Abstract
Medical imaging is becoming a vital component of war on cancer. Tremendous amounts of medical image data are captured and recorded in a digital format during cancer care and cancer research. Facing such an unprecedented volume of image data with heterogeneous image modalities, it is necessary to develop effective and efficient content-based medical image retrieval systems for cancer clinical practice and research. While substantial progress has been made in different areas of content-based image retrieval (CBIR) research, direct applications of existing CBIR techniques to the medical images produced unsatisfactory results, because of the unique characteristics of medical images. In this paper, we develop a new multimodal medical image retrieval approach based on the recent advances in the statistical graphic model and deep learning. Specifically, we first investigate a new extended probabilistic Latent Semantic Analysis model to integrate the visual and textual information from medical images to bridge the semantic gap. We then develop a new deep Boltzmann machine-based multimodal learning model to learn the joint density model from multimodal information in order to derive the missing modality. Experimental results with large volume of real-world medical images have shown that our new approach is a promising solution for the next-generation medical imaging indexing and retrieval system.
Keywords: content-based image retrieval, multi-modal and content-based medical image retrieval, extended probabilistic latent semantic analysis, deep learning, deep boltzmann machine
Introduction
Medical imaging has been ranked as one of the most important medical developments of the past 1,000 years.1 Over the last 10 years, tremendous amounts of medical image data have been captured and recorded in a digital format during daily clinical practice, medical research, and education.2–5 These data represent a rich source of information that is invaluable for diagnosis, treatment, recovery, rehabilitation, etc. This is particularly true for cancer-related research and clinical practice: with the advancement of medical imaging,6 health providers are able to not only investigate inside the body but also see deep within the chaos of cancer cells.6 For example, medical imaging is used to screen, diagnose, and stage cancer; to guide cancer treatments; to monitor cancer recurrence; and to support cancer research, such as drug discovery and therapeutic innovation.
Advancing the appropriate use of information technology in medical imaging as the newest frontier of medical imaging promises to contribute greatly to improving the quality of cancer care for each individual patient with lower cost. Common information technology and computational tasks related to medical imaging include image acquisition, image manipulation, image management, and image integration.7 Medical image retrieval is one of the few computational components that cover a broad range of tasks, including image manipulation, image management, and image integration. The goal of medical image retrieval is to find the most clinically relevant images in response to specific information needs represented as search queries.
Text-based information retrieval techniques are well researched. However, they are limited by the quality and quantity of the textual annotations of the images. Effective and efficient content-based approaches can be used in conjunction with text-based methods to improve the accuracy and completeness of the search results. Motivated by the important potential clinical benefits, content-based medical image retrieval (CBMIR) has become a very active research area over the last decade.8–13
Among many state-of-the-art techniques of CBMIR research, one of the most promising directions is to correlate multimodal information (eg, text and image) or to use the combinations of textual and visual analysis techniques for more effective and efficient CBMIR.14–19 In this paper, we focus on two modalities that are widely available in real-world clinic practice: visual information of a medical image and the corresponding text annotation of the medical image.
While substantial amount of research has been conducted in the area of medical image retrieval, real-world tools and applications that can access the medical image data by their content are rare in clinical practice.20–23 One of the major barriers we have identified is that the semantic gap exists between the low-level features (eg, low-level visual and textual features) and the high-level medical concepts. The key to addressing these issues is to develop new semantic meaningful features from multimodal information to bridge the semantic gap and to enable effective and efficient CBMIR. Another challenge is that the real-world data are very noisy and some modality information (eg, text annotation) may be missing from input. To address the missing modality issue, new algorithms that can derive the missing modality information from the existing known modality (eg, deriving missing text annotation from known visual content) are needed for making the system usable in clinical practice.
The goal of our research in this paper is to develop, evaluate, and demonstrate new image and textual analysis techniques for scalable semantic medical image content analysis and retrieval. In order to overcome the limitations introduced in the previous paragraph, our focus in this paper is to generate semantic features. Our new approach has great potential to substantially improve the performance of medical image retrieval. It also has good potential to be applied in clinical practice and healthcare applications. Our approach is inspired by recent advancement in statistical graphic models24,25 and deep learning.26 Specifically, we first develop a new extended probabilistic Latent Semantic Analysis (pLSA) model to integrate the visual and textual information from medical images to bridge the semantic gap. The proposed pLSA model is able to generate a representation space with the desired feature (eg, similarity in this feature space implies similarity in the corresponding medical concepts). While the proposed pLSA model is very good at bridging the semantic gap, we are still facing some additional issues when employing this model in real-world applications. In real-world clinical applications, the situations where some modalities are missing and are noisy happen frequently. We plan to develop our second model to address these issues. Specifically, we develop a new deep Boltzmann machine (DBM)-based27 multimodal learning model to learn the joint density model from multimodal information. The proposed DBM-based model can derive the missing modality information from known modality information. The combination of these two models, both of which are trained with a large volume of real-world medical image data, will enable us to search the most relevant images for a given query. Experimental results with large volume, real-world medical images have shown that our approach is a promising solution for the next-generation medical imaging indexing and retrieval system.
Motivations
As medical imaging is becoming an essential component for cancer care and research, many departments of cancer care and research would benefit directly from research efforts on multimodal CBMIR. Medical imaging is becoming even more important over the last ten years. One of the reasons behind this is the so-called big data in medical imaging: tremendous amounts of medical image data, in the last few years, are captured and recorded in a digital format during the daily clinical practice, medical research, and education. Driven by the aging population and technology advancements, the global diagnostic imaging market is expected to increase to $26.6 billion by 2016.2 In 2010, over 5 billion medical imaging studies had been conducted worldwide.3 In 2011, the number of US medical imaging procedures surpassed the 800 million mark.4 At Mayo Clinic’s Campus in Jacksonville, FL, USA, a radiologist viewed 1,500 cross-sectional images per day in 1994 compared to 16,000 images per day in 2004.5 The Radiology Department at University Hospital of Geneva, Geneva, Switzerland, produced over 12,000 images per day in 2004, 40,000 images per day in 2006, 70,000 images per day in 2007, and over 117,000 images per day in 2009.13 Images are ubiquitous in cancer care and research. The image viewers play a central role in many aspects of modern cancer care. These data provide an unprecedented opportunity for making smart and optimized cancer care decisions with improved outcomes while reducing costs.
Common computational tasks related to medical imaging include image acquisition, image manipulation, image management, and image integration.7 Medical image retrieval, with the goal of finding the most clinically relevant images in response to specific information needs represented as search queries, is one of the few computational components that cover a broad range of medical imaging computational tasks. Despite text-based information retrieval methods being both mature and well researched, they are limited by the quality of image annotations. Among other important limitations facing traditional text retrieval techniques are the fact that image annotations are subjective and context sensitive, and can be quite limited in scope or even completely absent. Manually annotating images is also label intensive and can be very error-prone. Image annotations are quite noisy if they are automatically extracted from the surrounding text using natural language processing techniques, and there is much more information in an image than can be extracted using a limited number of words. Effective and efficient content-based approaches can be used in conjunction with text-based methods to improve the accuracy and completeness of the search results.
Related Work
The related work to this paper falls under three categories. The first category is CBMIR research. The second category is multimodal fusion-based image retrieval research. The third category is deep learning research. We will introduce these three categories in more detail in the following paragraphs.
The first category, CBMIR, has been a very active field in recent years. CBMIR is rooted from content-based image retrieval (CBIR), which is any technology that in principle helps to organize digital picture archives by their visual content.28 CBIR has grown tremendously since 2000, and we refer interested readers to read the survey paper by Datta et al.28 to gain more detailed understanding for CBIR. CBMIR research, which is motivated by huge potential benefits and is one of the major applications of CBIR, has witnessed a large number of publications and explorations over the last decade.9–13 In Ref. 8, a database of computed tomography (CT) images of the chest called automated search and selection engine with retrieval tools was developed. Image retrieval in medical applications10 was a project that aims to develop high-level methods for CBIR with prototypical application to medical diagnostic tasks on a radiologic image archive. The medical GNU image finding tool (MedGIFT) project29 included several axes around the retrieval of medical images from a variety of databases and image types as well as several applications. Greenspan and Pinhas30 presented a representation and matching framework for image categorization in medical image archives. Napel et al.31 developed a system to facilitate radiologic image retrieval that contains similar-appearing lesions. System evaluation was performed with a CT image database of liver and an external standard of image similarity. In Ref. 32, Rahman et al proposed a unified medical image retrieval framework integrating visual and textual keywords using a novel multimodal query expansion. Quellec et al.33 introduced a content-based heterogeneous information retrieval framework. In this paper, they proposed a Bayesian network to recover missing information. The Medical Imaging Resource Center34 project was initiated by the RSNA Radiology Informatics Committee to construct a library of medical information globally accessible to the imaging community over the Internet. An example of evaluation projects is ImageCLEF Medical Image Retrieval Task,35 which is a task to benchmark and compare the performance of participating systems for medical image retrieval. Recently, researchers from US National Library of Medicine/National Institutes of Health (NLM/NIH) (36–38 developed new computer-aided techniques to identify and annotate the region of interests for a given medical image to facilitate biomedical document and image retrieval. Some medical image search prototypes, such as GoldMiner,39 were also developed. Kumar et al.40 presented a review of the state-of-the-art medical CBIR approaches in five main categories: (1) two-dimensional image retrieval, (2) retrieval of images with three or more dimensions, (3) the use of non-image data to enhance the retrieval, (4) multimodality image retrieval, and (5) retrieval from diverse data sets. Our system is different from the state-of-the-art medical CBIR approaches because our proposed statistic graphic model and deep learning model make it possible to develop semantic features for bridging the semantic gap.
The second category of related work is called multimodal fusion-based image retrieval. The research in this area is rooted in information fusion. Existing literature on multimodal retrieval can roughly be classified into two categories: feature fusion and retrieval fusion. The first strategy (feature fusion strategy) generates an integrated feature representation from multiple modalities. For example, in Ref. 24, the features from different modalities were normalized and concatenated to generate the feature vectors. Then, the latent semantic analysis (LSA) was applied on these features for image retrieval. Lienhart et al.25 proposed a multilayer pLSA to solve the multimodal image retrieval problem. The second strategy (retrieval fusion) refers to the techniques that merge the retrieval results from multiple retrieval algorithms. Our approach belongs to the first category (feature fusion). Our technique is different from Pham et al.24 in that we do not simply concatenate the features from different modalities. Instead, we represent the features from different modalities as a multidimensional matrix and incorporate these feature vectors using an extended pLSA model. Our method is also different from Lienhart et al.25 since we use a single pLSA model instead of multiple pLSA models.
The third category of related work, deep learning,26,41 aims to learn multiple levels of representation and abstraction that help infer knowledge from data such as images, videos, audios, and text. In the last five years, deep learning is making astonishing gains in computer vision, speech recognition, multimedia analysis, and drug designing. The impact of deep learning is far reaching on applications in medical, social, and commercial domains.42–44 In 2013, deep learning made MIT Technology Review’s list of top 10 breakthroughs of the year. Briefly speaking, there are two main classes of deep learning techniques: purely supervised learning algorithms (eg, convolutional neural network45,46) and unsupervised and semi-supervised learning algorithms (eg, denoising autoen-coders,47,48 restricted Boltzmann machines (RBMs),49,50 and DBMs27). Since this paper employs RBM and DBM heavily, we will mainly introduce these two techniques. RBM was first proposed as a significant improvement of Boltzmann machines (BMs).51 In the following introduction, we will follow the terms and conventions introduced in prior research.27,49–51 BM is a stochastic recurrent neural network, and it is named after the Boltzmann distribution in statistical mechanics. BM is a network of units with energy defined for the network. It also has binary units, but unlike Hopfield nets, BM units are stochastic. BM is a network of symmetrically coupled stochastic binary units. It includes a set of visible nodes v ∈ {0, 1}D and a set of hidden nodes h ∈ {0, 1}P. The state {v, h}’s energy is defined as follows: , where θ = {W, L, J} are the parameters of the model. W, L, and J indicate visible-to-hidden, visible-to-visible, and hidden-to-hidden symmetric interaction terms, respectively. In theory, the RM model is a general computational model that is suitable for many applications. However, in practice, learning in general BM is very inefficient. As a result, RBMs were proposed with a fast learning algorithm. Different from BM, RBM model does not allow visible-to-visible connections and hidden-to-hidden connections. In another word, in the energy model, we will set both L = 0 and J = 0. By setting both of them as zero, we can remove every intralayer connection. The inference in RBM is exact, which is also different from general BM’s inference. While exact maximum likelihood learning in RBM is intractable, new learning algorithms, such as contrastive divergence, has been proposed to carry out the learning process very efficiently. When one RBM is trained, we can treat the activities of its hidden nodes as inputs for a higher-level RBM. By stacking multiple RBMs together, we could train many layers of hidden units efficiently. This method of stacking RBMs will produce a new learning algorithm called DBMs. Among the state of the art of DBM techniques, the findings presented by papers52,53 are most similar to our proposed approach. In Ref. 52, the authors proposed a semi-supervised learning method for multimodal learning. The proposed model can use both labeled and unlabeled training data. Their work showed that learning model could be improved with unlabeled image data. It also showed that providing associated text tags may be incorporated as another modality to improve the performance. Most recently, Srivastava and Salakhutdinov53 presented a method utilizing a DBM to accomplish image recognition tasks for MIR Flickr data set54 using images and associated tags. In their paper,53 they demonstrated vast potential for DBMs and their competency for multimodal learning. In our paper, we will focus on developing new semi-supervised learning algorithms using DBMs.27,49,50 Our proposed solution is an extended model of DBM, which is a learning system through layers of binary stochastic variables. These layers are interconnected, but have no connections between nodes on the same layer. This allows for much faster processing of information than standard BMs, which are entirely interconnected.
Our Multimodal Approach
In this section, we will introduce our proposed multimodal approach in more detail. To the best of our knowledge, how to generate effective and efficient semantic features for large-scale medical image sets remains a challenging and unsolved problem. In this paper, we will focus on the development and evaluation of new semantic analysis techniques by investigating and evaluating innovative visual and textual modeling and analysis techniques for generating the semantic features. These semantic features have potential to bridge the semantic gap. Specifically, we develop two types of statistical graphic models that can fuse the distinct modalities to generate the semantic features. The newly generated semantic features are capable of capturing the real-world medical concepts effectively and efficiently. Furthermore, the proposed approach is able to handle the missing modality reliably. The semantic features are a new representation space with the following desired features: (1) the similarity in this representation space implies the similarity of the corresponding real-world medical concepts and (2) the representation space can be generated reliably even in the situations where there are missing and noisy modalities. In the following section, we will first introduce the proposed extended pLSA model to fuse the multimodal information (section Step 1: fusing the multimodal information). Then we will introduce our proposed DBM model (section Step 2: deriving missing modalities), which can be used to derive the missing modality. Lastly, we will discuss how we will utilize the two proposed models from Steps 1 and 2 for medical image retrieval (section Step 3: retrieval).
Step 1: fusing the multimodal information
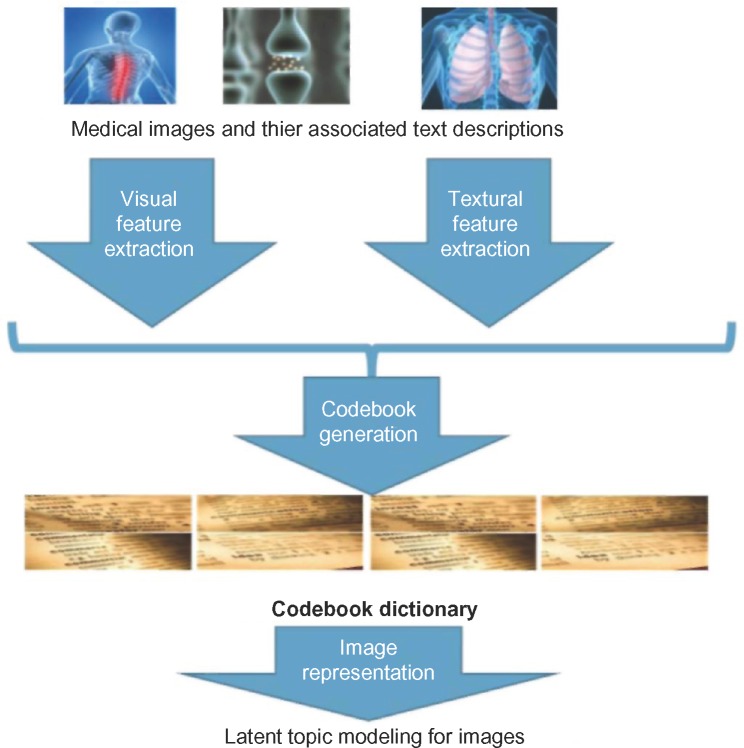
Figure 1 depicts an overview of the first step. Our goal in this step is to build the graphic model and to generate the latent topic representation for each image in the database. Given the images and their associated textual descriptions, our algorithms will generate a latent topic representation for each image.
Figure 1.
Overview architecture of the proposed model in Step 1.
We use an extended pLSA model to encode the visual and textual information for each image. The original pLSA method is based on an aspect model, which is a latent variable model for general co-occurrence data (eg, document-word frequency matrix). It models the distribution of words in the document as a mixture of a few aspects. It was recently employed by the computer vision community to solve the problems of image retrieval and object class recognition. We extend the pLSA model by employing two random variables to represent the visual and textual features, respectively. Please note, in our research, we employ the concept of visual bag-of-words (VBoW) model to extract the initial visual features from an image. In this VBoW model, an image is represented as a visual document composed of visual elements (a.k.a. visual words). This model has been very popular in the last few years55–60 because of its simplicity and scalability. Specifically, we first apply scale-invariant feature transform (SIFT)-based interesting point detection methods61 to identify the potential salient points from the image. For each interesting point identified by the SIFT method, we will extract the SIFT descriptor, which is a 128-dimensional vector. We then run k-means clustering algorithm for all the SIFT descriptors collected from each training image. The k centroids of the k-means algorithm are the visual words that can be used for late processing. For each image, we compare the SIFT interesting point and its SIFT descriptor with each visual word and find out the closet visual word. By this way, we can generate a histogram of visual words as a feature representation for each image. For textual feature extraction, we employ existing open source natural language processing package, Stanford NLP package,62 to extract textual features. Specifically, we employ the textual bag-of-words (BoW) model and a vocabulary of the 1,000 most frequently used medical terms. In the following description, we present the proposed extended pLSA model following the terms and conventions introduced in prior research.24,25,63
Suppose we have D (D = {d1 … dN}) images where di represents the ith image that contains both visual and textual information. We use two random variables wv and wt to represent the visual and textual words, respectively. We assume that the visual vocabulary is represented as WV = {wV_1, … wV_M}, while the textual vocabulary WT = {wT_1, … wT_K}. The corpus of the image database can be summarized in a three-dimensional co-occurrence matrix , whose degree is M × K × N. The entries n(wV_m, wT_k, dn) in this matrix represent how often the term wV_m and wT_k occurred in image dn. A latent topic variable z is used to associate the occurrence of words wV and wT to image d. The joint probability model over WV × WT × D is represented by the following equation:
| (1) |
From equation (1), we can perform further derivation by importing the latent variable
| (2) |
We employ the expectation–maximization (EM) algorithms for training. EM alternates two steps: (1) an expectation (E) step, where posterior probabilities are computed for the latent variables and (2) a maximization (M) step, where parameters are updated. In the final stage of the training component, we compute the value of P(zl|di) for each image di (l ∈ (1,L), where L is the number of latent topics). More specifically, in our extended pLSA model, the E-step equation is listed as follows:
| (3) |
The formulas for the M-step are listed as follows:
| (4) |
| (5) |
| (6) |
During the retrieval stage, similar operations are performed to the query image. More details are provided in the section Step 3: retrieval. Our proposed extended pLSA model, compared with the existing pLSA model, employs a three-dimension array. Therefore, compared with the original pLSA model, the number of parameters to be estimated during the EM algorithms is also increased. However, as indicated by equations (3)–(6), the increasing of parameters will not cause the computation intractable. Finally, we use a histogram intersection (or potentially other distance measures) to measure the similarity between the query image and the images in the database.
Step 2: deriving missing modalities
While our model (extended pLSA model) proposed in the first step is able to generate a representation space with desired characteristics (eg, similarity in this feature space implies the similarity in corresponding medical concepts), we are still facing some additional issues if employing this model in real-world applications. More specifically, in real-world clinical applications, the situations where some modalities are missing and noisy happen frequently. We plan to develop our second model to address these issues. Our proposed approach is rooted from the recent advances in deep learning.26,64,65 The main innovation in this step is to learn a joint probability density model from the visual and textual information with the capacity of filling in missing modalities.
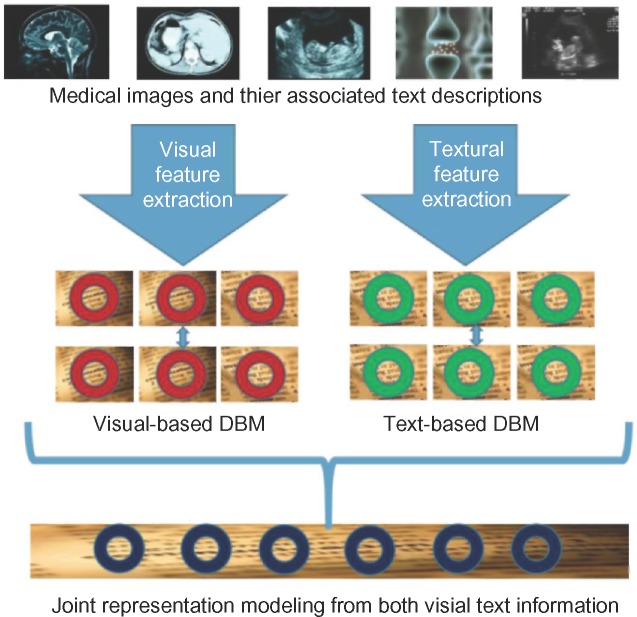
As shown in Figure 2, the novel part of our proposed approach is that of utilizing multimodal inputs for analysis. This joint model is accomplished by training two separate DBMs, with the top hidden layers connected to a combined hidden layer to act as a joint representation for the associated learning. As shown in Figure 2, we first extract both visual and textual features from the images. Then we train a visual-based DBM, as shown in the middle left of Figure 2. We also train a text-based DBM, as shown in the middle right of Figure 2. Both the DBMs have two hidden layers. In order to fuse the multimodal information, we add one additional layer on top of these two DBMs as the joint representation of multimodal data, as shown in the bottom of Figure 2.
Figure 2.
Overview architecture of the proposed model based on DBM.
As shown in Figure 2, our approach utilizes multiple layers of hidden variables, each layer connected to the neighboring layers through each and every node. One layer represents the visible data (in the case of image training, the image pixel data), and all subsequent layers are hidden. The connections between nodes are weighted according to a probability function to be evaluated during the training sessions.27 In order to derive the missing modality, we will first learn a joint density model from multimodal information using the proposed DBM. Specifically, the proposed DBM includes a set of visible nodes v ∈ {0, 1}D. It also includes several layers of hidden nodes , Please note, the basic building block of DBM is RBM, which is a special case of BM. In RBM, the connections exist only between nodes from different layers and there is no connection between nodes within the same layer. The joint distribution over the visible and hidden units is defined as P{v,h;θ) = {1/Z(θ))exp (–E(v,h;θ)), where Z(θ) is defined as the partition function and E(v,h;0) is the state {v,h}’s energy. Since there is no connection between the same layer, the state {v,h}’s energy is defined as follows: where θ = {W,L,J} are the parameters of the model. W, L, and J indicate visible-to-hidden, visible-to-visible, and hidden-to-hidden symmetric interaction terms sample the hidden modality from the conditional distributions, given the observed modalities. While there are many sampling algorithms that can obtain observations from probability distribution, we choose to employ the Gibbs sampling technique,66 used by recent deep learning research.27,67,68
Step 3: retrieval
Once the two models (extended pLSA model shown in Fig. 1 and the DBM model shown in Fig. 2) are trained, we will represent the visual and textual information using the trained models. Specifically, we will determine the distribution of the visual–textual words over the latent topic generated from the new pLSA model. We will also generate the missing data using conditional distribution over the observed data.
To obtain the visual features, we employ a BoW model.63,69 Textual features are extracted from the text annotations associated with the images. We apply the existing vector-space model to the textual annotations. Some necessary preprocessing (eg, removing stop words and stemming) is performed. Now, each image is represented by a two-dimensional matrix, which indicates the co-occurrence of the visual–textual words in this image. Therefore, a three-dimensional matrix represents the entire training data. Then we apply the EM algorithm to this three-dimensional co-occurrence table and obtain the model parameters.
The last step is to perform retrieval. The goal is to compute the similarity score between the database images and the query image. The first step is to extract the visual and textual features from the query image. Based on the features and the codebook (which is generated during the training stage), we could project the query image on the simplex spanned by the P(wV,wT|z), which is the visual–textual word distribution over a latent topic. Given a query image dq, we need to calculate the p(zk|dq) (k ∈ (1,L)), where L is the number of latent topics. To calculate p(zk|dq), we apply Bayes’ rule to generate the following equation:
| (7) |
In order to obtain the likelihood and the prior in Equation (7), an EM algorithm that is similar to the one used in the training stage is employed. Different from the EM method for training, which is introduced in section Step 1: fusing the multimodal information, the value of P(wV,wT|z) is fixed during the EM execution, and this value is obtained from the training stage, which is introduced in section Step 1: fusing the multimodal information. Once each p(zk|dq) is calculated, we generate a histogram representation for the query image by concatenating each p(zk|dq) value.
Distance metrics such as the histogram intersection are employed to compute the similarity between the query image and the database images. Finally, the database images are ranked based on the similarity score.
Experimental Results
In this section, we will introduce the data sets used in our experiments (section Data sets), the list of performance metrics (section List of definitions for performance metrics), as well as the detailed implementation and experimental results (section System implementation and detailed results). In the section Data sets, we introduce the characteristics of two data sets: ImageCLEF 2009 medical retrieval challenge and Image-CLEF 2013 medical retrieval challenge. Then we introduce the list of definitions of performance metrics at the section List of definitions for performance metrics. Some sample metrics, such as precision, recall, mean average precision (MAP), and ret_ret, used in our experiments are introduced in this section. Finally, we introduce our system implementation and present the experimental results with detailed analysis in the section System implementation and detailed results.
Data sets
We employ two data sets that have been widely used in medical image retrieval research.
The first data set is medical images from the ImageCLEF 2009 medical retrieval challenge. It contains 74,902 radiological images from two leading peer-reviewed journals (Radiology and RadioGraphics). These images are linked with their existing textual annotations (the captions of the images) extracted from the journal papers. Therefore, this image collection represents a wide range of medical knowledge. The ImageCLEF challenge also provides 25 realistic search topics. Each search topic contains both the textual keywords and the query images. In our implementation, we use these realistic search topics as our queries. Figure 3 illustrates some sample queries, including both textual keywords and the query images, used in this data set.
Figure 3.
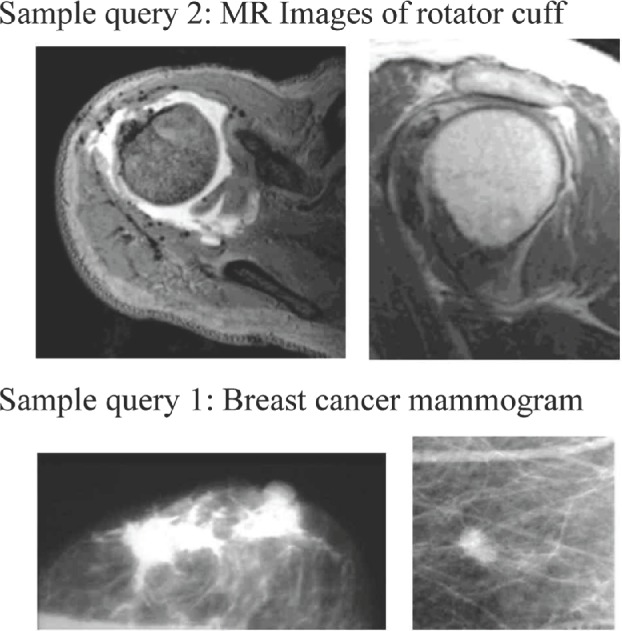
Sample queries (textual; keywords and the query images) from data set 1 (ImageCLEF 2009 medical retrieval challenge).
The second data set is medical images from the Image-CLEF 2013 medical image retrieval challenge. Similar to the ImageCLEF 2009 retrieval challenge, the images from 2013 challenge are also retrieved from open access biomedical literature. Instead of limiting the literature to the two radiology journals used in 2009 challenge, the 2013 challenge expands the literature to many other radiology journals in the PubMed Central. As a result, the 2013 retrieval challenge contains 305,000 medical images, which represent one of the largest medical image collections available to the research community. Similar to the 2009 challenge, the 2013 retrieval challenge also provides ad hoc image-based retrieval examples. The examples include 30 textual queries with two to three sample images for each query. We use these examples for our validation. Figure 4 illustrates some sample queries, including both textual keywords and the query images, used in this data set.
Figure 4.
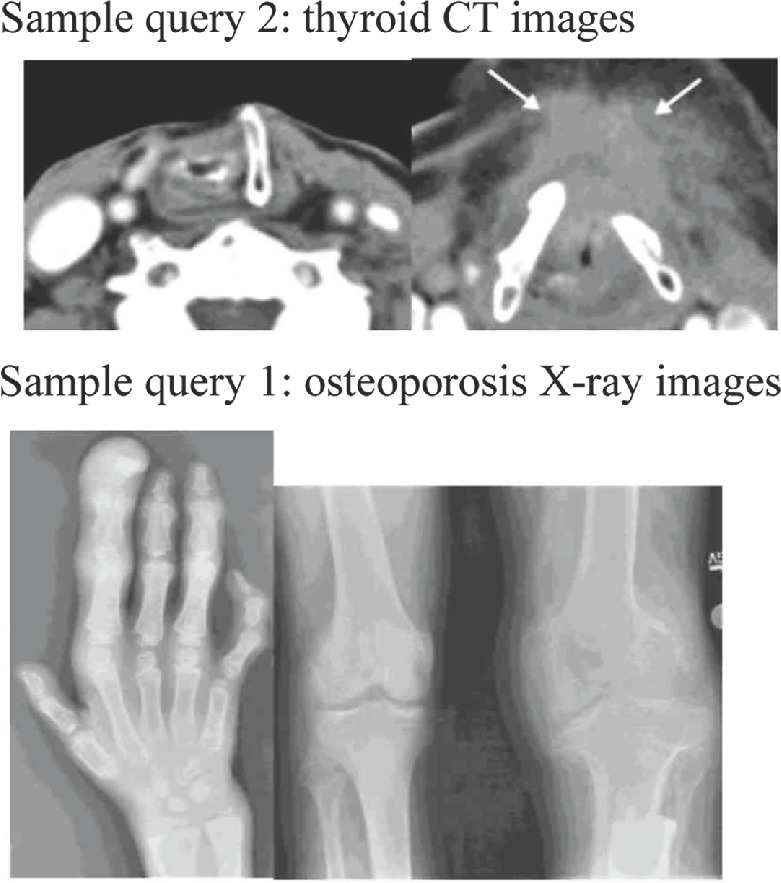
Sample queries (textual; keywords and the query images) from data set 2 (ImageCLEF 2013 medical retrieval challenge).
As shown in Figures 3 and 4, the number of keywords in most of the search topics in the ImageCLEFmed data sets is between 3 and 5. For example, sample query 1 in Figure 3 has three keywords – breast, cancer, and mammogram – and sample query 1 in Figure 4 (images from ImageCLEFmed 2013) has three keywords – osteoporosis, X-ray, and images.
Considering the total number of images (74,902 images from ImageCLEF 2009 and 305,000 images from Image-CLEF 2013) available in our data sets, the total number of queries (25 queries from ImageCLEF 2009 and 30 queries from ImageCLEF 2013) is relatively small. This is a normal setting for every participant in ImageCLEF 2009 and 2013.
List of definitions for performance metrics
In order to evaluate the results, we need to employ a sequence of metrics (a.k.a. performance measurements) to determine whether the returned results are relevant to a given query. The following performance measurements are used in our experiments: Precision, Recall, Average Precision, MAP, bpref, P5, P10, P30, and rel_ret.
Precision is defined as , where relevant documents are the returned results for this query from the ground truth and retrieved documents are the returned results for this query from the search algorithm. Therefore, precision is the number of elements in the array containing the intersection of ground truth results to our results divided by the number of elements in the array containing our results.
Recall is defined as , where relevant documents and retrieved documents have the same meaning as defined in the precision calculation (in the previous paragraph). Therefore, recall is equal to the number of elements in the array containing the intersection of ground truth results to our results divided by the number of elements in the array containing ground truth results.
Average precision is defined as , where relevant documents have the same definition as in precision and recall, r is the rank, N is the number retrieved, P(r) is the precision of result at rank r, and rel(r) is the relevance of result at rank r. In other words, to calculate average precision, at each result, if the current result is relevant, we calculate the precision for every result up to the current result. Then we divide that number by the amount of results so far. After we have performed this calculation for every retrieved document, we divide it by the number of relevant documents to obtain average precision for that query. Average precision is useful because it places more weight on relevant documents since irrelevant documents are considered zero in the calculation. To determine MAP, we calculate average precision for several different queries and then divide by the number of queries.
bpref is the number of times that non-relevant documents are retrieved before relevant documents. bpref is equal to , where r is the relevant retrieved documents, R is the relevant documents, and n is a member of the first R irrelevant retrieved documents.
Precision after 5, 10, and 30 retrieved results are represented by P5, P10, and P30, respectively. Performing these calculations illustrates how much weaker our returned results become as the rank decreases.
Finally, the rel_ret measure is simply another name for the number of relevant documents in the retrieved documents according to the ground truth. This is the same number used in precision and recall.
System Implementation and Detailed Results
Since both the first data set (ImageCLEF 2009 challenge) and the second data set (ImageCLEF 2013 challenge) provide sample queries (25 queries and 30 queries from 2009 challenge and 2013 challenge, respectively), we use them as the ground truth in our experiments. This ground truth is determined by a group of biomedical domain experts. Using the ground truth, we could measure the accuracy of the results of the 55 queries in our system.
For the purpose of training the retrieval model, following the experimental setting in the state of the art,35 we choose 25% of the images from ImageCLEF 2009 and 2013 data set, respectively, as the training data set. This means that the total number of images used for model training is around 100,000.
For visual features extraction, we follow the traditional VBoW approach. Briefly speaking, we first extract SIFT61 interesting point of the image and its corresponding SIFT descriptors from the 100,000 training images. We then apply the k-means clustering algorithm to all the SIFT descriptors. We experiment with different k values, and we choose k as 3,000 from the experimental results. Please note, based on the literature research28,40,70 and our own experiments, an optimal k value is largely application dependent. Next, we generate a histogram for each image. The number of the bins for the histogram is equal to the number of centroids from the k-means algorithm (in our context, the number of centroids is 3,000). The histogram is generated by comparing the SIFT interesting point and its SIFT descriptor with each centroid and identifying the closet centroid.
For textual feature extraction, we employ Stanford NLP package62 (an open source natural language processing package). Traditional textual BoW model was employed, and there were 1,000 most frequently used medical terms.
Our implementation run in a server equipped with 128 GB RAM, an eight-core Intel Xeon E5–2600 v2 Series CPU, and 2 NVIDIA K-40 GPU. The most time-consuming part is the training of DBM model. In our implementation, it took around 3 days to train the model. This is consistent with the state-of-the-art deep learning implementations, which usually take one week to train a deep model. The time for feature extraction, k-means clustering, and VBoW generation is relative short. For example, the average time for extracting visual feature for one image is around 50 milliseconds in our server.
Table 1 illustrates the results of the proposed approach when applied to the two data sets. The numbers in these tables are generated with the standard tool71 used by the Text REtrieval Conference (TREC) community for evaluating an ad hoc retrieval run, given the results file and a standard set of judged results. The overall performance is encouraging with an MAP at 0.2909. Other numbers, such as bpref, P_5, P_10, and ret_ret, are also equivalent or better than the results from the state-of-the-art. More detailed performance comparison between the proposed approach and the state-of-the-art is introduced in the next few paragraphs.
Table 1.
Results of the proposed approach for multimodal retrieval using the two data sets.
| rel_ret | map | gm_map | Rprec | Bpref | recip_rank |
|---|---|---|---|---|---|
| 1902 | 0.2909 | 0.2019 | 0.3101 | 0.3206 | 0.6421 |
| P_5 | P_10 | P_15 | P_20 | P_30 | P_100 |
| 0.5620 | 0.5510 | 0.5309 | 0.5270 | 0.4647 | 0.3281 |
For performance comparison, we implemented other retrieval algorithms with single modality. The first compared algorithm, defined as algorithm A, used similar visual features and learning framework as our approach. It did not use the textual information. In fact, algorithm A is a standard technique for CBIR using SIFT VBoW since the visual features used in algorithm A are based on SIFT feature extraction algorithms. The second algorithm being compared, defined as algorithm B, only used textual features. As shown in Table 2, the average MAP of algorithms A and B are 0.01 and 0.21, respectively. These experiments show that the proposed method is more effective because of the integration of both visual and textual features.
Table 2.
Performance comparisons between the proposed approach and the image retrieval techniques with single modality.
| Techniques | proposed multimodal approach | Algorithm A (only using visual modality) | Algorithm B (only using textual modality) |
|---|---|---|---|
| MAP | 0.2909 | 0.0101 | 0.2013 |
We further compared the proposed approach with the state-of-the-art. The first compared algorithm, defined as algorithm C, was developed using similar multimodal features and learning framework as introduced in Ref. 24. The second compared algorithm, defined as algorithm D, used similar multimodal features and learning framework as introduced in Ref. 25. The last compared algorithm, defined as algorithm E, was developed by researching and simulating the techniques used by the ImageCLEF medical retrieval challenge 2013 participant.72,73 We carefully researched paper from the best performer73 (in terms of MAP) and tried to simulating their proposed approach. The best performer in ImageCLEF 2013 is the ITI (Image and Text Integration Project) group from the Communications Engineering Branch of the Lister Hill National Center for Biomedical Communications. The Lister Hill National Center is a research division of the US National Library of Medicine. Table 3 shows the results (MAP) from different techniques. As shown in this table, our proposed approach outperforms algorithm C. This means that our proposed feature fusion techniques based on the extended pLSA model are more suitable than feature normalization and concatenation (which were used in algorithm C). The results of our proposed approach are only slightly better than the results from algorithm D. However, we should keep in mind that our proposed approach used a single pLSA model, but algorithm D employed multiple pLSA models. Therefore, the implementation of our proposed approach is much simpler than algorithm D. The average MAP in our approach is only slightly worse than algorithm E (best performer in the ImageCLEF medical retrieval challenge 2013). One of the possible reasons is the usage of the medical ontology (eg, Unified Medical Language System) by the best performer in the ImageCLEF challenge. We believe that further improvements can be achieved by employing a medical ontology. This will be one of our future works.
Table 3.
Performance comparisons between the proposed approach and the state-of-the-art image retrieval techniques.
One additional advantage of our approach, compared with the existing methodology, is able to derive the missing modality using the models we developed (as shown in the section Step 2: deriving missing modality). In order to verify the performance of the proposed model for deriving missing modality, we purposely removed part of the textual information. Specifically, we conducted the evaluation when 10%, 15%, 20%, 25%, and 30% of the textual information were missing from the training data set while keeping all the other conditions unchanged. The average MAP we received under these settings is listed in Table 4. In this table, the first row is the percentage of missing textual information. The second row in this table shows the results (MAP) under different missing rates. As shown in this table, the MAP values under different missing rates are just slightly worse than the MAP values with all the date sets ready. This verifies the effectiveness of our approach.
Table 4.
Results of the proposed approach when certain percentage of textual modality is missing.
| Percentage of missing textual modality | 10% | 15% | 20% | 25% | 30% |
|---|---|---|---|---|---|
| MAP | 0.2709 | 0.2601 | 0.2505 | 0.2459 | 0.2319 |
Conclusions
Our research aims to develop effective and efficient CBMIR systems for cancer clinical practice and research. This is very important because medical imaging is becoming a vital component of war on cancer. Direct applications of existing CBIR techniques to the medical images produced unsatisfactory results, because of the unique characteristics of medical images. In this paper, we developed a new multimodal medical image retrieval approach based on the recent advances in statistical graphic model and deep learning. We have investigated a new extended pLSA model to integrate the visual and textual information from medical images. We also developed a new DBM-based multimodal learning model to learn the joint density model from multimodal information in order to derive the missing modality. To verify the effectiveness of the proposed approach, we validated our system with a large volume of real-world medical images. The experimental results have shown that the proposed approach is a promising solution for next-generation medical imaging indexing and retrieval system. In the future, we plan to refine our proposed approach with larger data sets and to include medical ontology into our approach. We also plan to explore the possibility of integrating our proposed approach into clinical practice.
Acknowledgments
The authors would like to thank Chyeeka Brown and Huijuan Xu for contributing on part of the system implementation.
Footnotes
ACADEMIC EDITOR: J.T. Efird, Editor in Chief
FUNDING: The research is partially supported by National Science Foundation of the United States (Award No. 1156639, 1229213, 1415477, and 1440737) and National Natural Science Foundation of China (Award No. 61272147, 61272062, 61070127, 61102028, and 61100102). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: YC, JH, DX, CT. Analyzed the data: SS, JH, PC, HM. Wrote the first draft of the manuscript: YC, SS. Contributed to the writing of the manuscript: YC, JH, DX, CT, PC, HM. Agreed with manuscript results and conclusions: YC, JH, DX, CT, PC, HM. Jointly developed the structure and arguments for the paper: YC, JH, DX, CT, PC, HM. Made critical revisions and approved the final version: YC, SS, JH, DX, CT, PC, HM. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Marcia A, Kassirer P, Relman A. Looking back on the Millennium in medicine. N Engl J Med. 2000;342:42–9. doi: 10.1056/NEJM200001063420108. [DOI] [PubMed] [Google Scholar]
- 2.Diagnostic imaging market to increase to $26.6 billion by 2016 by Companies & Markets, a leading global aggregator of business information. 2013. Available at: http://www.companiesandmarkets.com/News/Healthcare-and-Medical/Diagnostic-imaging-market-to-increase-to-26-6-billion-by-2016/NI6386.
- 3.Roobottom C, Mitchell G, Morgan-Hughes G. Radiation-reduction strategies in cardiac computed tomographic angiography. Clin Radiol. 2010;65(11):859–67. doi: 10.1016/j.crad.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 4.AT&T, Accenture Service Stores Medical Images in Cloud. Available at: http://www.informationweek.com/news/healthcare/interoperability/232200581.
- 5.Morin RL. Transforming the radiological interpretation process (TRIP) J Digital Imaging. 2004;17(2):78–9. [Google Scholar]
- 6.Medical imaging in cancer care: charting the progress prepared by Polidais LLC for Philips Healthcare. Available at: http://www.healthcare.philips.com/pwc_hc/us_en/about/Reimbursement/assets/docs/cancer_white_paper.pdf.
- 7.Brinkley JF, Greenes RA. Imaging and structural informatics. In: Shortliffe EH, Cimino JJ, editors. Biomedical Informatics: Computer Applications in Health Care and Biomedicine. 3rd ed. New York: Springer; 2006. pp. 344–78. [Google Scholar]
- 8.Aisen AM, Broderick LS, Winer-Muram H, et al. Automated storage and retrieval of thin-section CT images to assist diagnosis: system description and preliminary assessment. Radiology. 2003;228(1):265–70. doi: 10.1148/radiol.2281020126. [DOI] [PubMed] [Google Scholar]
- 9.Müller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medicine – clinical benefits and future directions. Int J Med Inform. 2004;73:1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann TM, Güld MO, Thies C, et al. Content-based image retrieval in medical applications. Methods Inf Med. 2004;43(4):354–61. [PubMed] [Google Scholar]
- 11.Ruiz ME. Combining image features, case descriptions and UMLS concepts to improve retrieval of medical images; Paper presented at: Proceedings of American Medical Informatics Association Annual Symposium; 2006. [PMC free article] [PubMed] [Google Scholar]
- 12.Deserno TM, Antani S, Long R. Exploring access to scientific literature using content-based image retrieval; Paper presented at: Proceedings of the SPIE, Medical Imaging 2007: PACS and Imaging Informatics; San Diego, California, USA. 2007. [Google Scholar]
- 13.Müller H Kalpathy-Cramer J. Analyzing the content out of context – features and gaps in medical image retrieval. J Healthcare Inf Sys Inf. 2009;4(1):88–98. [Google Scholar]
- 14.Névéol A, Deserno TM, Darmoni SJ, Güld MO, Aronson AR. Natural language processing versus content-based image analysis for medical document retrieval. J Am Soc Inf Sci Technol. 2009;60(1):123–34. doi: 10.1002/asi.20955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalpathy-Cramer J, Hersh W. Medical image retrieval and automatic annotation: OHSU at ImageCLEF 2007; Paper presented at: Proceedings of 8th Workshop of the Cross-Language Evaluation Forum, CLEF; Budapest, Hungary. 2007. [Google Scholar]
- 16.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med. 1993;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demner-Fushman D, Antani S, Siadat M-R, Soltanian-Zadeh H, Fotouhi F, Elisevich AK. Automatically finding images for clinical decision support; Paper presented at: Proceedings of the Seventh IEEE International Conference on Data Mining (ICDM) Workshops; Omaha, Nebraska, USA. 2007. [Google Scholar]
- 18.Atmosukarto I, Travillian R, Franklin J, et al. A unifying framework for combining content-based image retrieval with relational database queries for biomedical applications; Paper presented at: Proceedings of Annual Meeting of the Society for Imaging Informatics in Medicine; 2008. [Google Scholar]
- 19.Syeda-Mahmood T, Wang F, Beymer D, Amir A, Richmond M, Hashmi S. AALIM: Multimodal Mining for healthcare decision support; Paper presented at: Proceedings of IEEE Conference on Computers in Cardiology (CinC), 2007–2009; Durham, North Carolina, USA. [Google Scholar]
- 20.Müller H Geissbuhler A. Medical Multimedia Retrieval 2.0. In: Murray P, editor. Year book of Medical Informatics – Methods of Information in Medicine. Vol. 3. Stuttgart, Germany: Schattauer Publishers; 2008. pp. 55–64. [Google Scholar]
- 21.Cao Y, Kalpathy-Cramer J, Ünay D. Medical multimedia analysis and retrieval; Paper presented at: Proceedings of the 19th ACM international conference on Multimedia (ACM MM 2011); Phoenix, AZ, U.S.A.. 2011. [Google Scholar]
- 22.Müller H Greenspan H. Medical Content-Based Retrieval for Clinical Decision Support. Berlin: Springer; 2012. Overview of the Third Workshop on Medical Content – Based Retrieval for Clinical Decision Support (MCBR—CDS 2012) pp. 1–9. [Google Scholar]
- 23.Benois-Pineau J, Briassouli A, Hauptmann A. ACM MM MIIRH 2013: workshop on multimedia indexing and information retrieval for healthcare; Paper presented at: Proceedings of the 21st ACM international conference on Multimedia; 2013. 2013. [Google Scholar]
- 24.Pham T-T, Maillot NE, Lim J-H, Chevallet JP. Latent semantic fusion model for image retrieval and annotation; Paper presented at: Proceedings of the sixteenth ACM conference on Conference on information and knowledge management (CIKM); Lisbon, Portugal. 2007. [Google Scholar]
- 25.Lienhart R, Romberg S, Hörster E. Multilayer pLSA for multimodal image retrieval; Paper presented at: Proceedings of the ACM International Conference on Image and Video Retrieval (CIVR); Island of Santorini, Greece. 2009. [Google Scholar]
- 26.Bengio Y. Learning Deep Architectures for AI Foundations and trends® in Machine Learning. Vol. 2. Boston: Now publishers inc; 2009. pp. 1–127. [Google Scholar]
- 27.Salakhutdinov R, Hinton GE. Deep Boltzmann machines; Paper presented at: International Conference on Artificial Intelligence and Statistics; 2009. [Google Scholar]
- 28.Datta R, Joshi D, Li J, Wang JZ. Image retrieval: Ideas, influences, and trends of the new age. ACM Comput Surv. 2008;40(2):5, 1–60. [Google Scholar]
- 29.Hidki A, Depeursinge A, Iavindrasana J, Pitkanen MJ, Zhou X, Müller H. The medGIFT project: perspective of a medical doctor. Jpn J Med Imaging Technol. 2007;25:356–61. [Google Scholar]
- 30.Greenspan H, Pinhas AT. Medical image categorization and retrieval for PACS using the GMM-KL framework. IEEE Trans Inf Technol Biomed. 2007;11(2):190–202. doi: 10.1109/titb.2006.874191. [DOI] [PubMed] [Google Scholar]
- 31.Napel SA, Beaulieu CF, Rodriguez C, et al. Automated retrieval of CT images of liver lesions on the basis of image similarity: method and preliminary results 1. Radiology. 2010;256(1):243–52. doi: 10.1148/radiol.10091694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman MM, Antani SK, Long RL, Demner-Fushman D, Thoma GR. Multi-Modal Query Expansion Based on Local Analysis for Medical Image Retrieval. Medical Content-Based Retrieval for Clinical Decision Support. Berlin: Springer; 2010. pp. 110–9. [Google Scholar]
- 33.Quellec G, Lamard M, Cazuguel G, Roux C, Cochener B. Case retrieval in medical databases by fusing heterogeneous information. IEEE Trans Med Imaging. 2011;30(1):108–18. doi: 10.1109/TMI.2010.2063711. [DOI] [PubMed] [Google Scholar]
- 34.Siegel E, Reiner B. The Radiological Society of North America’s medical image resource center: an update. J Digital Imaging. 2001;14(1):77–9. doi: 10.1007/BF03190302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalpathy-Cramer J, de Herrera AGS, Demner-Fushman D, Antani S, Bedrick S, Müller H. Evaluating performance of biomedical image retrieval systems – an overview of the medical image retrieval task at ImageCLEF 2004–2013. Comput Med Imaging Graph. 2015;39:55–61. doi: 10.1016/j.compmedimag.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson MS, You D, Rahman MM, Antani SK, Thoma GR, Demner-Fushman D. Towards the creation of a visual ontology of biomedical imaging entities; Paper presented at: AMIA Annual Symposium Proceedings; 2012. [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng B, Stanley RJ, De S, Antani S, Thoma GR. Automatic detection of arrow annotation overlays in biomedical images. IJHISI. 2011;6(4):23–41. [Google Scholar]
- 38.You D, Simpson M, Antani S, Demner-Fushman D, Thoma GR. Annotating image ROIs with text descriptions for multimodal biomedical document retrieval; Paper presented at: IS&T/SPIE Electronic Imaging; 2013. [Google Scholar]
- 39.Charles E, Kahn J, Thao C. GoldMiner: a radiology image search engine. Am J Roentgenol. 2007;188:1475–8. doi: 10.2214/AJR.06.1740. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A, Kim J, Cai W, Fulham M, Feng D. Content-based medical image retrieval: a survey of applications to multidimensional and multimodality data. J Digital Imaging. 2013;26(6):1025–39. doi: 10.1007/s10278-013-9619-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–7. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 42.Scientists see promise in deep-learning programs, by John Markoff. New York Times. 2012. Available at: http://www.nytimes.com/2012/11/24/science/scientists-see-advances-in-deep-learning-a-part-of-artificial-intelligence.html.
- 43.Deep learning makes MIT Tech review list of top-10 breakthroughs of 2013 by MIT Tech Review. 2013. Available at: http://www.technologyreview.com/featuredstory/513696/deep-learning/
- 44.NYU Deep Learning Professor LeCun Will Head Facebooks New Artificial Intelligence Lab, by Josh Constine, Ntechcrunch.com. 2013. Available at: http://techcrunch.com/2013/12/09/facebook-artificialintelligence-lab-lecun/
- 45.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks; Paper presented at: NIPS; 2012. [Google Scholar]
- 46.Eigen D, Rolfe J, Fergus R, LeCun Y. Understanding Deep Architectures Using a Recursive Convolutional Network. arXiv. Ithaca: 2013. preprint arXiv:1312.1847. [Google Scholar]
- 47.Vincent P, Larochelle H, Bengio Y, Manzagol PA. Extracting and composing robust features with denoising autoencoders; Paper presented at: Proceedings of the 25th international conference on Machine learning; 2008. [Google Scholar]
- 48.Bengio Y, Yao L, Alain G, Vincent P. Generalized denoising auto-encoders as generative models; Paper presented at: Advances in Neural Information Processing Systems; 2013. [Google Scholar]
- 49.Salakhutdinov R, Mnih A, Hinton G. Restricted Boltzmann machines for collaborative filtering; Paper presented at: Proceedings of the 24th international conference on Machine learning; 2007. [Google Scholar]
- 50.Hinton GE. A Practical Guide to Training Restricted Boltzmann Machines. Neural Networks: Tricks of the Trade. Berlin: Springer; 2012. pp. 599–619. [Google Scholar]
- 51.Hinton GE, Sejnowski TJ, Ackley DH. Boltzmann Machines: Constraint Satisfaction Networks that Learn. Pittsburgh, PA: Carnegie-Mellon University, Department of Computer Science; 1984. [Google Scholar]
- 52.Guillaumin M, Verbeek J, Schmid C. Multimodal semi-supervised learning for image classification; Paper presented at: Computer Vision and Pattern Recognition (CVPR), 2010 IEEE Conference; 2010. [Google Scholar]
- 53.Srivastava N, Salakhutdinov R. Multimodal learning with deep Boltzmann machines; Paper presented at: NIPS; 2012. [Google Scholar]
- 54.The MIRFLICKR retrieval evaluation. Available at: http://press.liacs.nl002Fmirflickr/
- 55.Zheng Y-T, Zhao M, Neo S-Y, Chua T-S, Tian Q. Visual Synset: Towards a higher-level visual representation; Paper presented at: Proceedings of IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Anchorage, AK, USA. 2008. [Google Scholar]
- 56.Liu D, Hua G, Viola P, Chen T. Integrated feature selection and higher-order spatial feature extraction for object categorization; Paper presented at: Proceedings of IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Anchorage, AK. 2008. [Google Scholar]
- 57.Zhang S, Tian Q, Hua G, Huang Q, Li S. Descriptive visual words and visual phrases for image applications; Paper presented at: Proceedings of the seventeen ACM international conference on Multimedia (ACM MM); Beijing, China. 2009. [Google Scholar]
- 58.Lazebnik S, Schmid C, Ponce J. Spatial pyramid matching. In: Dickinson S, Leonardis A, Schiele B, Tarr M, editors. Object Categorization: Computer and Human Vision Perspectives. Cambridge, UK: Cambridge University Press; 2009. pp. 401–5. [Google Scholar]
- 59.Lazebnik S, Raginsky M. Supervised learning of quantizer codebooks by information loss minimization. IEEE Trans Pattern Anal Mach Intell. 2009;31(7):1294–309. doi: 10.1109/TPAMI.2008.138. [DOI] [PubMed] [Google Scholar]
- 60.Moosmann F, Nowak E, Jurie F. Randomized clustering forests for image classification. Trans Pattern Anal Mach Intell. 2008;30(9):1632–46. doi: 10.1109/TPAMI.2007.70822. [DOI] [PubMed] [Google Scholar]
- 61.Lowe DG. Distinctive image features from scale-invariant keypoints. Int J Comput Vis. 2004;60(2):91–110. [Google Scholar]
- 62.Manning CD, Surdeanu M, Bauer J, Finkel J, Bethard SJ, McClosky D. The Stanford CoreNLP natural language processing toolkit; Paper presented at: Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics System Demonstrations 2014; 2014. [Google Scholar]
- 63.Sivic J, Russell B, Efros A, Zisserman A, Freeman W. Discovering object categories in image collections; Paper presented at Proceedings of the IEEE International Conference on Computer Vision (ICCV); Beijing, P.R. China. 2005. [Google Scholar]
- 64.Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18(7):1527–54. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 65.Arel I, Rose DC, Karnowski TP. Deep machine learning-a new frontier in artificial intelligence research [research frontier] IEEE Comput Intell Mag. 2010;5(4):13–8. [Google Scholar]
- 66.Casella G, George EI. Explaining the Gibbs sampler. Am Stat. 1992;46(3):167–74. [Google Scholar]
- 67.Ngiam J, Khosla A, Kim M, Nam J, Lee H, Ng AY. Multimodal deep learning; Paper presented at: Proceedings of the 28th International Conference on Machine Learning (ICML-11); 2011. [Google Scholar]
- 68.Srivastava N, Salakhutdinov R. Multimodal learning with deep Boltzmann machines; Paper presented at: Advances in Neural Information Processing Systems 25; 2012. [Google Scholar]
- 69.Lazebnik S, Schmid C, Ponce J. Beyond bags of features: Spatial pyramid matching for recognizing natural scene categories; Paper presented at: Proceedings of the 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR); New York, NY, USA. 2006. [Google Scholar]
- 70.Akgül CB, Napel DLRS, Beaulieu CF, Greenspan H, Acar B. Content-based image retrieval in radiology: current status and future directions. J Digital Imaging. 2011;24(2):208–22. doi: 10.1007/s10278-010-9290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.trec_eval A standard tool used by the TREC community for evaluating an ad hoc retrieval run. 2010. [Accessed February 1, 2010]. Available at: http://trec.nist.gov/trec_eval/2010.
- 72.de Herrera AGS, Kalpathy—Cramer J, Demner-Fushman D, Antani S, Müller H. Working notes of CLEF. 2013. Overview of the ImageCLEF 2013 medical tasks. [Google Scholar]
- 73.Simpson MS, You D, Rahman MM, Demner-Fushman D, Antani S, Thoma G. Working Notes of CLEF. 2013. ITI’s participation in the 2013 medical track of ImageCLEF. [Google Scholar]