Abstract
Development and progression of type 2 diabetes is a complex interaction between genetics and environmental influences. High dietary fat is one environmental factor that is conducive to the development of insulin-resistant diabetes. In the present report, we compare the responses of lean poly-genic, diabetic Goto-Kakizaki (GK) rats to those of control Wistar-Kyoto (WKY) rats fed a high fat diet from weaning to 20 weeks of age. This comparison included a wide array of physiological measurements along with gene expression profiling of abdominal adipose tissue using Affymetrix gene array chips. Animals of both strains fed a high fat diet or a normal diet were sacrificed at 4, 8, 12, 16, and 20 weeks for this comparison. The microarray analysis revealed that the two strains developed different adaptations to increased dietary fat. WKY rats decrease fatty acid synthesis and lipogenic processes whereas GK rats increase lipid elimination. However, on both diets the major differences between the two strains remained essentially the same. Specifically relative to the WKY strain, the GK strain showed lipoatrophy, chronic inflammation, and insulin resistance.
Keywords: diabetes, high fat diet, gene expression, microarray
Introduction
High dietary fat is one of the most common and important environmental risk factors associated with metabolic-related diseases such as type 2 diabetes mellitus (T2DM), hypertension, stroke, and coronary artery disease.1,2 High fat diets can induce weight gain and adiposity in both animals and humans. High dietary fat is often associated with hyperglycemia, hyperinsulinemia, hyper-triglyceridemia, high VLDL (very low-density lipoprotein) and LDL (low-density lipoprotein) in circulation.3,4 Such diets can also have extensive effects on insulin-responsive tissues.5 This is particularly true of adipose tissue, which is central to lipid homeostasis and secretes adipokines and bioactive lipids that regulate systemic energy balance. High fat diets may induce alterations in levels of secretion and action of adipokines and lipids as well as obesity-related macrophage invasion in adipose tissue.6–8 In addition, high fat diets may change the expression of a great number of genes in adipose tissue, decreasing the expression of many genes involved in lipid metabolism and detoxification, adipocyte differentiation, and cytoskeleton components, but increasing inflammatory markers.9,10 Several signaling pathways such as PPAR (peroxisome proliferator-activated receptor) signaling and Wnt signaling may also be modulated by high fat feeding.11–14
However, genetic background can greatly influence the response of both humans and experimental animals to a high fat diet. The differences in response may include not only the development of disease processes but also the effect of diet on gene expression. Rodent models are extensively employed to study the development and progression of a variety of human pathologies including those caused by high fat diet. Strain differences in the response to diet can provide important insight into the link between diet, gene expression, and diet-induced pathological conditions such as insulin- resistant diabetes.
The Goto-Kakizaki (GK) rat is a spontaneous diabetic animal model with a polygenic background. Their polygenetic mode of inheritance and non-obese phenotype make this animal model a useful surrogate for the study of human T2DM without the confounding influence of increased obesity- related factors. GK rats were originally developed in Japan by generations of breeding Wistar rats, selecting for high blood glucose.15 These rats exhibit a spontaneous form of diabetes, with elevated blood glucose, peripheral insulin resistance, and a non-obese phenotype being consistent features of these animals.15,16 Previous work demonstrated that disease progression involves both increased insulin resistance and beta-cell failure in these animals. While genetic factors play a major role in the etiology of disease in these non-obese animals, it is unclear if superimposing environmental factors such as high fat diet will impact disease progression in these animals.
Previously, we described an extensive analysis of adipose tissue from male diabetic GK and non-diabetic control Wistar-Kyoto (WKY) rats.17 In that study, rats from weaning through 20 weeks of age were fed a standard rodent diet containing 10% fat. Our analysis, which involved the substantial application of gene arrays, suggested that there is extensive inflammation caused by chronic activation of innate immunity in adipose tissue of the diabetic GK rats that is not evident in the adipose tissue of non-diabetic WKY animals. These data also demonstrated that there is an adipose tissue deficiency in the GK strain fed a normal diet. An unanswered question is whether chronic high fat feeding will influence adipose tissue mass, diabetes, and/or indices of heightened inflammation in this lean animal model of T2DM. In the present report, we used gene arrays to examine the response of adipose tissue from both strains to high fat feeding from weaning to 20 weeks of age with subgroups sacrificed at 4, 8, 12, 16, and 20 weeks. Within the context of extensive physiological measurements, we were able to identify genes whose expression is different in adipose tissue between the two strains regardless of diet. In addition, we identified genes that responded to high fat diet in both strains as well as those whose response to high fat diet was unique to either GK or WKY animals.
Materials and Methods
Experimental design
A more extensive description of this experiment can be found in our previously published report describing the array results on the livers from these animals.18 In brief, this study involved 25 GK spontaneously diabetic and 25 WKY non-diabetic male rats obtained from Taconic Farms (Germantown, NY) and maintained on a high fat diet (HFD: Harlan Teklad TD.06415, 45% energy from fat) from 3 weeks of age onward. Our research protocol adheres to the “Principles of Laboratory Animal Care” (NIH publication 85–23, revised in 1985) and was approved by the University at Buffalo Institutional Animal Care and Use Committee. Animals were received at 21 ± 3 days of age. For experimental purposes, animals were considered to be 22 days old at the time of arrival. They were maintained in our animal facilities under stringent environmental conditions with strict adherence to 12 hour:12 hour light:dark cycles. All animal care, manipulations, and sacrifices were carried out between 1.5 and 3.5 hours after lights on. Animals were housed in individual cages with free access to HFD and water. Food intake and body weights were measured twice weekly on all animals. Five animals from each strain were sacrificed at 4, 8, 12, 16, and 20 weeks of age by aortic exsanguination using EDTA as anticoagulant. Discrete abdominal fat pads (perirenal/retroperitoneal fat) as well as epididymal fat pads were harvested, weighed, rapidly frozen in liquid nitrogen, and warehoused at −80° C. Comparison was made to equivalent animals fed a normal diet (ND: Harlan Teklad 2016, 10% energy from fat).
Blood and plasma measurements
Blood glycosylated hemoglobin (HbA1C) was measured using A1cNOW InView HbA1C test meters (Metrika) from whole blood at sacrifice. Plasma glucose was measured by the glucose oxidase method (Sigma GAGO-20) modified such that the assay was carried out in a 1-mL assay volume. Plasma leptin, insulin, and adiponectin were measured by commercial ELISA assays (Rat Leptin TiterZyme EIA, Assay Designs; Ultra Sensitive Rat Insulin ELISA kit, Crystal Chem Inc; Rat Adiponectin EIA, ALPCO Diagnostics). All plasma assays were carried out according to manufacturer’s directions with standards run in duplicate and experimental samples run in triplicate. Two experimental samples were selected as “quality controls (QCs)” for all assays to control possible inter-assay variations. All inter- and intra-assay variations were within 10%.
RNA preparation
Abdominal adipose tissue samples from each animal were ground to a fine powder in a mortar cooled by liquid nitrogen and tissue was added to pre-chilled QIAzol Lysis Reagent (QIAGEN Sciences) in a weight/volume ratio of 1:10. Total RNAs were extracted according to manufacturer’s directions and further purified using RNeasy mini columns (RNeasy Mini Kit, QIAGEN Sciences). Final RNA preparations were eluted in RNase-free water, aliquoted and stored at −80° C. RNAs were quantified spectrophotometrically and exhibited 260/280 absorbance ratios of approximately 2.0. Purity and integrity were assessed by formaldehyde/agarose gel electrophoresis and all samples showed intact ribosomal 28S and 18S RNA bands in an approximate ratio of 2:1.
qRT-PCR
For validation purposes, gene-specific fluorescence-based real-time qRTPCR assays were developed. Our approach to quantitative RT-PCR involves use of in vitro transcribed cRNA standards, gene-specific TAQMAN-based probes, and a single-step assay with gene-specific mRNA normalized to total RNA in the assay.
Microarrays
Isolated RNA from each sample was used to prepare target according to manufacturer’s protocols. The biotinylated cRNAs were hybridized to 50 individual Affymetrix GeneChips Rat Genome 230–2 (Affymetrix, Inc.) which contains 31,099 different probe sets.
Data mining
Affymetrix Microarray Suite 5.0 (Affymetrix) was used for initial data acquisition and analysis. The signal intensities were normalized for each chip using a distribution of all genes around the 50th percentile. The generated dataset was submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/projects/geo/) database (GSE 13271). Gene-Spring 7 (Silicon Genetics) was employed for further analysis.
In order to objectively identify probe sets of interest, the entire dataset was subjected to similar analyses procedures and filtered with the same criteria as those applied to previous gene array datasets for an identical experiment where the animals had received a ND.17,19,20 These previous results were presented as a comparison between GK-ND and WKY-ND animals. Here with datasets from animals on HFD, three comparisons were made as follows: 1) GK-HFD vs WKY-HFD, 2) GK-HFD vs GK-ND, and 3) WKY-HFD vs WKY-ND. This approach does not select for probe sets but rather eliminates those probe sets that do not meet certain criteria, leaving the remainder for further consideration. In brief, the first filter eliminated genes not expressed in adipose tissue, ie, those not having a call function of “P” (present) in at least five of the 25 chips. The second level of filtering eliminated probe sets that could not meet the basic criterion of having a two-fold expression difference between compared groups.
Statistics
For statistical comparisons, two-way ANOVAs were carried out on raw or rank transformed data as appropriate using SigmaStat 3.5 software (Systat Software) with Tukey’s post hoc tests.
Results
Postnatal growth of GK and WKY on ND and HFD
As previously published,18 body weights of non-diabetic WKY animals were significantly heavier than those of diabetic GK animals from 8 weeks of age onward regardless of diet. High fat feeding caused an increase in body weights in both strains. By the end of the study, the effect of high fat feeding was fractionally similar in both strains, with animals on high fat being about 20% heavier than the same strain on the ND.
Adipose tissue development
Figure 1 presents adipose tissue weights taken at sacrifice. We have previously shown that there is almost a linear increase in abdominal fat from four weeks to 20 weeks of age in the non-diabetic WKY animals fed a ND,17 whereas the amount of adipose tissue ceases to increase in ND-GK animals starting at approximately 12 weeks of age. In contrast, fat mass in GK-HFD animals was significantly different than that of GK-ND animals (P < 0.001) from eight weeks onward. Essentially, high fat feeding resulted in abdominal fat masses in GK-HFD equivalent to those in WKY-ND animals (Fig. 1A). In contrast, the two WKY populations did not diverge until 20 weeks when the high fat fed animals showed an abrupt increase in adipose tissue. Therefore in the high fat fed animals, there was only a significant difference in abdominal adipose tissue weight between GK and WKY (P < 0.05) at 20 weeks, which appears to be because of an abrupt increase in the WKY population. However, the effect of high fat feeding was more pronounced in epididymal fat, as WKY-HFD had increased epididymal fat compared to GK-HFD animals from 12 weeks onward (Fig. 1B).
Figure 1.
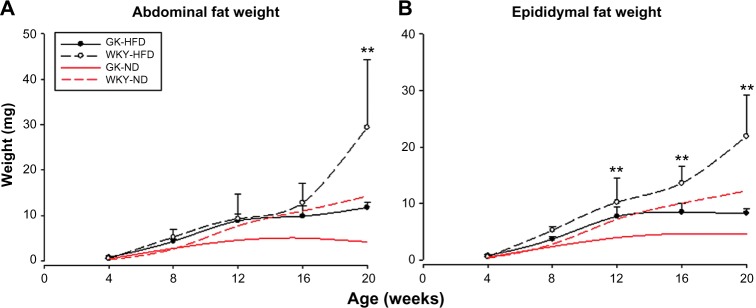
Adipose tissue weights at sacrifice as a function of age in GK animals (closed circles) and WKY animals (open circles) when animals are fed a high fat diet (black lines). Symbols represent means and error bars of one standard deviation. Red lines represent comparable data obtained from animals fed a normal diet and are provided for reference. (A) Abdominal fat pads; (B) Epididymal fat pads.
Notes: **WKY-HFD vs GK-HFD, P < 0.001.
Indices of diabetes
As we reported previously,18 the GK population had significantly higher plasma glucose as well as HbA1c than WKY (P < 0.001) from 4 weeks throughout the 20-week experimental period regardless of diet. Diet had no significant effect on plasma glucose or HbA1c in either strain (Supplementary Fig. 1). However, modest effects on plasma insulin were observed. There was a dramatic increase in plasma insulin between four and eight weeks in GK-ND animals, such that at eight weeks, insulin was significantly higher in GK relative to WKY (P < 0.001) after which it began to decline. In contrast, in the WKY-ND population, there was a slow increase in plasma insulin to a plateau after 12 weeks of age. High fat feeding did affect insulin dynamics in both strains. In the GK-HFD population, the increase in plasma insulin occurred more slowly, such that the peak before the decline occurred at 12 weeks instead of eight weeks. In the WKY-HFD population, plasma insulin showed an abrupt increase at 20 weeks suggesting that the animals are becoming hyperinsulinemic.
Adipose tissue derived hormones
As previously reported, when fed a ND plasma adiponectin decreased from four to 12 weeks in both GK and WKY populations with a trend for higher adiponectin in GK animals at all ages.17 In both strains, high fat feeding resulted in lower plasma adiponectin at early ages. However, high fat feeding magnified the difference in plasma adiponectin between GK and WKY animals. Figure 2A demonstrates that in high fat fed animals, plasma adiponectin was significantly higher in the GK population than in the WKY population throughout the entire experimental period (P < 0.001). Likewise, the higher plasma leptin concentrations in GK animals was magnified by high fat feeding (Fig. 2B), which reflected primarily an effect of diet on GK animals with no significant effects in control WKY animals.
Figure 2.
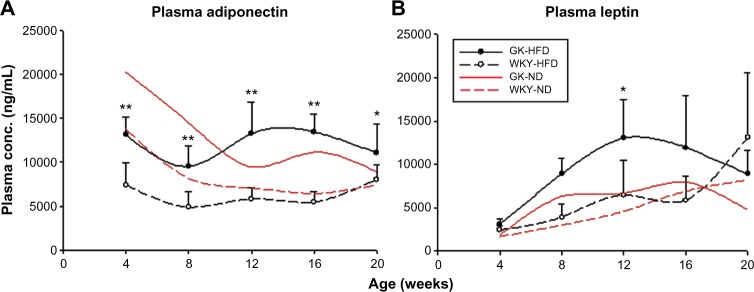
Comparison of plasma adipokine concentrations in GK-HFD and WKY-HFD animals as a function of age. Symbols, error bars, and lines are as described in Figure 1. (A) Adiponectin; (B) Leptin.
Notes: *P < 0.05; **P < 0.001.
Gene Expression – GK-HFD compared to WKY-HFD animals
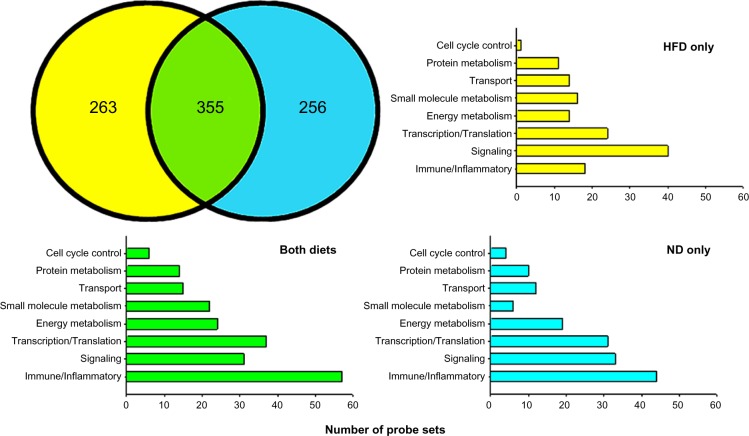
Based on our filtering approach, there were 618 probe sets that were differentially expressed in abdominal adipose tissue between GK-HFD and WKY-HFD animals. Previously using the same mining approach, we identified 611 probe sets that were differentially expressed in animals fed a ND.17 As illustrated in Figure 3, 355 of the probe sets were differentially expressed on both diets (green) while 263 were unique to HFD (yellow). These 618 probe sets are listed in Supplementary Table 1, with probe sets differentially regulated only with HFD highlighted in bold font. Of the 611 differentially regulated probe sets from ND fed animals, 256 of these were not differently expressed on a HFD (Fig. 3, blue). These probe sets are listed in Supplementary Table 2.
Figure 3.
Venn diagram showing the numbers of differentially regulated probe sets between strains regardless of diet (green), and those regulated between strains only when animals are fed a high fat diet (yellow). Probe sets regulated only when animals are fed a normal diet are depicted in blue.
Using the NCBI Basic Local Alignment Search Tool (BLAST) and the accession number for the probe set provided by Affymetrix, we identified corresponding genes to differentially regulated probe sets. There were 169 probe sets whose gene could not be identified by the BLAST program, leaving 449 identifiable individual genes that exhibit differential expression in abdominal adipose tissue of GK-HFD vs WKY-HFD animals. We then used NCBI “across database search” to identify all aliases and alternate symbols. Next, we applied NCBI AceView and extensive PubMed Boolean logic searches to ascertain the function of the gene in adipose tissue. Based on this information, we separated genes into groups based on their function in adipose tissue. These groups were as follows: Immune/Inflammatory (75 probe sets); Signal Transduction (71 probe sets); Transcription/Translation (61 probe sets); Energy Metabolism (38 probe sets); Small Molecule Metabolism (38 probe sets); Transport (29 probe sets); Protein Metabolism (25 probe sets); and Cell Cycle Control (seven probe sets). Figure 3 also presents a breakdown of the number of probe sets in each functional category which are differentially expressed on both diets, on HFD only and on ND only. Genes which did not readily fit into these categories were grouped as Other, and probe sets not identifiable by BLAST were listed as ESTs on Supplementary Table 1.
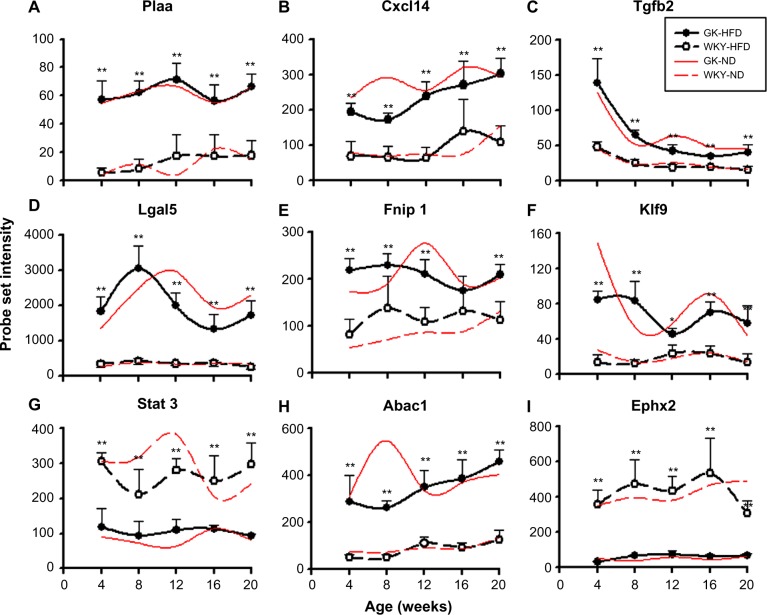
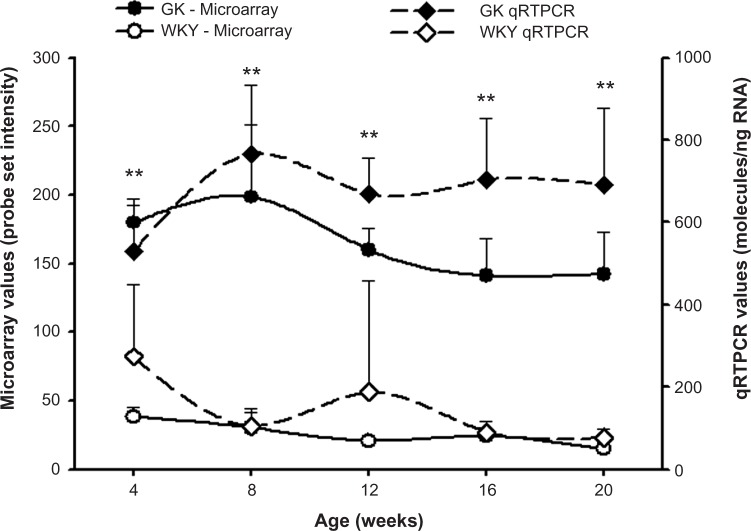
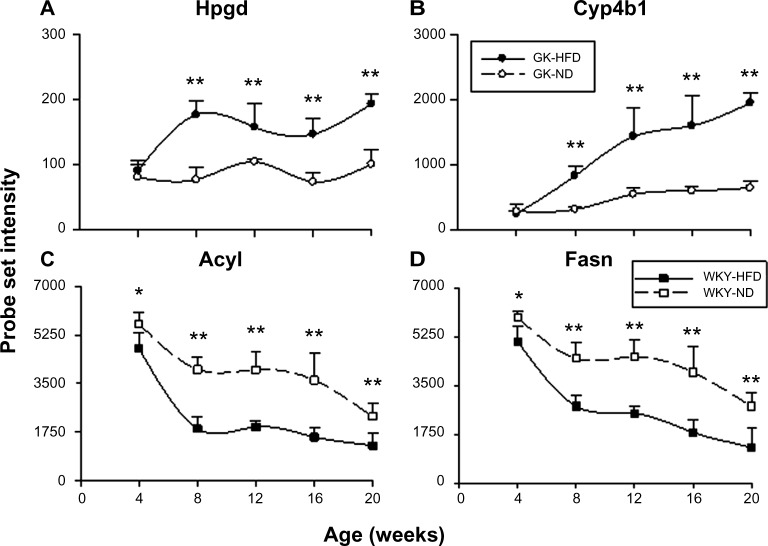
Four of the most populated functional categories (Immune/Inflammatory; Signal Transduction; Transcription/Translation; and Energy Metabolism) included many genes relevant to diabetes. Figure 4 presents representative examples of probe sets from these categories, which were expressed to different degrees in GK and WKY animals when fed a high fat diet. These exhibited similar differential regulation when animals were fed a normal rodent diet. In addition, Figure 5 provides an example of a direct comparison of microarray and qRTPCR data. Figure 6 presents examples of two diabetes-related genes exhibiting different expression levels only when animals are fed a high fat diet.
Figure 4.
Representative probe sets which are differentially regulated between strains regardless of diet. Comparisons are made between GK animals (closed circles) fed a high fat diet (black lines) and WKY animals (open circles) fed a high fat diet (black lines), with symbol representing means and error bars of one standard deviation. Comparable data from animals fed a normal diet (red lines) are provided for reference.
Notes: *P < 0.05; **P < 0.001.
Figure 5.
Ifit expression in GK-HFD animals (closed symbols) and WKY-HFD animals (open symbols) measured by microarray analysis (circles) and qRT-PCR (diamonds).
Figure 6.
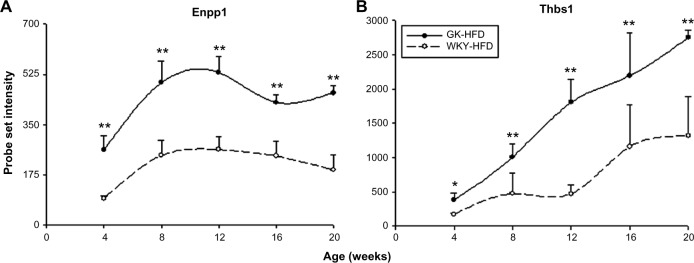
Representative probe sets which are differentially regulated between strains only when animals are fed a high fat diet. Symbols, error bars, and lines are as described in Figure 4.
Notes: *P < 0.05; **P < 0.001.
Gene expression changes in response to HFD by strain
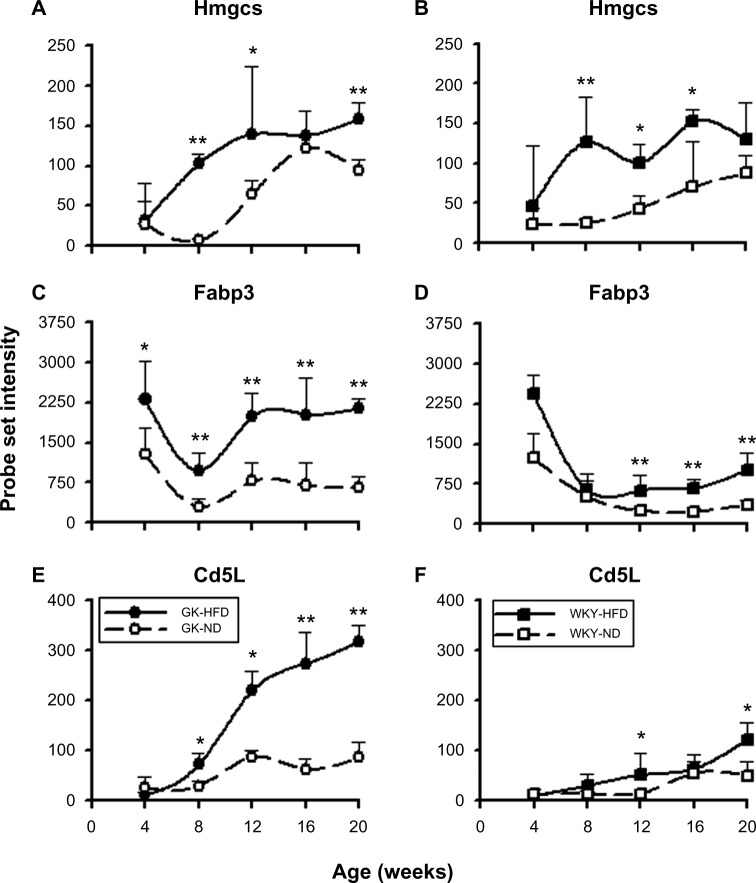
A different perspective can be gained by identifying genes whose expression changed in response to high fat feeding compared to the same strain fed a ND. A total of 262 probe sets were identified as being regulated by diet. Of these 33 were regulated by diet in both strains (Supplementary Table 3), 159 were differentially expressed in HFD compared to ND only in GK animals but not in WKY animals (Supplementary Table 4), and 70 were different only in WKY but not in GK (Supplementary Table 5). Figure 7 presents examples of probe sets whose expression levels differed with diet in both GK and WKY animals. In contrast to genes regulated by diet in both rat strains, Figure 8 presents probe sets regulated by diet either in GK animals only (A,B) or in WKY animals only (C,D).
Figure 7.
Representative probe sets which are differentially regulated by diet in both strains. Panels A, C, and E (left side) illustrate regulation in GK animals, while panels B, D, and F (right side) present data from WKY animals. Comparisons are made between high fat diet (closed symbols) and normal diet (open symbols).
Notes: *P < 0.05; **P < 0.001.
Figure 8.
Representative probe sets which are differentially regulated by diet only in GK animals (A, B) or regulated by diet only in WKY animals (C,D). Comparisons are made between high fat diet (closed symbols) and normal diet (open symbols).
Notes: *P < 0.05; **P < 0.001.
Discussion
A diet high in fat is one factor driving the increase in obesity in western society. Attendant on this increase in obesity is an increase in the incidence of T2DM. It is thought that as adipose tissue mass increases, macrophages invade causing a state of chronic systemic inflammation that disrupts the metabolic balance causing T2DM.21–24 While obesity is clearly a risk factor for diabetes, the relationship between obesity and diabetes is not absolute. This suggests that genetic background in association with environmental risk factors plays an important role in the development of the disease. This conclusion is reinforced by the fact that about 60% of Asian diabetics are lean.25 High fat feeding is an important approach to developing T2DM in animal models. However, whether or not increased dietary fat leads to the development of rodent diabetes is strongly dependent on genetic background. In the present study, we evaluated the effect of a HFD on development of adipose tissue from four weeks to 20 weeks after birth in both the diabetic GK and non-diabetic WKY control strains. We used this evaluation to conduct a within strain and cross strain analysis with the same time series progression when animals were fed a ND.
High fat feeding had only minimal effects on plasma glucose and insulin. The acute elevation of plasma insulin after four weeks of age in the GK strain regardless of diet probably reflects an attempt by β islet cells to compensate for hyperglycemia. The only effect of high fat feeding on the GK animals is that the increase in insulin is less acute and peaks later on HFD than on ND before beginning to decline. The decline in plasma insulin after the peak supports the observations of others that with time there is β-cell failure in the GK strain.16 HFDs are considered a risk factor for diabetes in both humans and animals. However, the significantly lower plasma insulin at eight weeks in the GK animals on HFD relative to ND along with the tendency toward lower plasma glucose and HbA1c in the HFD animals (Supplementary Fig. 1) suggests that the HFD may actually provide some mitigation for the diabetes. It has been reported that in both humans and animals, individuals are often hyperinsulinemic for a period of time prior to becoming hyperglycemic.26 One hypothesis for these observations is that chronic elevation in insulin output from the β-cells is a compensation for developing insulin resistance which eventually leads to β-cell failure and the expression of T2DM. The significant elevation of plasma insulin at 20 weeks in the WKY animals on HFD suggests the possibility that these animals are becoming insulin resistant because of increased fat consumption.
Previously, we reported that the diabetic GK strain was deficient in adipose tissue relative to the control WKY strain when both are fed a normal rodent diet (10% calories derived from fat).17 Both of these inbred strains of rat were derived from an outbred colony of Wistar rats at Kyoto University. WKY rats fed a ND had continuous increases in adipose mass, while the increase halted at around 12 weeks of age in GK animals. In the rat, the number of mature adipocytes expands for about the first 12 weeks of life. After that time, any increase in adipose tissue mass involves the expansion of the triglyceride content of individual mature adipocytes. Our results are consistent with the histological results reported by Barbu et al showing that white adipose tissue from the mature GK strain contained an unusually large number of immature adipocytes.27 The present study addressed the question of whether high fat feeding affected body weight and fat mass in the lean GK animals. High fat diet increased both body weight and adipose mass in GK animals. High fat diet causes a significant increase in adipose accumulation in the GK animals from eight weeks on such that adipose weights were comparable to those in WKY animals, while any effect of HFD on the control WKY animals is only observed at 20 weeks (Fig. 1). Since in rats, increases in adipose tissue mass after 12 weeks of age are because of an increase in triglyceride content of mature adipocytes,28 these results may indicate that the increased weight of white adipose tissue in GK animals that have fewer mature adipocytes reflects increased triglyceride content of the fewer than normal mature adipocytes that are present. Epididymal fat development shows both similarities and differences to abdominal adipose tissue development. Like abdominal fat, on ND epididymal fat accumulation ceases after 12 weeks in the GK population while continues to increase in WKY animals. In contrast to abdominal fat, epididymal fat seems to be more responsive to HFD as indicated by a greater increase in both strains.
Adiponectin is an adipocyte-derived hormone. In our previous work, we observed no significant circadian rhythm in the plasma concentration of adiponectin, so time of measurement may not be relevant.29 In adult animals, plasma adiponectin concentrations are generally inversely proportional to the amount of body fat.30 From that perspective, our results showing that plasma adiponectin is significantly higher in the GK strain regardless of diet (Fig. 2A) is consistent with the literature. However, since adiponectin is reported to be insulin sensitizing, our results suggests that either the insulin-resistant GK animals are insensitive to the hormone or incapable of responding to this hormone. Leptin is another adipocyte-derived hormone. In contrast to adiponectin, our previous work demonstrated a distinct circadian rhythm in plasma leptin, with peak concentration around the dark-to-light transition, and nadir around the light-to-dark transition.29 Therefore, the time at which measurements were taken in the current study would reflect concentrations near the high point of the circadian rhythm in a normal Wistar rat on a 12:12 light:dark regimen. In adult rats plasma leptin is proportional to the amount of fat. Leptin acts on the hypothalamus to suppress hunger and is a potent anorexic and energy-enhancing hormone associated with decreased appetite and increased metabolic rate. It increases fatty acid oxidation, stimulates adipose tissue lipolysis and inhibits lipogenesis. Leptin production increases proportionally with adiposity and is higher in rodent and human models of diet-induced or adult-onset obesity.31 In our data, although there is no difference at four weeks, plasma leptin is significantly higher in GK rats compared to WKY control rats at eight weeks on both diets and even higher at 12 weeks in GK rats on HFD (Fig. 2B). Combined with the fact that both abdominal and epididymal adipose tissue in GK rats are less than or not different from control rats at these times and the leptin level should be proportional to the adipose tissue weight, it is interesting that plasma leptin is two-fold higher in GK rats. This hyperleptinemia likely indicates leptin resistance in GK rats. High fat diet significantly increases plasma leptin in GK rats from eight weeks onward, but not in control rats. This result is consistent with literature reports, since HFD significantly increased both abdominal fat and epididymal fat in GK rats from eight weeks to 20 weeks. High fat diet increased adipose weight in WKY control rats at 20 weeks which may explain the abrupt increase of plasma leptin in control rats on HFD at this age.
Data mining of array data yielded 355 probe sets differentially regulated between GK and WKY animals regardless of diet (Fig. 3). A group of genes that is consistently differentially expressed between the two strains on both diets also lead us to conclude that there is a defect in adipocyte maturation in the GK strain. For example, KLF9 is a transcription factor whose expression is transiently increased during preadipocyte differentiation but decreased in mature adipocytes.32 Klf9 is involved in the middle stage of adipocyte differentiation and is involved in the expression of PPARγ2.32 Klf9 expression is higher on both diets in GK (Fig. 4F). Similarly, the observation that Tnrc6 is higher in WKY controls on both diets is consistent with the defect in adipocyte differentiation in the GK strain (Supplementary Table 1). TNRC6 is a component of the miRNP silencing complex and miRNA-mediated gene silencing is important in preadipocyte differentiation.33 Another important gene whose expression is higher in GK regardless of diet is transforming growth factor, beta 2 (Tgfb2, Fig. 4C). Increased expression of TGFb2 inhibits adipogenesis.34,35
In addition, the lipoatrophy in GK rats also involves genes related to lipogenesis. STAT3 (Fig. 4G), which is stimulated by leptin, is a transcription factor responsible for increased lipogenesis and GUCY1B3 (Supplementary Table 1) is involved in nitric oxide-mediated adipogenesis.14,36 Both of these genes have lower expression in GK rats, implying that the adipose tissue of GK rats may also have defects in both lipogenesis and adipogenesis. In contrast, folliculin-interacting protein 1 (Fnip1, Fig. 4E), which may be involved in energy and/or nutrient sensing through the AMPK (AMP-activated protein kinase) and mTOR (mammalian target of rapamycin) signaling pathways37 is more highly expressed in GK on both diets. A gene whose expression was higher in GK at all ages under both diets is ATP-binding cassette, sub-family A, member 1 (Abca1, Fig. 4H). ABCA1 is a cholesterol efflux pump in the cellular lipid removal pathway.38 Taken together, these results suggest that the lipoatrophy in GK rats is independent of diet and multifactorial involving defect in lipogenesis, inability of lipid storage and accumulation, and impaired preadipocyte differentiation.
Notable among those genes whose expression is different between the two strains on both diets are several genes related to inflammation. For example, Cxcl14 is consistently higher in GK on both diets (Fig. 4B). CXCL14 is a secreted protein that is involved in attracting macrophages into WAT.39 It is interesting that this message is significantly higher in the leaner GK animals. Other examples include Ifit1, which is associated with chronic inflammation and insulin resistance40,41 (Fig. 5), and Iigp1 (Supplementary Table 1). The higher expression of these genes in GK on both diets indicates that chronic inflammation is an intrinsic difference between the two strains. This conclusion is reinforced by the observation that Plaa is more highly expressed in adipose tissue in GK at all ages on both diets (Fig. 4A). PLAA mediates the endothelial and smooth muscle response to inflammation. Also consistent with this conclusion is the lower expression of Otub1 in GK on both diets (Supplementary Table 1). OTUB1 is a cytokine signaling pathway inhibitor involved in T cell anergy.42 In addition lectin, galactose binding, soluble 5 (Lgals5, Fig. 4D) which is involved in the regulation of NF-κB (nuclear factor κB) signaling43 is higher in GK on both diets. Our time series analysis of gene expression in tissues from the GK strain, which included not only adipose tissue but also liver and skeletal muscle, indicated that there is a heightened expression of genes involved in natural immunity in all three tissues,17,19,20 which is reinforced here in adipose tissue taken from animals fed a HFD. Additional genes related to immune/inflammatory activity can be found in Supplementary Table 1. The heightened natural immune-mediated inflammation is most likely the cause of diabetes in the GK strain. This conclusion is supported by a recent report from our group demonstrating the anti- inflammatory drug salsalate which inhibits NF-κB activation greatly decreases hyperglycemia in the GK strain.44,45
Besides the strain difference common to both diets, we also noted strain difference unique to high fat fed animals (263 probe sets – Fig. 3). We mined the array data for genes differentially expressed between GK and WKY controls only when fed on HFD, which provides insight into the underlying difference unique to high fat feeding. Interestingly, there are two diabetes biomarker genes present on this list (Fig. 6). ENPP1 is a membrane glycoprotein ectoenzyme and the high expression of this gene is associated with type 2 diabetes, hyperglycemia, and hyperinsulinemia in both human and experimental animals.46 The mechanism of ENPP1 in the disease is to inhibit the insulin signaling pathway and it also inhibits preadipocyte differentiation in adipose tissue.47,48 Thus, the higher expression of Enpp1 in GK rats is an indicator of diabetes in these animals (Fig. 6A). THBS1 is another biomarker of insulin resistance and diabetes, and is a novel adipokine primarily produced by visceral adipocytes.49 Highly expressed Thbs1 is noted in adipose tissue of obese insulin-resistant subjects and diabetic obese rodents and THBS1 blocking therapy is a promising treatment for diabetes.50,51 Adipose tissue expression of Thbs1 in GK rats increases with age and is significantly higher than that in WKY control rats at all 5 ages, which strongly supports the diabetic status of GK rats (Fig. 6B). However, it is interesting that the differential expression of these two genes are only exhibited on HFD, although we do observe the existence of diabetes in GK rats on a ND as indicated by the elevated plasma glucose. The reason may be either there is no difference in the expression of these genes between strains on ND or the difference is not large enough to pass our filtering criteria (two-fold difference). These results suggest that the exposure to HFD augments the difference between these two strains, and may exacerbate the diabetic state or accelerate disease progression in GK rats.
Additionally, data mining was utilized to extract probe sets differentially regulated by diet within strain. A comparison of diet within strain indicated 262 probe sets were differentially regulated by HFD in at least one of the two strains studied. Of these, only 33 were regulated in both GK and WKY control strains, suggesting that they are regulated by food composition with little influence of genetic background. In contrast, 159 probes sets were regulated by diet only in GK animals, and 70 were regulated only in WKY control animals. Thus, the diabetic GK animals were more highly impacted at the transcriptional level than normoglycemic WKY controls by increased dietary fat.
Although both strains show increased adiposity and similar adaptive responses, WKY and GK rats also develop different strategies to deal with increased fat consumption, as suggested by the genes differentially regulated by HFD in the two different strains. Differentially expressed genes unique to non-diabetic WKY rats mainly are those involved in fatty acid synthesis and lipogenesis, including Fasn (Fig. 8D, Acly (Fig. 8C), Me1 (Supplementary Table 5), and Elovl6 (Supplementary Table 5). For example, Fasn encodes a lipo-genic enzyme that catalyzes the synthesis of palmitate from acetyl-CoA into long-chain saturated fatty acids.52,53 ME1 converts malate to pyruvate, which links the glycolytic and citric acid cycles, and thus is involved in fatty acid synthesis.54 The expressions of all these genes are downregulated by HFD and show a very similar pattern to each other. Since those genes are exclusively differentially regulated in WKY rats, it indicates that these rats uniquely decrease fatty acid synthesis and lipogenesis by coordinately downregulating lipogenic genes as an adaptive response to high fat feeding. In contrast, GK rats develop a different metabolic strategy to handle the HFD as suggested by genes showing differential expression unique to GK rats. Among those genes are Hpgd (Fig. 8A) and Cyp4b1 (Fig. 8B). HPGD is a prostaglandin receptor and is involved in free fatty acid metabolism and degradation, especially prostaglandins, which are also inflammatory mediators.55 CYP4B1 is a member of the cytochrome P450 family, responsible for regulating PPARγ ligand homeostasis by promoting their elimination. FFA and eicosanoids are both PPARγ ligands and eicosanoids are also inflammatory mediators.56 The expression of both genes is significantly increased by HFD from eight weeks onward, suggesting that GK rats adapt to the increased dietary fat content by accelerating the lipid elimination process instead of decreasing lipogenesis. In addition, since HPGD and CYP4B1 are both involved in inflammatory mediator degradation, higher expression on HFD indicates suppressed inflammatory responses which may be a unique adaptation in GK to what would otherwise be an even more elevated inflammatory state induced by high fat feeding.
In our study, we also made extensive measurements related to postnatal growth, glucose levels, plasma hormones, and lipids. Although we did not observe HFD-induced hyperglycemia, the increased body weight, fat weight, and elevated circulating free fatty acids (FFAs) in both strains are consistent with literature reports. Microarray data of Hmgcs2 and Cd5l, which are differentially regulated by HFD regardless of strain (Fig. 7), reflect these changes in adipose tissue. HMGCS2 is a mitochondrial enzyme involved in ketone body synthesis and increased ketogenesis.57,58 Since the ketone bodies are metabolic products of mitochondrial FFA β-oxidation, the elevated expression of Hmgcs2 in both strains indicates an increase in the utilization of lipids in adipose tissue, which may be an adaptive response to HFD-induced plasma FFA elevation. In addition, the magnitude of increase in Hmgcs2 expression and the difference of food consumption between diets are almost the same, but weight gain and fat accumulation in GK rats are much more prominent than those in control rats, suggesting that GK rats are more capable of using lipids over carbohydrates than WKY control rats. Similarly, high fat consumption increased the expression of fatty acid binding protein 3 (Fabp3) in both strains (Fig. 7C, D). FABP3 is involved in the intracellular transport and metabolism of long-chain fatty acids.59
Also related to high fat-induced weight gain and fat accumulation is the adipose tissue inflammation capacity. Recent studies show that obesity is associated with macrophage infiltration into adipose tissue and heightened inflammatory cytokine levels, such as TNF-α (tumor necrosis factor α) and IL-6 (interleukin 6), in both adipose tissue and circulation, indicating a chronic inflammatory state.24,60 In our study, we did not observe an elevation of either circulating TNF-α or tissue mRNA expression of TNF-α or IL-6 by HFD (data not shown). White blood cell (WBC) counts were not different between diets, but significantly higher in GKs than controls on comparable diets. Expressions of interferon-induced genes, such as Ifit1 and Iigp1 in adipose tissue, are also not significantly regulated by diet, although higher in GKs than in controls regardless of diet. However, as indicated by the higher expression of Cd5l on HFD in both strains (Fig. 7), which is a well-known macrophage survival protein and whose origin is macrophages, recruitment and infiltration of macrophage into adipose tissue does occur along with fat expansion and precedes other indications of chronic inflammation in adipose tissue.
Although HFD has some beneficial effects of suppressing inflammatory response on GK rats as indicated by the higher expression of Hpgd and Cyp4b1 (Fig. 8), we do observe the significantly higher expression of diabetes biomarkers Enpp1 and Thbs1 in diabetic GK rats compared to non-diabetic WKY rats on HFD (Fig. 6). Taken together, these results suggest that high fat consumption may alter diabetes disease progression before the signs are manifested phenotypically as elevated blood glucose. Thus, the involvement of genes associated with adaptive responses may contribute to a delay in deterioration of the disease symptoms and indicate a degree of self- adjustment by the system.
In conclusion, HFD feeding induces increased lipid utilization as an adaptive response to elevated circulating free fatty acid and macrophage invasion associated with adiposity regardless of strain. In addition, microarray analysis also reveals that WKY rats and GK rats develop different adaptations to increased dietary fat. WKY rats decrease the fatty acid synthesis and lipogenesis processes, whereas GK rats increase lipid elimination. Furthermore, the ability of GK rats to increase the degradation of inflammatory mediators indicates a unique adaptation to what otherwise may be the elevated inflammation induced by HFD. Comparisons of strains on HFD reinforce the differences observed on ND, especially presence of lipoatrophy, chronic inflammation, and insulin resistance in GK rats. The molecular mechanism behind the lipoatrophy of GK rats may involve three aspects: a defect in lipogenesis, an inability of lipid storing and accumulation, and an impaired preadipocyte differentiation. It is of special interest that several diabetes biomarkers were only differentially expressed between strains in high fat fed animals, which may suggest that exposure to HFD perhaps does influence the disease in GK rats subclinically, either exacerbating the diabetic state or accelerating disease progression.
Supplementary Material
Supplementary Figure 1. Plasma glucose (A) and insulin (B) in GK and WKY animals as a function of age. Symbols are defined in Figure 1.
Notes: ##GK-ND vs WKY-ND, P < 0.001. *GK-HFD vs WKY-HFD, P < 0.05. **GK-HFD vs WKY-HFD, P < 0.001.
Supplementary Table 1. Differentially expressed genes in GK versus WKY animals fed hfd.
Supplementary Table 2. Differentially expressed genes in GK versus WKY animals fed nd.
Supplementary Table 3. Differentially regulated by diet in both strains.
Supplementary Table 4. Differentially regulated by diet only in GK animals.
Supplementary Table 5. Differentially regulated by diet only in WKY animals.
Footnotes
ACADEMIC EDITOR: James Willey, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,709 words, excluding any confidential comments to the academic editor.
FUNDING: This work was partly supported by grant GM 24211 from the National Institute of General Medical Sciences, NIH, Bethesda, MD, and by funds from the UB-Pfizer Strategic Alliance. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties.
Author Contributions
Conceived and designed experiments: RRA, DCD, WJJ. Analyzed the data: RRA, BX, DCD. Wrote first draft: RRA, DCD. Contributed to writing: BX, JN, XW, DCD, WJJ, RRA. Agree with results and conclusions: BX, JN, XW, DCD, WJJ, RRA. Jointly developed structure: BX, JN, XW, DCD, WJJ, RRA. Made critical revisions: BX, JN, XW, DCD, WJJ, RRA. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Nutritional recommendations and principles for individuals with diabetes mellitus: 1986. American Diabetes Association. Diabetes Care. 1987;10(1):126–32. doi: 10.2337/diacare.10.1.126. [DOI] [PubMed] [Google Scholar]
- 2.Storlien LH, James DE, Burleigh KM, Chisholm DJ, Kraegen EW. Fat feeding causes widespread in vivo insulin resistance, decreased energy expenditure, and obesity in rats. Am J Physiol. 1986;251(5 pt 1):E576–83. doi: 10.1152/ajpendo.1986.251.5.E576. [DOI] [PubMed] [Google Scholar]
- 3.Reaven GM. Banting lecture 1988. Role of insulin resistance in human disease. 1988. Nutrition. 1997;13(1):65. doi: 10.1016/s0899-9007(96)00380-2. [DOI] [PubMed] [Google Scholar]
- 4.Reaven GM, Lerner RL, Stern MP, Farquhar JW. Role of insulin in endogenous hypertriglyceridemia. J Clin Invest. 1967;46(11):1756–67. doi: 10.1172/JCI105666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tannenbaum BM, Brindley DN, Tannenbaum GS, Dallman MF, McArthur MD, Meaney MJ. High-fat feeding alters both basal and stress-induced hypothalamic-pituitary-adrenal activity in the rat. Am J Physiol. 1997;273(6 pt 1):E1168–77. doi: 10.1152/ajpendo.1997.273.6.E1168. [DOI] [PubMed] [Google Scholar]
- 6.Barnea M, Shamay A, Stark AH, Madar Z. A high-fat diet has a tissue-specific effect on adiponectin and related enzyme expression. Obesity. 2006;14(12):2145–53. doi: 10.1038/oby.2006.251. [DOI] [PubMed] [Google Scholar]
- 7.Scarpace PJ, Zhang Y. Leptin resistance: a prediposing factor for diet-induced obesity. Am J Physiol Regul Integr Comp Physiol. 2009;296(3):R493–500. doi: 10.1152/ajpregu.90669.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Heek M, Compton DS, France CF, et al. Diet-induced obese mice develop peripheral, but not central, resistance to leptin. J Clin Invest. 1997;99(3):385–90. doi: 10.1172/JCI119171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller MP, Attie AD. Physiological insights gained from gene expression analysis in obesity and diabetes. Annu Rev Nutr. 2010;30:341–64. doi: 10.1146/annurev.nutr.012809.104747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moraes RC, Blondet A, Birkenkamp-Demtroeder K, et al. Study of the alteration of gene expression in adipose tissue of diet-induced obese mice by micro array and reverse transcription-polymerase chain reaction analyses. Endocrinology. 2003;144(11):4773–82. doi: 10.1210/en.2003-0456. [DOI] [PubMed] [Google Scholar]
- 11.Bunger M, Hooiveld GJ, Kersten S, Muller M. Exploration of PPAR functions by microarray technology – a paradigm for nutrigenomics. Biochim Biophys Acta. 2007;1771(8):1046–64. doi: 10.1016/j.bbalip.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294(5548):1866–70. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 13.Christodoulides C, Lagathu C, Sethi JK, Vidal-Puig A. Adipogenesis and WNT signalling. Trends Endocrinol Metab. 2009;20(1):16–24. doi: 10.1016/j.tem.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engeli S, Janke J, Gorzelniak K, et al. Regulation of the nitric oxide system in human adipose tissue. J Lipid Res. 2004;45(9):1640–8. doi: 10.1194/jlr.M300322-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Goto Y, Suzuki K, Ono T, Sasaki M, Toyota T. Development of diabetes in the non-obese NIDDM rat (GK rat) Adv Exp Med Biol. 1988;246:29–31. doi: 10.1007/978-1-4684-5616-5_4. [DOI] [PubMed] [Google Scholar]
- 16.Portha B. Programmed disorders of beta-cell development and function as one cause for type 2 diabetes? The GK rat paradigm. Diabetes Metab Res Rev. 2005;21(6):495–504. doi: 10.1002/dmrr.566. [DOI] [PubMed] [Google Scholar]
- 17.Xue B, Sukumaran S, Nie J, Jusko WJ, Dubois DC, Almon RR. Adipose tissue deficiency and chronic inflammation in diabetic Goto-Kakizaki rats. PLoS One. 2011;6(2):e17386. doi: 10.1371/journal.pone.0017386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almon RR, Dubois DC, Sukumaran S, et al. Effects of high fat feeding on liver gene expression in diabetic goto-kakizaki rats. Gene Regul Syst Bio. 2012;6:151–68. doi: 10.4137/GRSB.S10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almon RR, DuBois DC, Lai W, Xue B, Nie J, Jusko WJ. Gene expression analysis of hepatic roles in cause and development of diabetes in Goto-Kakizaki rats. J Endocrinol. 2009;200(3):331–46. doi: 10.1677/JOE-08-0404. [DOI] [PubMed] [Google Scholar]
- 20.Nie J, Xue B, Sukumaran S, Jusko WJ, Dubois DC, Almon RR. Differential muscle gene expression as a function of disease progression in Goto-Kakizaki diabetic rats. Mol Cell Endocrinol. 2011;338(1–2):10–7. doi: 10.1016/j.mce.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehses JA, Perren A, Eppler E, et al. Increased number of islet-associated macrophages in type 2 diabetes. Diabetes. 2007;56(9):2356–70. doi: 10.2337/db06-1650. [DOI] [PubMed] [Google Scholar]
- 22.Kraakman MJ, Murphy AJ, Jandeleit-Dahm K, Kammoun HL. Macrophage polarization in obesity and type 2 diabetes: weighing down our understanding of macrophage function? Front Immunol. 2014;5:470. doi: 10.3389/fimmu.2014.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navarro E, Funtikova AN, Fito M, Schroder H. Can metabolically healthy obesity be explained by diet, genetics, and inflammation? Mol Nutr Food Res. 2015;59(1):75–93. doi: 10.1002/mnfr.201400521. [DOI] [PubMed] [Google Scholar]
- 24.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116(7):1793–801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunetti P. The lean patient with type 2 diabetes: characteristics and therapy challenge. Int J Clin Pract. 2007;153:3–9. doi: 10.1111/j.1742-1241.2007.01359.x. [DOI] [PubMed] [Google Scholar]
- 26.Tirone TA, Brunicardi FC. Overview of glucose regulation. World J Surg. 2001;25(4):461–7. doi: 10.1007/s002680020338. [DOI] [PubMed] [Google Scholar]
- 27.Barbu A, Hedlund GP, Lind J, Carlsson C. Pref-1 and adipokine expression in adipose tissues of GK and Zucker rats. Mol Cell Endocrinol. 2009;299(2):163–71. doi: 10.1016/j.mce.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78(3):783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- 29.Sukumaran S, Xue B, Jusko WJ, Dubois DC, Almon RR. Circadian variations in gene expression in rat abdominal adipose tissue and relationship to physiology. Physiol Genomics. 2010;42 A(2):141–52. doi: 10.1152/physiolgenomics.00106.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swarbrick MM, Havel PJ. Physiological, pharmacological, and nutritional regulation of circulating adiponectin concentrations in humans. Metab Syndr Relat Disord. 2008;6(2):87–102. doi: 10.1089/met.2007.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arch JR. Central regulation of energy balance: inputs, outputs and leptin resistance. Proc Nutr Soc. 2005;64(1):39–46. doi: 10.1079/pns2004407. [DOI] [PubMed] [Google Scholar]
- 32.Hackl H, Burkard TR, Sturn A, et al. Molecular processes during fat cell development revealed by gene expression profiling and functional annotation. Genome Biol. 2005;6(13):R108. doi: 10.1186/gb-2005-6-13-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279(50):52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 34.Ali AT, Hochfeld WE, Myburgh R, Pepper MS. Adipocyte and adipogenesis. Eur J Cell Biol. 2013;92:6–7. 229–36. doi: 10.1016/j.ejcb.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Du B, Cawthorn WP, Su A, et al. The transcription factor paired-related homeo-box 1 (Prrx1) inhibits adipogenesis by activating transforming growth factor-beta (TGFbeta) signaling. J Biol Chem. 2013;288(5):3036–47. doi: 10.1074/jbc.M112.440370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cernkovich ER, Deng J, Bond MC, Combs TP, Harp JB. Adipose-specific disruption of signal transducer and activator of transcription 3 increases body weight and adiposity. Endocrinology. 2008;149(4):1581–90. doi: 10.1210/en.2007-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baba M, Hong SB, Sharma N, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103(42):15552–7. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Lay S, Robichon C, Le Liepvre X, Dagher G, Ferre P, Dugail I. Regulation of ABCA1 expression and cholesterol efflux during adipose differentiation of 3T3-L1 cells. J Lipid Res. 2003;44(8):1499–507. doi: 10.1194/jlr.M200466-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Nara N, Nakayama Y, Okamoto S, et al. Disruption of CXC motif chemokine ligand-14 in mice ameliorates obesity-induced insulin resistance. J Biol Chem. 2007;282(42):30794–803. doi: 10.1074/jbc.M700412200. [DOI] [PubMed] [Google Scholar]
- 40.Pei L, Castrillo A, Tontonoz P. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol Endocrinol. 2006;20(4):786–94. doi: 10.1210/me.2005-0331. [DOI] [PubMed] [Google Scholar]
- 41.Sugerman PB, Faber SB, Willis LM, et al. Kinetics of gene expression in murine cutaneous graft-versus-host disease. Am J Pathol. 2004;164(6):2189–202. doi: 10.1016/S0002-9440(10)63776-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soares L, Seroogy C, Skrenta H, et al. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat Immunol. 2004;5(1):45–54. doi: 10.1038/ni1017. [DOI] [PubMed] [Google Scholar]
- 43.Anand PK. Exosomal membrane molecules are potent immune response modulators. Commun Integr Biol. 2010;3(5):405–8. doi: 10.4161/cib.3.5.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Y, Dubois DC, Sun H, Almon RR, Jusko WJ. Modeling diabetes disease progression and salsalate intervention in Goto-Kakizaki rats. J Pharmacol Exp Ther. 2011;339(3):896–904. doi: 10.1124/jpet.111.185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, DuBois DC, Cao Y, Jusko WJ, Almon RR. Diabetes disease progression in Goto-Kakizaki rats: effects of salsalate treatment. Diabetes Metab Syndr Obes. 2014;7:381–9. doi: 10.2147/DMSO.S65818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bacci S, De Cosmo S, Prudente S, Trischitta V. ENPP1 gene, insulin resistance and related clinical outcomes. Curr Opin Clin Nutr Metab Care. 2007;10(4):403–9. doi: 10.1097/MCO.0b013e3281e386c9. [DOI] [PubMed] [Google Scholar]
- 47.Maddux BA, Chang YN, Accili D, McGuinness OP, Youngren JF, Goldfine ID. Overexpression of the insulin receptor inhibitor PC-1/ENPP1 induces insulin resistance and hyperglycemia. Am J Physiol Endocrinol Metab. 2006;290(4):E746–9. doi: 10.1152/ajpendo.00298.2005. [DOI] [PubMed] [Google Scholar]
- 48.Liang J, Fu M, Ciociola E, Chandalia M, Abate N. Role of ENPP1 on adipocyte maturation. PLoS One. 2007;2(9):e882. doi: 10.1371/journal.pone.0000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varma V, Yao-Borengasser A, Bodles AM, et al. Thrombospondin-1 is an adipokine associated with obesity, adipose inflammation, and insulin resistance. Diabetes. 2008;57(2):432–9. doi: 10.2337/db07-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniel C, Schaub K, Amann K, Lawler J, Hugo C. Thrombospondin-1 is an endogenous activator of TGF-beta in experimental diabetic nephropathy in vivo. Diabetes. 2007;56(12):2982–9. doi: 10.2337/db07-0551. [DOI] [PubMed] [Google Scholar]
- 51.Tan BK, Adya R, Chen J, et al. Metformin decreases angiogenesis via NF-kappaB and Erk1/2/Erk5 pathways by increasing the antiangiogenic thrombospondin–1. Cardiovasc Res. 2009;83(3):566–74. doi: 10.1093/cvr/cvp131. [DOI] [PubMed] [Google Scholar]
- 52.Berndt J, Kovacs P, Ruschke K, et al. Fatty acid synthase gene expression in human adipose tissue: association with obesity and type 2 diabetes. Diabetologia. 2007;50(7):1472–80. doi: 10.1007/s00125-007-0689-x. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Jones Voy B, Urs S, et al. The human fatty acid synthase gene and de novo lipogenesis are coordinately regulated in human adipose tissue. J Nutr. 2004;134(5):1032–38. doi: 10.1093/jn/134.5.1032. [DOI] [PubMed] [Google Scholar]
- 54.Dahlman I, Linder K, Arvidsson Nordström E, et al. Changes in adipose tissue gene expression with energy-restricted diets in obese women. Am J Clin Nutr. 2005;81(6):1275–85. doi: 10.1093/ajcn/81.6.1275. [DOI] [PubMed] [Google Scholar]
- 55.Diggle CP, Carr IM, Zitt E, et al. Common and recurrent HPGD mutations in Caucasian individuals with primary hypertrophic osteoarthropathy. Rheumatology. 2010;49(6):1056–62. doi: 10.1093/rheumatology/keq048. [DOI] [PubMed] [Google Scholar]
- 56.Way JM, Harrington WW, Brown KK, et al. Comprehensive messenger ribonucleic acid profiling reveals that peroxisome proliferator-activated receptor gamma activation has coordinate effects on gene expression in multiple insulin-sensitive tissues. Endocrinology. 2001;142(3):1269–77. doi: 10.1210/endo.142.3.8037. [DOI] [PubMed] [Google Scholar]
- 57.Wentz AE, d’Avignon DA, Weber ML, et al. Adaptation of myocardial substrate metabolism to a ketogenic nutrient environment. J Biol Chem. 2010;285(32):24447–56. doi: 10.1074/jbc.M110.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yi W, Fu P, Fan Z, et al. Mitochondrial HMG-CoA synthase partially contributes to antioxidant protection in the kidney of stroke-prone spontaneously hypertensive rats. Nutrition. 2010;26:11–12. 1176–80. doi: 10.1016/j.nut.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 59.Hageman RS, Wagener A, Hantschel C, Svenson KL, Churchill GA, Brockmann GA. High-fat diet leads to tissue-specific changes reflecting risk factors for diseases in DBA/2J mice. Physiol Genomics. 2010;42(1):55–66. doi: 10.1152/physiolgenomics.00072.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cawthorn WP, Sethi JK. TNF-alpha and adipocyte biology. FEBS Lett. 2008;582(1):117–31. doi: 10.1016/j.febslet.2007.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Plasma glucose (A) and insulin (B) in GK and WKY animals as a function of age. Symbols are defined in Figure 1.
Notes: ##GK-ND vs WKY-ND, P < 0.001. *GK-HFD vs WKY-HFD, P < 0.05. **GK-HFD vs WKY-HFD, P < 0.001.
Supplementary Table 1. Differentially expressed genes in GK versus WKY animals fed hfd.
Supplementary Table 2. Differentially expressed genes in GK versus WKY animals fed nd.
Supplementary Table 3. Differentially regulated by diet in both strains.
Supplementary Table 4. Differentially regulated by diet only in GK animals.
Supplementary Table 5. Differentially regulated by diet only in WKY animals.