming incomplete dominance, we tested heterozygous vs. wildtype; variant vs. wildtype. The variant form of metabolism SNP ALDH2 rs2238151 in Aldehyde Dehydrogenase was associated with a shorter time to recurrence (overall HR 2.03, FDR adjusted P-value <0.001) (Table 2, Figure 1a). Because alcohol metabolism is a major role of ALDH2, we investigated the possibility that this SNP modified the effect of alcohol exposure. We observed an interaction between ALDH2 rs2238151 variant genotype and drinking at least 31 alcoholic drinks per month during the 5-year period prior to diagnosis, compared to never drinkers (interaction P value 0.011). Among ALDH2 variants, drinking alcohol over the 5 years prior to diagnosis lowered the risk of recurrence compared to never drinking (HR 0.11 95%CI 0.031 – 0.38). Restricting to those who did not consume alcohol, ALDH2 variant genotype was associated with increased risk of recurrence compared to wildtype, however the confidence intervals were imprecise (HR 12.89 95%CI 4.00-41.49).
Fig. 1.
Bladder cancer recurrence by SNP genotype. Kaplan-Meier plots depict the percent of patients by the time to their first bladder cancer recurrence in years. Shaded lines separate patients by genotype for a) ALDH2 rs2238151 among all patients, logrank P=0.0001 b) IGF1 rs5742714 among all patients, log-rank P=0.0018. Vertical hatches represent censoring due to death or end of follow-up period.
For non-invasive bladder cancer cases, participants with the heterozygous form of proliferation SNP IGF1 rs5742714 in Insulin-like Growth Factor 1 had shorter time to recurrence compared with wildtype (HR 1.61, FDR 0.20) (Table 2, Figure 1b). Despite limited statistical power, we also evaluated the recurrence – associated main effect SNPs in relation to tumor progression. IGF1 rs5742714 heterozygotes showed a non-significant trend towards an increased risk of tumor progression compared to wildtype (adjusted HR 2.24 95%CI 0.83-6.05).
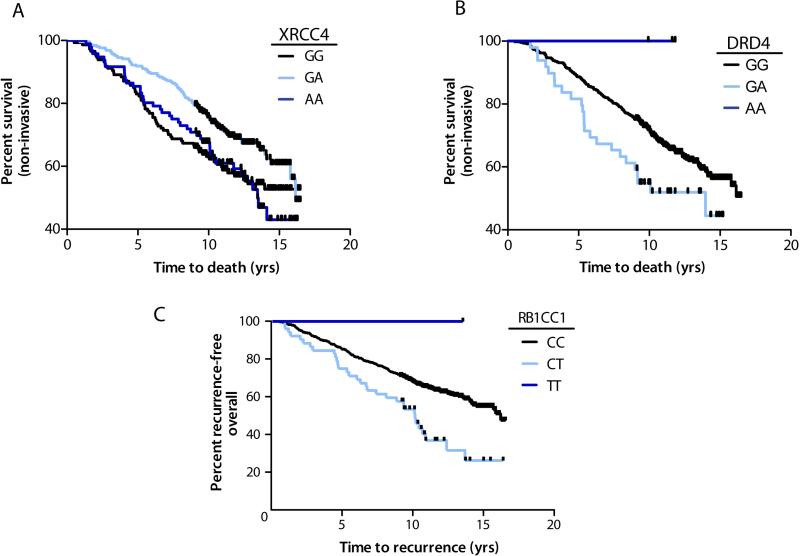
Table 3 shows the survival – associated SNPs based on analysis of the time from the initial bladder cancer diagnosis to death or censoring. For non-invasive cases, the double-strand DNA repair SNP XRCC4 rs2662238 was associated with longer survival among heterozygotes (HR 0.52, FDR <0.001) (Figure 2a). Heterozygotes for neural SNP in the dopamine receptor D4 DRD4 rs4987059 had shorter survival times (HR 1.83, FDR 0.02) (Figure 2b). A SNP in the proliferation pathway RB1-inducible coiled-coil 1 gene (RB1CC1 rs35402311) conferred shorter survival overall (HR 2.29 FDR <0.001) and for both non-invasive (HR 2.13) and invasive cancers (HR 3.86) (Figure 2c).
Fig. 2.
Bladder cancer survival by SNP genotype. Kaplan-Meier plots depict the percent of patients by the time to death in years. Shaded lines separate patients by genotype for a) XRCC4 rs2662238 restricted to patients with non- invasive tumors, logrank P=0.018, b) DRD4 rs4987059, restricted to patients with non- invasive tumors, log-rank P=0.041, and c) RB1CC1 rs35402311 among all patients, log-rank P=0.001. Vertical hatches represent censoring due to end of follow-up period.
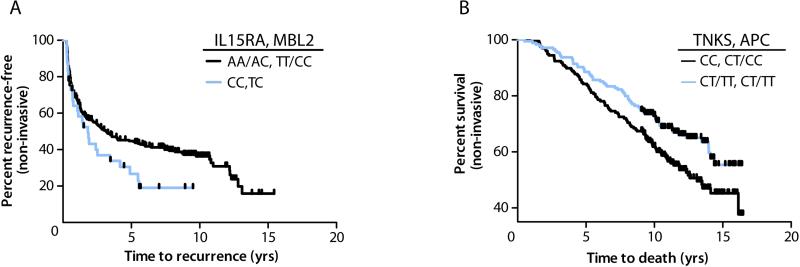
We also ran ‘Surv-MDR’ algorithms to detect potential gene-gene interactions in time-to-event data using log-rank statistics [19]. We ranked the two SNP model combinations within each of the major carcinogenesis processes in relation to recurrence and survival of non-invasive bladder cancer cases. Cox-regression analysis confirmed a multiplicative interaction for recurrence between the top model, including the variant forms of immune genes IL15RA rs2228059 and MBL2 rs2099902 heterozygosity (interaction P<0.0001) (Table 4). For survival, the combination of proliferation SNPs TNKS rs34206126 heterozygous/variant and APC rs2229992 heterozygous/variant showed a multiplicative association by Cox-regression analysis (interaction P=0.001) (Table 4).
Receiver operating characteristic curves demonstrate the predictive value of the models with and without the inclusion of SNP genotype. In Figure 4, the blue line shows the area under the curve (AUC) for the ‘Base’ models containing age, gender, smoking, stage, grade, treatment for non-muscle invasive urothelial cell carcinoma recurrence (Fig 4a,) and survival (Fig 4b). The addition of genotype parameters for ALDH2 rs2238151, IGF1 rs5742714, and IL15RA rs2228059 / MBL2 rs2099902 significantly improved the prediction of 5-year recurrence (AUC 0.71 to 0.75, chi-square P =0.02). Likewise, the AUC for 10-year survival went from 0.76 to 0.80 with the addition of SNP information for XRCC4 rs2662238 DRD4 rs4987059 RB1CC1 rs35402311 TNKS rs34206126*APC rs2229992 (P =0.008).
We then assessed the SNPs grouped into sets using Gene Set Enrichment Analysis. Using the nine major carcinogenesis pathway groups, we observed enrichment scores indicating shorter time to bladder cancer recurrence with the variant form of ‘immune response’ gene SNPs (FDR 0.06) and longer time to recurrence with ‘hormone regulation’ variant gene SNPs (FDR 0.11) (Table 5). We also tested for enrichment of the 600 GeneOntology (GO) Biological Processes in relation to recurrence and survival, however all FDR adjusted p-values exceeded 0.25.
We then tested the hypothesis that the effectiveness of immunotherapy treatment would be modified by SNPs in immune-related genes. We performed likelihood ratio testing for interactions between immune-related SNPs and immunotherapy in relation to recurrence and survival. The lowest interaction P-values was observed for VCAM1 rs3176879 heterozygous or variant genotype shortening time to recurrence for cases receiving immunotherapy treatment (interaction P<0.001, FDR 0.06) (Table 6). There was also a three-way multiplicative interaction between IL1B rs16944, IFNG_07 rs1861494 variant alleles and immunotherapy treatment (interaction P=0.003, HRs with immunotherapy: IL1B GG vs. GA/AA 0.34 (95%CI 0.17- 0.71), IFNG TT vs. TC/CC 0.40 (95%CI 0.22 - 0.75)), with longer time to recurrence for immunotherapy treated patients who were carriers of at least one variant allele for both genes (median recurrence time 9.76 years), compared to patients who were wildtype for both genes (median time to recurrence 1.30 years).
Analyses of the prognostic SNPs within each of the cohorts of bladder cancer patients separately demonstrate concordant results across the two different New Hampshire populations. Study phase I included n=303 genotyped patients, while study phase II included n=260. For recurrence for non-muscle invasive urothelial tumors, adjusted HRs were: ALDH_02 var., phase I HR 1.61 95%CI 0.93-2.78, phase II 2.58 95%CI 1.47-4.53; IGF1 rs5742714 het. Phase I HR 1.80 95%CI 1.16-2.80, phase II HR 1.78 95%CI 1.13-2.80). For survival, adjusted HRs were: XRCC4 rs2662238 het., phase I 0.48 95%CI 0.31-7.4, phase II 0.62 95%CI 0.34-1.12; DRD4 rs4987059 het. phase I HR 2.06 95%CI 1.16-3.68. phase II HR 1.52 95%CI 0.72-3.22; RB1CC1 rs35402311 het phase I HR 2.68 95%CI 1.42-5.04, phase II HR 1.44 95%CI 0.70-2.99. The interaction P-values were: phase I P<0.0001, phase II P=0.008 for IL15RA rs2228059/MBL2 rs2099902; phase I P=0.02, phase II P=0.01 TNKS rs34206126/APC rs2229992.
Discussion
We observed that genetic variations impacting several major carcinogenesis processes modified time to bladder cancer recurrence, progression and length of survival. We also identified SNP-SNP and SNP-treatment interactions significantly associated with these events. To our knowledge, our unique study has one of the only population-based datasets with detailed follow-up of patients for bladder cancer recurrence and survival information in the U.S.
We observed a higher rate of recurrence associated with variants in the Aldehyde dehydrogenase (ALDH2) enzyme, which performs oxidative catalysis of alcohol. ALDH2 SNPs have been associated with higher levels of smoking-DNA adducts in blood [22], with higher rates of esophageal cancer [23] and with lung cancer in smokers [24], possibly involving the DNA damaging effects of acetaldehyde [25]. Among cases with the variant ALDH2 rs2238151 genotype, alcohol consumption was associated with a longer time to tumor recurrence, possibly due to decreased enzymatic activity, which reduces formation of reactive intermediates. Treatment with the ALDH2 inhibitor, Disulfiram, lowers bladder cancer risk in rats exposed to nitrosamines, because the inhibitor prevents ALDH2 from converting the chemicals into reactive intermediates that are excreted through the bladder [26] (Suppl. Figure 4a).
We also observed a shorter time to first recurrence associated with a cell proliferation – related polymorphism in the Insulin-like Growth Factor 1 (IGF1). IGF1 activates AKT signaling to promote cell cycle, proliferation and inhibit apoptosis. The same SNP that we observed (rs5742714 C/G) in the 3’UTR was associated with survival for non-small cell lung cancer cases [27]. SNPs in this 3’UTR region could function by disrupting binding of a microRNA. IGF1 is a target of miR-1 and miR-206 [28] (Suppl. Figure 4b).
In the DNA repair pathway, heterozygotes for SNP rs2662238 in X-ray repair cross-complementing protein 4 (XRCC4) experienced longer survival. XRCC4 is involved in the ligation step of DNA double-strand break by non-homologous end joining (Suppl. Figure 4c). While XRCC4 SNPs have previously been associated with increased bladder cancer risk, our study is the first to report a relationship with prognosis. SNPs identified in prior studies of bladder cancer incidence include the XRCC4 SNP G-1394T rs6869366 in a Taiwanese study of n=158 cases and controls (low LD with XRCC4 rs2662238: R2 0.019) [29], intron 7 SNP rs1805377 in a Spanish study of 1150 cases and controls [30], and in 211 cases, 250 controls in North India and (low LD with XRCC4 rs2662238: R2 0.005) [31]. The observed contrast (increasing cancer risk, but lowering survival time) may be explained by compromised repair of DNA promoting mutations in tumor suppressor genes. Once a tumor has formed, however, it is conceivable that tumor cells with damaged DNA are more easily recognized for elimination by immunoediting machinery and compromised DNA repair increases chemotherapy efficacy, as observed for cisplatin treatment of lung cancer [32].
We also found an association between heterozygous genotype for the dopamine receptor D4 (DRD4) and 2-fold shorter survival. DRD4 polymorphisms are associated with increased novelty seeking behavior, including a propensity to take up smoking as an adolescent and with a higher intensity of smoking overall [33, 34].
RB1CC1 rs35402311 in the 5’ region near the RB1-inducible coiled-coil 1 gene was also associated with a 2.29 fold shorter survival time. RB1CC1 is a tumor suppressive transcription factor that regulates cell growth, cell proliferation, as well as apoptosis and cell migration by controlling RB1 (retinoblastoma 1) expression. RB1CC1 binds and stabilizes the tumor suppressor p53, is mutated in tumors, and was associated with multi-drug resistance to chemotherapy agents [35] (Suppl. Figure 4d).
Survival was lengthened for cases with variant forms of Wnt pathway regulators Tankyrase (TNKS) and Adenomatous polyposis coli (APC) gene, which is mutated in colon cancers. APC normally holds the cell cycle promoting transcription factor B-catenin in a proteolytic degradation complex, keeping levels low, however APC alterations compromise this control [36] (Suppl. Figure 4e). Tankyrase (TNKS) is a poly-ADP ribosylase that normally degrades Axin. Inhibiting TNKS stabilizes Axin, facilitating degradation of B-catenin, and preventing stem cell proliferation [37]. Levels of TNKS mRNA in urine are associated with bladder cancer recurrence [38]. The combination of TNKS and APC variations that both increase B-catenin degradation would block proliferation.
The Gene Set Enrichment Analysis identified clusters of SNPs related to longer time to recurrence in the ‘hormone’ and ‘immune’ sets. The predominance of bladder cancer in males, as well as other studies implicate a specific role for androgens and the androgen receptor in bladder cancer [40]. In the immune set, a SNP in the cytokine receptor antagonist IL15RA interacted with a SNP in mannose-binding lectin (protein C) 2 (MBL2) to increase bladder cancer recurrence rates (HR 4.32). MBL2 activates the complement pathway when it recognizes mannose and N-acetylglucosamine on microorganisms and also activates macrophages to destroy late apoptotic cells. MBL SNPs were previously associated with lung cancer survival [41].
We specifically hypothesized that immune response genotypes would modify the response to immunotherapy treatment. In a study of 99 patients randomized to several different BCG immunotherapy regimens, Chiong et al. found shorter time to bladder cancer recurrence associated with SNPs in the natural resistance-associated macrophage protein 1 (NRAMP1) gene{Chiong, 2011 #8867}. We observed a higher risk of recurrence among vascular cell adhesion molecule 1 (VCAM1) heterozygous or variant patients who received immunotherapy treatment (HR 5.0). VCAM1 is a cell surface glycoprotein involved in the development of lymphoid tissues. BCG treatment upregulated VCAM1 in mouse bladder tissue. VCAM1 upregulation could also increase lymphangiogenesis in the bladder [42].
The population-based nature of this study is an advantage for the potential generalizability of the results, allowing us to investigate genetic variations in relation to recurrence of non-muscle invasive lesions. Limitations include small numbers of highly invasive lesions and the possibility of false positive results. We have used the FDR method to reduce the false positive findings associated with statistical comparisons across many SNPs and also looked at the consistency of our results across two subsequent cohorts. Replication studies in other population-based studies of non-muscle invasive tumor recurrence are needed.
Our data suggest novel associations between SNPs and bladder cancer recurrence that merit future investigation. Prognostic variations will help identify sub-groups of bladder cancer cases at high risk of tumor recurrence and progression for more intensive tumor surveillance and targeted treatment regimens.
Supplementary Material
Fig. 3.
Bladder cancer prognosis by SNP genotype combination. Kaplan-Meier plots depict the percent of patients by the time to recurrence or death in years. Shaded lines separate patients with non-invasive tumors by genotype for a) IL15RA rs2228059/ MBL2 rs2099902 in relation to recurrence, log-rank P=0.05 and b) TNKS rs34206126/ APC rs2229992 in relation to survival, log-rank P=0.016. Vertical hatches represent censoring due to end of follow-up period.
Acknowledgments
This publication was funded in part by grant numbers 5P20RR024475-02, 8P20GM103534-02, RR028309, LM009012, CA102327, CA121382, CA099500, CA82354, CA57494, CA078609, ES00002, 5 P42 ES05947, RR018787, and ES07373 from the National Cancer Institute, NIH, from the National Institute of Environmental Health Sciences, NIH, the National Center for Research Resources, NIH, and the National Institute of General Medical Sciences, NIH. The New Hampshire State Cancer Registry is supported by the Centers for Disease Control and Prevention's National Program of Cancer Registries (NPCR) through cooperative agreement U58/DP000798 awarded to the New Hampshire Department of Health and Human Services, Division of Public Health Services, Bureau of Public Health Statistics & Informatics, Health Statistics and Data Management Section. The authors wish to thank the staff and participants of the New Hampshire Health Study and the New Hampshire State Cancer Registry for making this project possible.
Footnotes
Conflicts of interest: none reported
Conflict of Interest: None declared
Supplemental Data: Supplemental data includes Supplemental Figures 4a-e, and Supplemental Table 1.
References
- 1.Burger M, Catto JW, Dalbagni G, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013 Feb;63(2):234–41. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 2.Honma I, Masumori N, Sato E, et al. Local recurrence after radical cystectomy for invasive bladder cancer: an analysis of predictive factors. Urology. 2004 Oct;64(4):744–8. doi: 10.1016/j.urology.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R. The health economics of bladder cancer: a comprehensive review of the published literature. Pharmacoeconomics. 2003;21(18):1315–30. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 4.Strope SA, Montie JE. The causal role of cigarette smoking in bladder cancer initiation and progression, and the role of urologists in smoking cessation. J Urol. 2008 Jul;180(1):31–7. doi: 10.1016/j.juro.2008.03.045. discussion 7. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz-Drager BJ. Identifying risk factors in patients with non-muscle-invasive bladder cancer: clinical implications. Eur Urol. 2011 Oct;60(4):721–3. doi: 10.1016/j.eururo.2011.07.065. [DOI] [PubMed] [Google Scholar]
- 6.Habuchi T. Common genetic polymorphisms and prognosis of sporadic cancers: prostate cancer as a model. Future Oncol. 2006 Apr;2(2):233–45. doi: 10.2217/14796694.2.2.233. [DOI] [PubMed] [Google Scholar]
- 7.Sancar A, Lindsey-Boltz LA, Unsal-Kaccmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004:7339–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 8.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11(8):558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 9.Dudas A, Chovanec M. DNA double-strand break repair by homologous recombination. Mutat Res. 2004 Mar;566(2):131–67. doi: 10.1016/j.mrrev.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Lei AQ, Cheng L, Pan CX. Current treatment of metastatic bladder cancer and future directions. Expert Rev Anticancer Ther. 2011 Dec;11(12):1851–62. doi: 10.1586/era.11.181. [DOI] [PubMed] [Google Scholar]
- 11.Groth A, Kloss S, von Strandmann EP, Koehl U, Koch J. Mechanisms of tumor and viral immune escape from natural killer cell-mediated surveillance. J Innate Immun. 2011;3(4):344–54. doi: 10.1159/000327014. [DOI] [PubMed] [Google Scholar]
- 12.de Visser KE. Spontaneous immune responses to sporadic tumors: tumor-promoting, tumor-protective or both? Cancer Immunol Immunother. 2008 Oct;57(10):1531–9. doi: 10.1007/s00262-008-0501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simons MP, Nauseef WM, Griffith TS. Neutrophils and TRAIL: insights into BCG immunotherapy for bladder cancer. Immunol Res. 2007;39(1-3):79–93. doi: 10.1007/s12026-007-0084-1. [DOI] [PubMed] [Google Scholar]
- 14.Bartsch H, Rojas M, Nair U, Nair J, Alexandrov K. Genetic cancer susceptibility and DNA adducts: studies in smokers, tobacco chewers, and coke oven workers. Cancer Detect Prev. 1999;23(6):445–53. doi: 10.1046/j.1525-1500.1999.99055.x. [DOI] [PubMed] [Google Scholar]
- 15.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 16.Karagas MR, Tosteson TD, Blum J, Morris JS, Baron JA, Klaue B. Design of an epidemiologic study of drinking water arsenic exposure and skin and bladder cancer risk in a U.S. population. Environ Health Perspect. 1998;(Suppl 4):1047–50. doi: 10.1289/ehp.98106s41047. 1998/08//106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrew AS, Gui J, Sanderson AC, et al. Bladder cancer SNP panel predicts susceptibility and survival. Hum Genet. 2009 Mar 1;125(5-6):527–39. doi: 10.1007/s00439-009-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dennis G, Jr., Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(9):R60.1 – R.11. [PubMed] [Google Scholar]
- 19.Gui J, Moore JH, Kelsey KT, Marsit CJ, Karagas MR, Andrew AS. A novel survival multifactor dimensionality reduction method for detecting gene-gene interactions with application to bladder cancer prognosis. Hum Genet. 2011 Jan;129(1):101–10. doi: 10.1007/s00439-010-0905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coffey CS, Hebert PR, Ritchie MD, et al. An application of conditional logistic regression and multifactor dimensionality reduction for detecting gene-gene interactions on risk of myocardial infarction: the importance of model validation. BMC Bioinformatics. 2004 Apr 30;:549–59. doi: 10.1186/1471-2105-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coffey CS, Hebert PR, Krumholz HM, Morgan TM, Williams SM, Moore JH. Reporting of model validation procedures in human studies of genetic interactions. Nutrition. 2004 Jan;20(1):69–73. doi: 10.1016/j.nut.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Ichiba M, Iyadomi M, Zhang J, Tomokuni K. Effects of genetic polymorphism of metabolic enzymes, nutrition, and lifestyle factors on DNA adduct formation in lymphocytes. Ind Health. 1998 Oct;36(4):337–46. doi: 10.2486/indhealth.36.337. [DOI] [PubMed] [Google Scholar]
- 23.Itoga S, Nomura F, Makino Y, et al. Tandem repeat polymorphism of the CYP2E1 gene: an association study with esophageal cancer and lung cancer. Alcohol Clin Exp Res. 2002 Aug 26;(8 Suppl):15S–9S. doi: 10.1097/01.ALC.0000026828.13868.B5. [DOI] [PubMed] [Google Scholar]
- 24.Park JY, Matsuo K, Suzuki T, et al. Impact of smoking on lung cancer risk is stronger in those with the homozygous aldehyde dehydrogenase 2 null allele in a Japanese population. Carcinogenesis. 2010 Apr;31(4):660–5. doi: 10.1093/carcin/bgq021. [DOI] [PubMed] [Google Scholar]
- 25.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011 Jul 7;475(7354):53–8. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 26.McGregor NR. Pueraria lobata (Kudzu root) hangover remedies and acetaldehyde-associated neoplasm risk. Alcohol. 2007 Nov;41(7):469–78. doi: 10.1016/j.alcohol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M, Hu Z, Huang J, et al. A 3′-untranslated region polymorphism in IGF1 predicts survival of non-small cell lung cancer in a Chinese population. Clin Cancer Res. 2010 Feb 15;16(4):1236–44. doi: 10.1158/1078-0432.CCR-09-2719. [DOI] [PubMed] [Google Scholar]
- 28.Shan ZX, Lin QX, Fu YH, et al. Upregulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochem Biophys Res Commun. 2009 Apr 17;381(4):597–601. doi: 10.1016/j.bbrc.2009.02.097. [DOI] [PubMed] [Google Scholar]
- 29.Chang CH, Chang CL, Tsai CW, et al. Significant association of an XRCC4 single nucleotide polymorphism with bladder cancer susceptibility in Taiwan. Anticancer Res. 2009 May;29(5):1777–82. [PubMed] [Google Scholar]
- 30.Figueroa JD, Malats N, Rothman N, et al. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007 Aug;28(8):1788–93. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- 31.Mittal RD, Gangwar R, Mandal RK, Srivastava P, Ahirwar DK. Gene variants of XRCC4 and XRCC3 and their association with risk for urothelial bladder cancer. Mol Biol Rep. 2011 May 27;:391667–75. doi: 10.1007/s11033-011-0906-z. [DOI] [PubMed] [Google Scholar]
- 32.Su D, Ma S, Liu P, et al. Genetic polymorphisms and treatment response in advanced non-small cell lung cancer. Lung Cancer. 2007 May;56(2):281–8. doi: 10.1016/j.lungcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Laucht M, Becker K, El-Faddagh M, Hohm E, Schmidt MH. Association of the DRD4 exon III polymorphism with smoking in fifteen-year-olds: a mediating role for novelty seeking? J Am Acad Child Adolesc Psychiatry. 2005 May;44(5):477–84. doi: 10.1097/01.chi.0000155980.01792.7f. [DOI] [PubMed] [Google Scholar]
- 34.Das D, Tan X, Easteal S. Effect of model choice in genetic association studies: DRD4 exon III VNTR and cigarette use in young adults. Am J Med Genet B Neuropsychiatr Genet. 2011 Jan 13;:156346–51. doi: 10.1002/ajmg.b.31169. [DOI] [PubMed] [Google Scholar]
- 35.Chano T, Ikebuchi K, Ochi Y, et al. RB1CC1 activates RB1 pathway and inhibits proliferation and cologenic survival in human cancer. PLoS One. 2010;5(6):e11404. doi: 10.1371/journal.pone.0011404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phelps RA, Chidester S, Dehghanizadeh S, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009 May 15;137(4):623–34. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouelaa-Benslama R, Emami S. Pinworm and TNKS inhibitors, an eccentric duo to derail the oncogenic WNT pathway. Clin Res Hepatol Gastroenterol. 2011 Sep;35(8-9):534–8. doi: 10.1016/j.clinre.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 38.Gelmini S, Quattrone S, Malentacchi F, et al. Tankyrase-1 mRNA expression in bladder cancer and paired urine sediment: preliminary experience. Clin Chem Lab Med. 2007;45(7):862–6. doi: 10.1515/CCLM.2007.133. [DOI] [PubMed] [Google Scholar]
- 39.Mirabello L, Yu K, Kraft P, et al. The association of telomere length and genetic variation in telomere biology genes. Hum Mutat. 2010 Sep;31(9):1050–8. doi: 10.1002/humu.21314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyamoto H, Yang Z, Chen YT, et al. Promotion of bladder cancer development and progression by androgen receptor signals. J Natl Cancer Inst. 2007 Apr 4;99(7):558–68. doi: 10.1093/jnci/djk113. [DOI] [PubMed] [Google Scholar]
- 41.Pine SR, Mechanic LE, Ambs S, et al. Lung cancer survival and functional polymorphisms in MBL2, an innate-immunity gene. J Natl Cancer Inst. 2007 Sep 19;99(18):1401–9. doi: 10.1093/jnci/djm128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saban MR, O'Donnell MA, Hurst RE, et al. Molecular networks discriminating mouse bladder responses to intravesical bacillus Calmette-Guerin (BCG), LPS, and TNF-alpha. BMC Immunol. 2008:94. doi: 10.1186/1471-2172-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahirwar DK, Agrahari A, Mandhani A, Mittal RD. Cytokine gene polymorphisms are associated with risk of urinary bladder cancer and recurrence after BCG immunotherapy. Biomarkers. 2009 Jun;14(4):213–8. doi: 10.1080/13547500902818246. [DOI] [PubMed] [Google Scholar]
- 44.Taniguchi K, Koga S, Nishikido M, et al. Systemic immune response after intravesical instillation of bacille Calmette-Guerin (BCG) for superficial bladder cancer. Clin Exp Immunol. 1999 Jan;115(1):131–5. doi: 10.1046/j.1365-2249.1999.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.