SUMMARY
Cancer cells adapt their metabolic processes to support rapid proliferation, but less is known about how cancer cells alter metabolism to promote cell survival in a poorly vascularized tumor microenvironment1–3. Here, we identify a key role for serine and glycine metabolism in the survival of brain cancer cells within the ischemic zones of gliomas. In human glioblastoma multiforme (GBM), mitochondrial serine hydroxymethyltransferase (SHMT2) and glycine decarboxylase (GLDC) are highly expressed in the pseudopalisading cells that surround necrotic foci. We find that SHMT2 activity limits that of pyruvate kinase (PKM2) and reduces oxygen consumption, eliciting a metabolic state that confers a profound survival advantage to cells in poorly vascularized tumor regions. GLDC inhibition impairs cells with high SHMT2 levels as the excess glycine not metabolized by GLDC can be converted to the toxic molecules aminoacetone and methylglyoxal. Thus, SHMT2 is required for cancer cells to adapt to the tumor environment, but also renders these cells sensitive to glycine cleavage system inhibition.
Many inborn disorders of amino acid metabolism lead to severe impairment of the developing nervous system, at least in part through toxic effects on neural stem cells4,5. As brain cancer cells with high tumorigenic potential share characteristics with neural stem cells6, we wondered whether they might have similar metabolic vulnerabilities. To begin to test this idea, we identified a set of amino acid catabolism genes whose loss causes developmental brain toxicity (Supplemental Table 1) and identified those with elevated expression in glioma compared to normal brain (Supplemental Table 2). This analysis yielded seven genes (Fig. 1a), and we focused on glycine decarboxylase (GLDC) because its expression was also highly enriched in neural stem cells (Fig. 1a). Previous work shows that elevated GLDC expression in non-small cell lung cancer tumor initiating cells promotes oncogenesis by upregulating pyrimidine biosynthesis7. GLDC encodes the central component of a four-protein complex (glycine cleavage complex) that catalyzes the degradation of glycine into ammonia, carbon dioxide, and a methylene group that enters the folate pool, and its loss causes nonketotic hyperglycinemia (NKH), a disorder that severely affects the developing brain5,8.
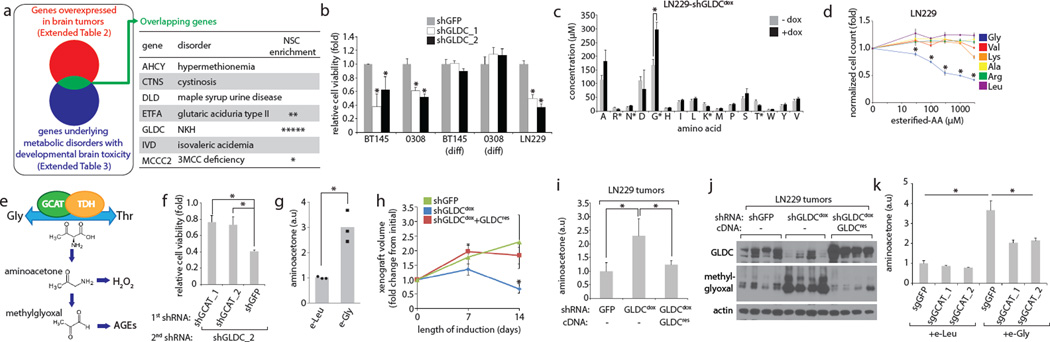
Figure 1. GLDC is required to prevent glycine accumulation and its conversion to aminoacetone and methylglyoxal.
a, Candidate gene identification scheme. Each asterisk in the “NSC enrichment” column indicates that the given gene was significantly overexpressed (over 2-fold, p < 0.05) in neural stem cells compared to differentiated controls (Methods; total of 5 microarray studies). b, Viability of cells expressing the indicated shRNAs for 6 days. Values are relative to that of cells expressing shGFP. c, Amino acid analysis of LN229 cells with or without doxycycline induction of shGLDCdox for 5 days. d, Cell numbers following treatment with indicated doses of esterified amino acids for 5 days. Values are relative to the cell number counts of untreated controls. e, Diagram depicting glycine/threonine interconversion. f, Viability of LN229 cells first transduced with control (shGFP) or GCAT shRNAs, then transduced with shGLDC_2 shRNA for 5 days. Values are relative to that of the same cells secondarily transduced with shGFP instead of shGLDC_2. g, Aminoacetone levels in LN229 cells treated with 1 mM esterified leucine or glycine for 3 days. h, Volumes of xenografts formed from LN229 cells expressing shGFP (n=5), shGLDCdox (n=8), or shGLDCdox plus shRNA-resistant GLDC (n=8). Tumors were allowed to form for two weeks prior to dox induction (Methods). Volumes are shown as relative to the starting volume (at beginning of induction) for each tumor. Error bars are SE. i, Aminoacetone levels, normalized to tumor weight, from xenograft tumors shown in h, n=4 per group. j, Immunoblots from xenograft tumors shown in h. Methylglyoxal levels are indicated by argpyrimidine antibody which recognizes proteins modified by methylglyoxal. k, Aminoacetone levels in cells stably transduced with Cas9 and single guide RNA against GCAT or control (GFP), and treated (4 days and 2 days prior to collection) with 1 mM esterified leucine or glycine. For b,c,d,f,g, and k, n=3 independent biological replicates; error bars are SD. For h, and i, each n is a xenograft tumor; error bars are SD. For all panels, *P<0.05 (student’s t test).
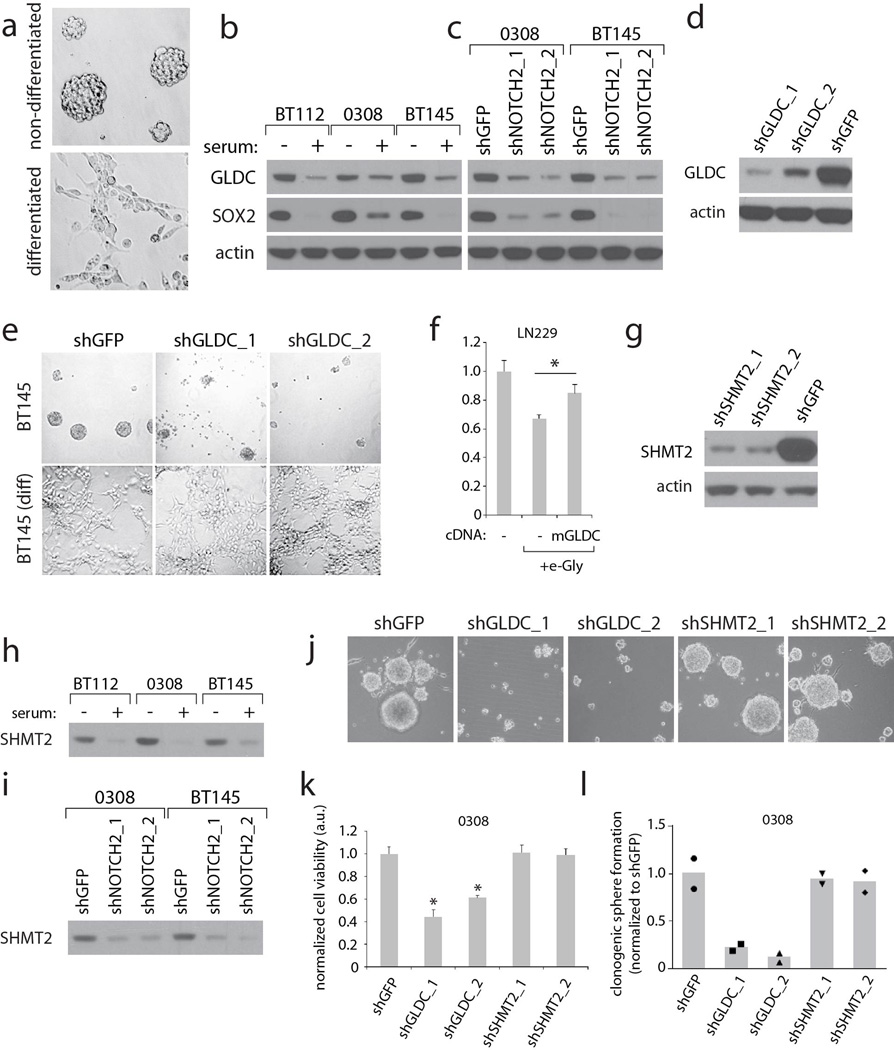
Consistent with the bioinformatic analysis, GLDC protein was highly expressed in tumorigenic9,10 glioblastoma-derived neurosphere-forming cell lines BT145 and 0308, but not in their differentiated, non-tumorigenic counterparts (Extended Data Fig. 1a–c). RNAi-mediated inhibition of GLDC caused loss of viability and breakdown of neurospheres, but did not affect the differentiated cells (Fig. 1b, Extended Data Fig. 1d and e). GLDC suppression was also toxic to LN229 cells, an adherent GBM cell line. Thus, loss of GLDC function has toxic consequences on a subset of GBM cell lines in culture.
We hypothesized that loss of GLDC may lead to the accumulation of toxic amounts of glycine. Indeed, in LN229 cells GLDC suppression raised the levels of intracellular glycine (Fig. 1c), as has been observed in the plasma in NKH5. Interestingly, esterified glycine, which readily crosses cellular membranes and is processed into glycine11, caused dose dependent toxicity to the cells while other esterified amino acids did not (Fig. 1d), and this toxicity was reduced by overexpression of GLDC (Extended Data Fig. 1f).
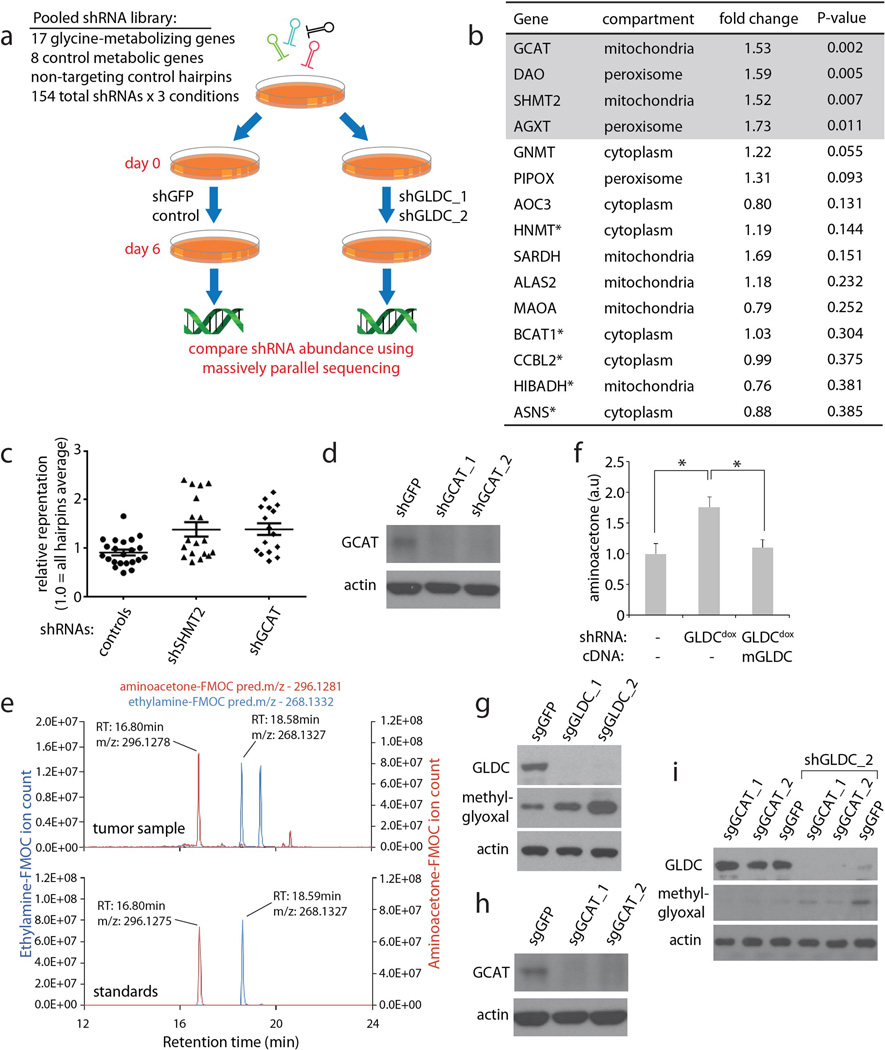
To understand why excess glycine may be toxic to cells, we considered possible alternative fates for glycine not degraded by GLDC, its primary route of catabolism. Based on the KEGG database, there are at least 17 metabolic enzymes that process glycine, and thus we examined whether disruption of any of these other metabolic routes may affect cell sensitivity to GLDC suppression, using a pooled shRNA approach (Extended Data Fig. 2a–c). We found that suppression of glycine C-acetyltransferase (GCAT) protects against the toxicity of GLDC knockdown (Fig. 1f, Extended Data Fig. 2c and d). GCAT is part of a pathway that interconverts glycine and threonine in the mitochondria12,13 (Fig. 1e) via 2-amino-3-ketobutyrate, an unstable intermediate that is spontaneously decarboxylated to form the toxic pro-oxidant metabolite aminoacetone14, which itself is readily metabolized to methylglyoxal, a toxic, highly reactive aldehyde implicated in the pathology of diabetes and other disorders15.
This raised the possibility that the glycine that is metabolized by GCAT, instead of GLDC, can be converted to aminoacetone and methylglyoxal. Indeed, GLDC knockdown or esterified glycine overload led to aminoacetone formation in LN229 cells grown in culture or as a xenografted tumor (Fig. 1g–i, Extended Data Fig. 2e and f). GLDC knockdown also increased methylglyoxal levels, as indicated by increases in argpyrimidine, a methylglyoxal-derived advanced glycation end product (Fig. 1j, Extended Data Fig. 2g). Importantly, these changes were suppressed by silencing of GCAT (Fig. 1k, Extended Data Fig. 2i). Thus, glycine accumulation is deleterious, at least in part, because it is converted via GCAT to aminoacetone and methylglyoxal when not sufficiently catabolized by GLDC. Recent work shows that in the absence of serine large quantities of glycine can be toxic by causing a depletion of the one-carbon pool that is rescued by formate supplementation16. Formate does not rescue glycine toxicity under our conditions (data not shown), arguing that additional toxicities from excess glycine beyond depletion of one-carbon units contribute to the deleterious effects of GLDC inhibition in these cells.
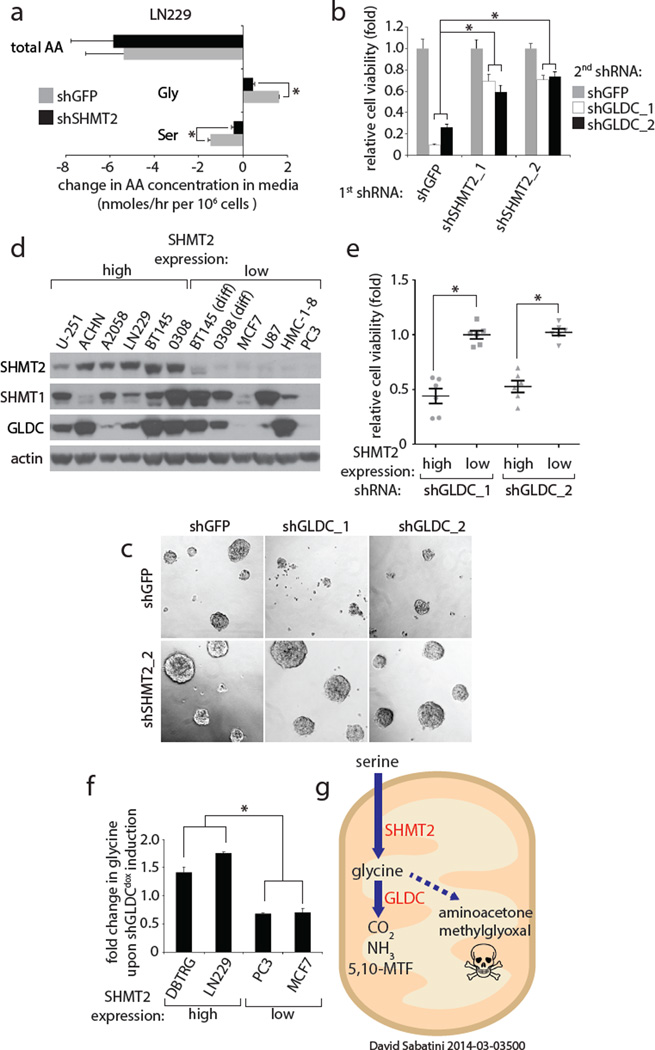
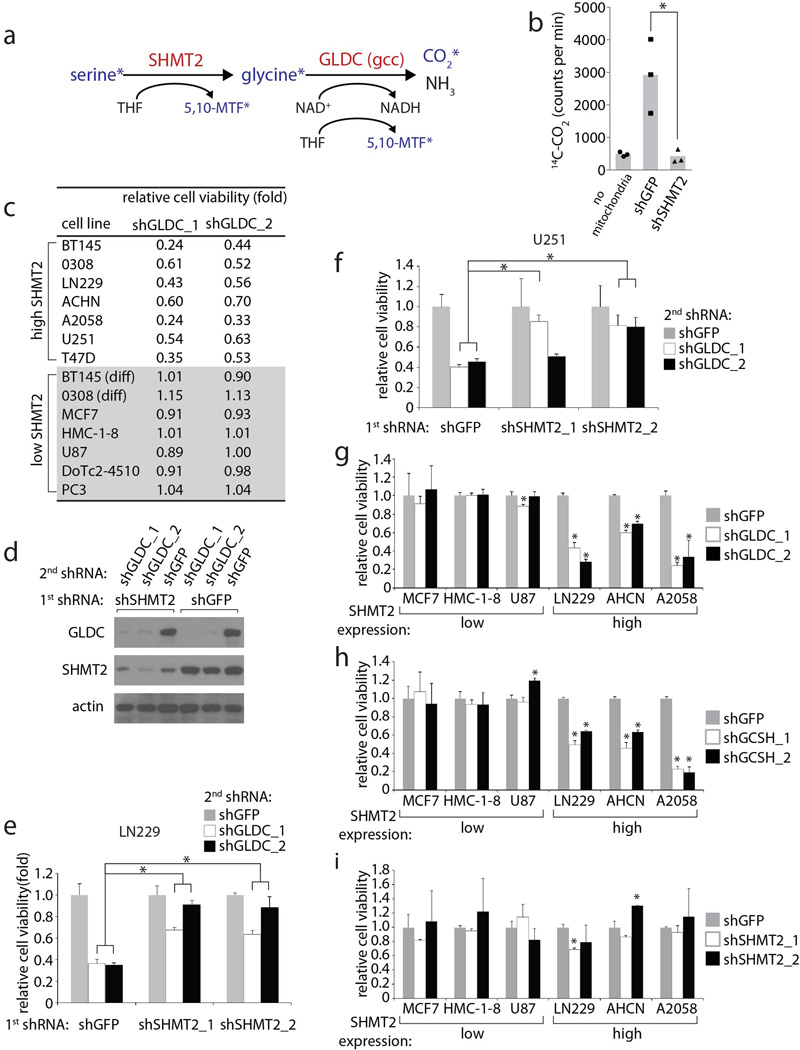
To more rigorously test the idea that GLDC inhibition impairs cell viability by causing the accumulation of glycine, we suppressed the upstream enzyme serine hydroxymethyltransferase (SHMT2) (Extended Data Fig. 3a). While SHMT2 is a mitochondrial enzyme that converts serine to glycine and acts as a key source of glycine in proliferating cells, the cytoplasmic SHMT1 enzyme does not significantly contribute to glycine production17,18. Consistent with SHMT2 functioning upstream of GLDC, suppression of SHMT2 (Extended Data Fig. 1g) decreased both net serine consumption and glycine production in LN229 cells (Fig. 2a) and completely prevented glycine cleavage activity in isolated mitochondria as measured by 14C-CO2 release (Extended Data Fig. 3a and b). Importantly, the preemptive knockdown of SHMT2 protected BT145, LN229, and U251 (a GBM line) cells against the detrimental effects of GLDC knockdown (Fig. 2b and c, Extended Data Fig. 3d–f). These results strongly suggest that the toxicity caused by GLDC suppression is due to an accumulation of the GLDC substrate glycine instead of the depletion of 5,10-methylenetetrahydrofolate (5,10-MTF) and NADH, metabolites produced by the glycine cleavage reaction (Extended Data Fig. 3a). Furthermore, this may explain why the differentiated BT145 and 0308 cells, which express low levels of SHMT2 (Extended Data Fig. 1h and i), are insensitive to suppression of GLDC. In a panel of cancer cell lines, we found a striking correlation between SHMT2 expression levels and sensitivity to GLDC silencing (Fig. 2d and e and Extended Data Fig. 3g–i), a pattern that also matched their intracellular glycine accumulation (Fig. 2f). Collectively, these findings reveal a conditionally lethal relationship between SHMT2 and GLDC, in which SHMT2-mediated production of glycine necessitates its clearance by GLDC so as to prevent its conversion to toxic metabolites such as aminoacetone and methylglyoxal (Fig. 2g). As seen in the panel of cell lines, this relationship is likely relevant across multiple cancer cell types and is not limited to GBM cells.
Figure 2. SHMT2 activity renders cells liable to toxic accumulation of glycine upon GLDC loss.
a, Changes in serine, glycine, and total amino acid levels over 84 hours in media of LN229 cells expressing shGFP or shSHMT2_1, measured using absolute quantitative CE-MS (Methods). Positive values (right of the y axis) indicate a net accumulation in the media, while negative values indicate net consumption from the media. b, Viability of BT145 cells first transduced with shGFP or SHMT2 shRNAs, then with shGFP or GLDC shRNAs for 5 days. Values are relative to that of cells secondarily transduced with shGFP. c, Representative micrographs of b. d, Immunoblots in a panel of cell lines with high or low SHMT2 expression. e, Viability of cell lines in the high and low expression groups expressing shGLDC_1 and shGLDC_2 for 6–7 days. Values are relative to the viability of the same cells secondarily transduced with shGFP; individual results shown in Extended Data Fig. 3a. f, Glycine levels upon doxycycline-induced expression of shGLDC_2 for 5 days in different cell lines; values are relative to cells without induction; 1.0 indicates no change. g, Schematic of serine/glycine metabolism and cell survival in cancer cells. For a,b, and f, n=3 independent biological replicates; error bars are SD. For e, each point (n=6) represents a single cell line from g. Bars are mean ± SEM. For all panels, *P<0.05 (student’s t test).
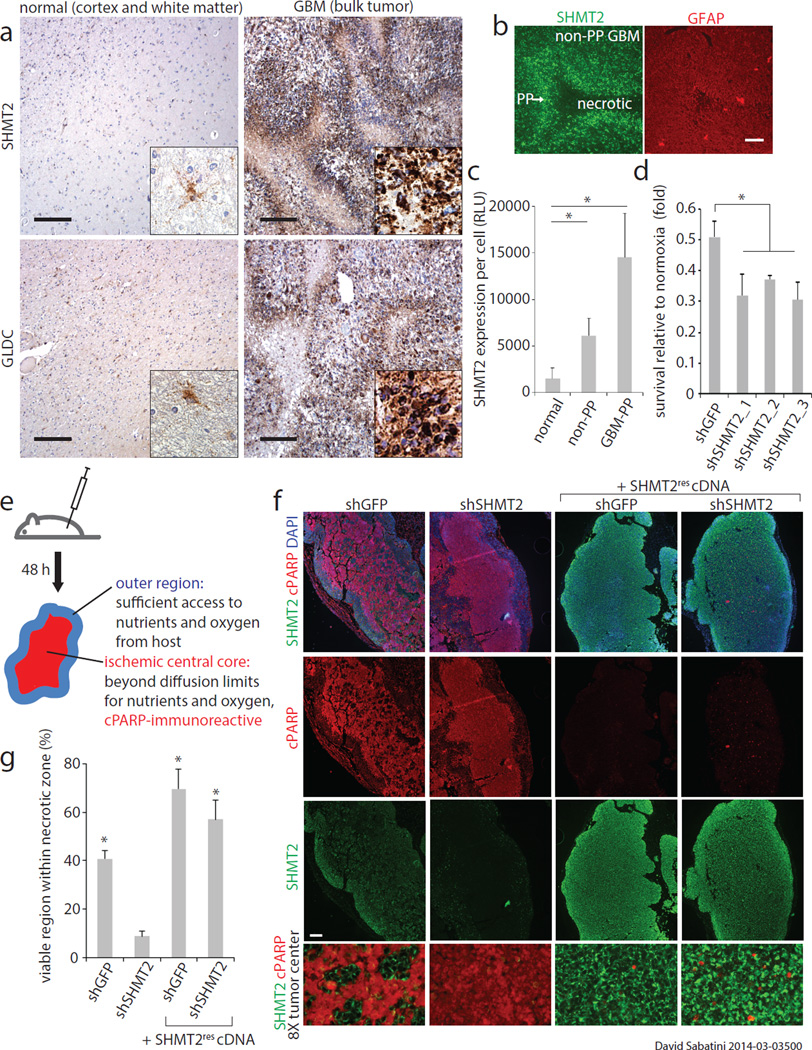
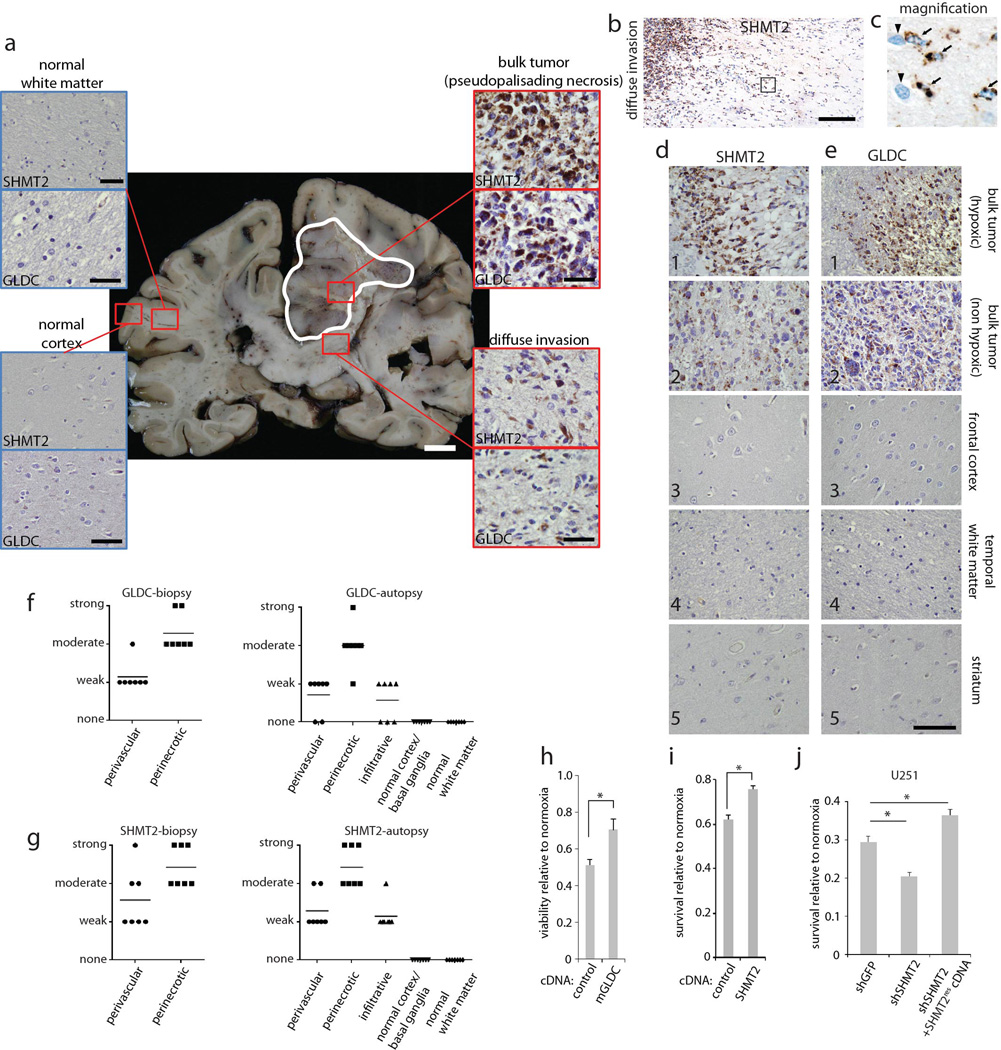
In contrast to the toxic effects GLDC knockdown, that of SHMT2 did not affect the proliferation or survival of multiple cell lines under normal culture conditions (Extended Data Fig. 3i). Furthermore, SHMT2 was not necessary for the proliferation or self-renewal of neurosphere forming cells (Extended Data Fig.1 j–l). As it seemed unlikely that cancer cells would obtain high SHMT2 expression if it did not provide a benefit, we considered that SHMT2 might have a context-dependent role and examined SHMT2 and GLDC expression in sections of human GBM tumors. In normal brains SHMT2 and GLDC expression was not detected in most cells but was at low levels in astrocytes and vessels (Fig. 3a, Extended Data Fig. 4a,d,e). In GBM tumors, however, both SHMT2 and GLDC were expressed at high levels (Fig. 3a, Extended Data Fig. 4a,d, and e) that even allowed the detection of individual cancer cells migrating into the brain parenchyma (Extended Data Fig. 4b and c). Interestingly, the highest levels of SHMT2 and GLDC expression were distinct bands surrounding necrotic and acellular regions, highlighting cells of what is referred to as the pseudopalisading necrosis (Fig. 3a–c, Extended Data Fig. 4a,f, and g). This feature, which is unique to glioblastomas, consists of a dense layer of “pseudopalisading” viable cells that outline an ischemic tumor region that is thought to form upon the collapse or occlusion of an intratumoral vessel19.
Figure 3. SHMT2 expression provides a survival advantage in the ischemic tumor microenvironment.
a, SHMT2 and GLDC expression in normal human brains and GBM tumors. Insets are 5× fold magnifications. Representative images are shown; comprehensive histological analyses are in Extended Data Figure 5. Scale bar = 200 µm. b, SHMT2 immunofluorescence in the cells in the pseudopalisades (PP) and in the non-pseudopalisade GBM regions. Glial fibrillary acidic protein (GFAP), a general GBM cell marker, does not show increased signal in pseudopalisades. c, Quantification of SHMT2 expression, measured as fluorescence intensity per cell, in normal brain regions, non-pseudopalisade GBM regions, and pseudopalisade regions (n=5 patient samples per group). Error bars are SD. d, Cell number counts from LN229 cells expressing shRNAs against GFP or SHMT2 and cultured in 0.5% hypoxia for 8 days. Values are relative to the counts of the same cells cultured in parallel in normoxia; n=3 independent biological replicates; error bars are SD. e, Schematic of experimental design for rapid xenograft model. f, Representative micrographs of rapid xenograft tumors formed by LN229 cells transduced with indicated shRNAs and cDNAs, and immunostained for SHMT2 and cleaved PARP. Bottom row shows 8× magnified, merged images of the central tumor region, displayed without DAPI channel for clarity. Scale bar = 200 µm. g, Quantification of the percentage of cleaved PARP-negative, viable area within the central necrotic region of xenografts of as shown in f. Error bars are SEM (shGFP, n=10; shSHMT2_1, n=9; shGFP+SHMT2rescDNA, n=8; shSHMT2_1 +SHMT2rescDNA, n=8; each n is a xenograft tumor). For all panels, *P<0.05 (student’s t test).
The expression of SHMT2 in ischemic tumor zones suggested that it might have a key role in cells in environments with limited oxygen or nutrient levels. Indeed, under hypoxic conditions (0.5% oxygen), SHMT2 suppression impaired and SHMT2 overexpression enhanced LN229 cell proliferation (Fig. 3d, Extended Data Fig. 4i). As these effects were relatively modest, we set out to more closely recapitulate conditions of tumor ischemia by using a previously described rapid xenograft model20. In this heterotopic model, a large bolus of cells is injected subcutaneously and the tumor collected prior to angiogenesis. Thus, the tumor core experiences oxygen and nutrient deprivation, which frequently results in extensive cell death, while the outermost regions of the tumor receive sufficient oxygen and nutrients and are completely viable (Fig. 3e). In such xenografts, LN229 cells, which express high levels of SHMT2 (Fig. 2d), formed a tumor with a heterogeneous central region that contained both dying cells (labeled by cleaved-PARP) and numerous ‘islands’ of viable cells lacking cleaved PARP (Fig. 3f and g). On the other hand, tumors formed from LN229 cells expressing a SHMT2 shRNA had a uniformly barren, cleaved-PARP immunoreactive central region that was almost completely devoid of any surviving cells. Importantly, overexpression of an RNAi-resistant SHMT2 cDNA not only rescued the effects of SHMT2 knockdown but also had a strong protective effect, in some cases resulting in central tumor regions that were almost entirely viable (Fig. 3f and g). While this model does not directly mimic pseudopalisading necroses, it indicates that SHMT2 expression is an important determinant of cancer cell survival within an ischemic tumor context.
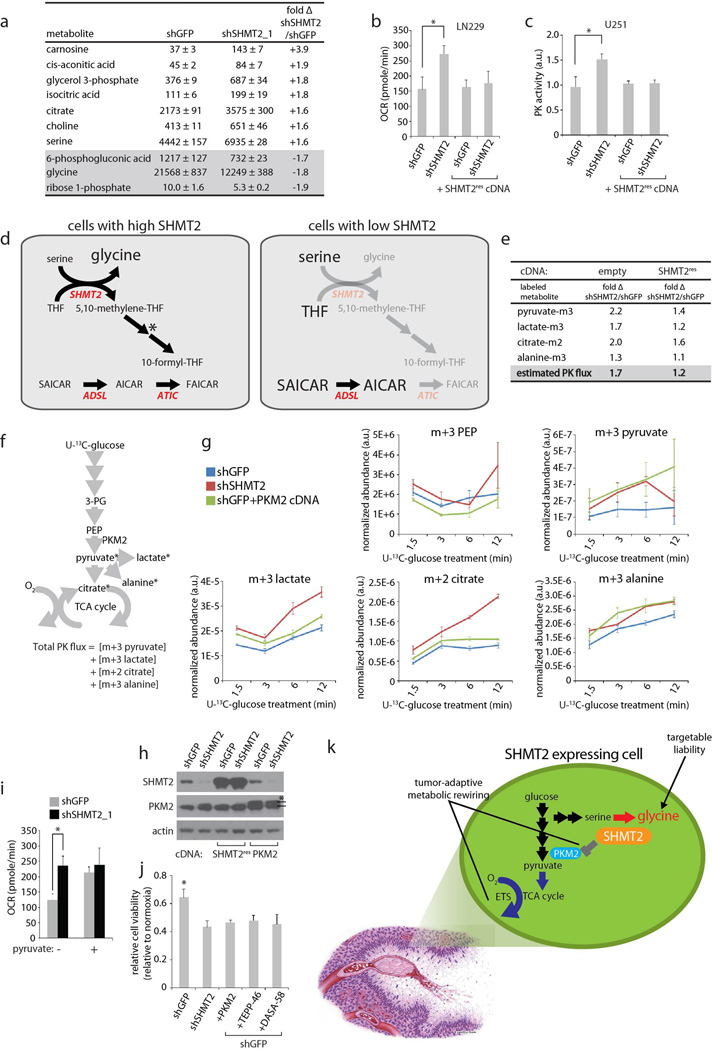
To begin to understand why this might be, we surveyed the metabolic consequences of SHMT2 suppression in LN229 cells. Quantitative central carbon metabolism profiling revealed that in addition to the expected accumulation of serine and depletion of glycine, SHMT2 suppression increased the levels of TCA cycle intermediates and decreased those of the pentose phosphate pathway (Extended Data Fig. 5a). SHMT2 suppression also increased cellular oxygen consumption (Extended Data Fig. 5b), which may reflect increased TCA cycle activity driving NADH into the oxidative phosphorylation pathway. Furthermore, untargeted metabolite profiling identified AICAR, SAICAR, and fructose bisphosphate (FBP) as amongst the most highly elevated metabolites (Fig 4a and Supplemental Tables 6 and 7). The increase in the sequential intermediates SAICAR and AICAR can be explained because 10-formyl-THF, a downstream product of SHMT2 and SHMT1 activity, is required for the conversion of AICAR to FAICAR during de novo purine biosynthesis (Extended Data Fig. 5d). While a link between SHMT2 and FBP is less clear, we nonetheless noted that the suppression of SHMT2 significantly increases levels of all three known activators of pyruvate kinase isoform M2 (PKM2) –serine, FBP, and SAICAR21–23– raising the possibility that SHMT2 antagonizes PKM2 activity by decreasing the levels of its activators.
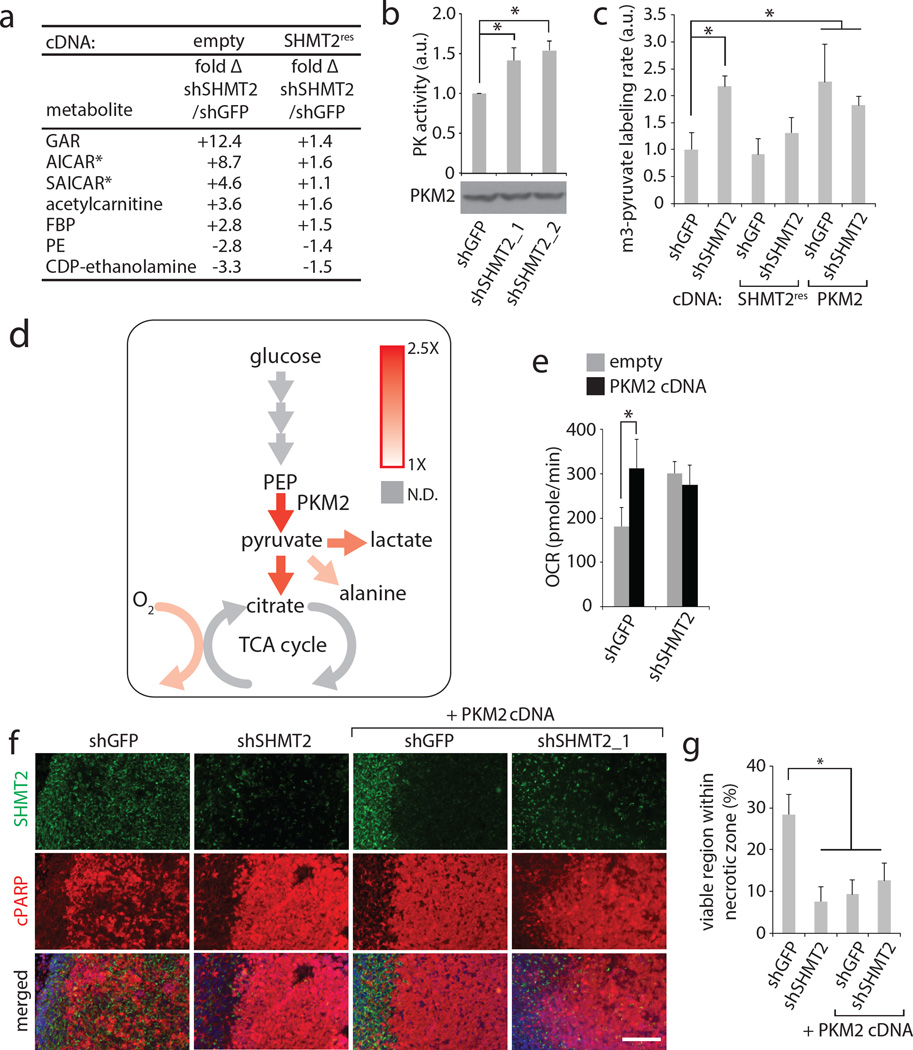
Figure 4. SHMT2 elicits a PKM2-dependent metabolic rewiring that is advantageous to cancer cells in an ischemic environment.
a, Liquid chromatography-mass spectrometry (LC-MS) based, untargeted discovery of metabolites that change in abundance following SHMT2 knockdown in LN229 cells. GAR- glycineamideribotide; FBP – fructose bisphosphate (either 1,6 or 2,6; cannot be distinguished by LC-MS); PE- phosphoethanolamine. Differential peaks were identified and quantified as described in the Methods, and the metabolites with largest change are listed. Metabolite levels are relative and are expressed as fold change in cells transduced with shSHMT2_1 versus cells transduced with shGFP, with or without the RNAi-resistant SHMT2 cDNA. All differences in first column are significant (P<0.05). b, Pyruvate kinase activity assay from lysates of LN229 cells transduced with indicated shRNAs. c, m3-pyruvate labeling rates in LN229 cells transduced with shRNAs and cDNAs as indicated, and fed [U-13C]-glucose media. d, Summary diagram of labeling rate changes seen as a result of SHMT2 silencing. Colored arrows indicate increased flux according to the heat map, while gray arrows indicate non-determined labeling rates. Detailed analyses are in Extended Data Fig. 5. e, Oxygen consumption in LN229 cells (in RPMI) expressing shGFP or shSHMT2_1 with or without PKM2 cDNA. Error bars are SD (n=5 technical replicates). f, Representative micrographs of rapid xenograft tumors formed from LN229 cells stably expressing indicated shRNAs and cDNAs, and immunostained for SHMT2 and cleaved PARP. Viable regions are oriented on the left, and the central ischemic regions on the right. Scale bar = 100 µm. g, Quantification of the percentage of cleaved PARP-negative, viable area within the central necrotic region of xenografts of as shown in f. Error bars are SEM (shGFP, n=10; shSHMT2_1, n=8; shGFP+PKM2 cDNA, n=10; shSHMT2_1 + PKM2 cDNA, n=6; each n is a xenograft tumor). For a,b, and c, n=3 independent biological replicates; error bars are SD. For all panels, *P<0.05 (student’s t test).
Pyruvate kinase catalyzes the conversion of phosphoenolpyruvate to pyruvate in glycolysis, and PKM2 is the isoform associated with proliferating cells1,2. PKM2 has regulated activity, unlike the constitutively active PKM1. Decreasing PKM2 activity can allow redistribution of glycolytic carbons in a manner advantageous for cancer cell proliferation in tumors3,24,25, and either pharmacological PKM2 activation or PKM1 expression can impair tumor growth25,26. Consistent with the increase in metabolites known to activate PKM2 (Fig. 2a and 4a), PKM2 activity was significantly increased in cells with suppressed SHMT2, despite no change in PKM2 protein levels (Fig. 4b, Extended Data Fig. 5c). To determine whether SHMT2 silencing induces changes in central carbon metabolism that are consistent with increased pyruvate kinase (PK) activity, we measured kinetic flux through glycolysis in live cells using 13C-stable isotope labeled glucose (U-13C-glucose) (Fig. 4c–d, Extended Data Fig. 5e–h, Supplemental Table 8 and 9). The 13C labeling rate of pyruvate, the product of PKM2, was elevated in cells with suppressed SHMT2, indicating increased PKM2 activity, which was also confirmed in cells overexpressing PKM2 (Fig. 4c). By calculating the sum 13C labeling of lactate, citrate, and alanine, the major downstream fates of pyruvate27, we estimate that the total pyruvate kinase flux is increased by ~70% following SHMT2 knockdown (Extended Data Fig. 5e–g). Furthermore, these changes as well as changes in metabolite levels and oxygen consumption were suppressed by overexpression of an RNAi-resistant SHMT2 cDNA (Fig. 4c, Extended Data Fig. 5e), arguing against off-target RNAi effects. These results support a model in which SHMT2 suppression leads to increased pyruvate kinase activity and carbon flux into the TCA cycle, while cells that express high levels of SHMT2 limit PKM2 activity and flux into the TCA cycle (Fig. 4d). This may confer a survival benefit in ischemic tumor contexts, as it has been shown that limiting pyruvate entry into TCA cycle, and thus limiting oxygen consumption, provides a survival advantage under hypoxia28.
If the effects of SHMT2 on oxygen consumption and survival within an ischemic microenvironment occur via suppression of PKM2 activity, then forced activation of PKM2 should antagonize these effects. Indeed, either overexpression of PKM2 or the addition of the PKM2 product pyruvate to the media increased the oxygen consumption rate in LN229 cells to the equivalent levels observed following SHMT2 knockdown (Fig. 4e and Extended Data Fig. 5i). Thus, pyruvate kinase activity may be a determinant of oxygen consumption in these cells. Furthermore, overexpression of PKM2, or the pharmacological activation of PKM2 using TEPP-46 or DASA-5826 reduced LN229 survival in 0.5% hypoxia to a similar extent as SHMT2 suppression (Extended Data Fig. 5j). Finally, in the rapid xenograft model, PKM2 overexpression, like SHMT2 loss, reduced the survival of LN229 cells (Fig. 4f and g). These findings support a model in which high SHMT2 expression rewires metabolism to suppress PKM2 activity and promote survival in the ischemic tumor environment (Extended Data Fig. 5k).
In summary, we identified toxic glycine accumulation following loss of GLDC as a metabolic liability in cells expressing high levels of SHMT2. Thus, in nonketotic hyperglycinemia, preventing endogenous glycine production via SHMT2 inhibition may be the desired route of therapy, as current treatment options targeting exogenous glycine, such as dietary restriction or plasma glycine conjugation, are largely ineffective5.
On the other hand, SHMT2 is elevated in a subset of cancer cells and promotes changes in metabolism that allow cells to survive in an ischemic tumor microenvironment. It is observed that hypoxia/ischemia selects for cancer cells with increased tumorigenicity and therapy-resistance, and manifestations of tumor ischemia, such as pseudopalisading necrosis, are associated with poor prognoses29. Thus, our findings raise the possibility that GLDC inhibition may be exploited to specifically target malignant and refractory subpopulations of cells expressing high levels of SHMT2.
METHODS
Materials
The following antibodies were used: antibodies to GLDC (HPA002318), SHMT2 (HPA020549) from Sigma; antibodies to actin (sc-1616), SHMT1 (sc-100849), and GCAT(sc-86466) from Santa Cruz; anti-GCSH (H00002653-A01)from Abnova; anti-SOX-2 (MAB2018) from R & D systems; anti-GFAP (IF03L) from Calbiochem; anti-cleaved-PARP (19F4) and anti-PKM2 (D78A4) from Cell Signaling Technologies; anti-GCAT (ab85202) from Abcam; anti-methylglyoxal antibody (MMG-030) from Genox; HRP-conjugated anti-mouse, anti-rabbit, and anti-goat secondary antibodies from Santa-Cruz Biotechnology.
The following cell culture reagents were used: Neurobasal medium, N-2 and B-27 supplements from Invitrogen; Recombinant Human FGF basic (4114-TC) and EGF (236-EG) from R & D systems; DMEM and RPMI-1640 media, doxycycline (D9891) from Sigma; Leucine ethyl ester hydrochloride (61850), Arginine ethyl ester hydrochloride (A2883), Alanine ethyl ester hydrochloride (855669), Valine ethyl ester hydrochloride (220698), Lysine ethyl ester dihydrochloride (62880), Ethylamine (395064) from Sigma; Glycine ethyl ester hydrochloride (sc-295020) and Polybrene (sc-134220) from Santa Cruz.
Additional materials used: formalin from VWR; Borg Decloaker RTU solution and pressurized Decloaking Chamber from Biocare Medical; Prolong Gold Antifade reagent from Invitrogen; CellTiter-Glo Luminescent Assay from Promega; U14C-Serine from MP Biomedicals; Matrigel (356230) from BD Biosciences.
Cell lines, tissue culture, and media
The neurosphere forming lines lines 0308, BT145, and BT112 were established as described10,30,31, kindly provided by Howard Fine and Keith Ligon, and maintained as tumorigenic neural stem cell-like neurospheres in NBE medium (Neurobasal medium containing N-2 and B-27 supplements, Epidermal Growth Factor, basic fibroblast growth factor, L-glutamine, and Penicillin-Streptomycin) as described10. When passaging, spheres were manually broken into smaller spheres and single cells by trypsinization and pipetting. For differentiation experiments, neurospheres were broken into single cells and grown in DMEM (containing 10% inactivated fetal bovine serum and Penicillin-Streptomycin) for at least 1 week.
All other cell lines (LN229, ACHN, A2058, U251, T47D, MCF7, HMC-1–8, U87, DoTc2-4510, and PC3) were obtained from Broad Institute Cancer Cell Line Encyclopedia, and cultured as adherent cell lines in DMEM with exceptions noted below. Cell lines were verified to be free of mycoplasma contamination. Cell line origins are as follows: 0308, BT145, BT112, LN229, U251, U87 (glioblastoma), ACHN (renal cell adenocarcinoma), A2058 (melanoma), T47D (breast ductal carcinoma), MCF7 (breast pleural effusion), HMC-1–8 (breast pleural effusion), DoTc2-4510 (cervical carcinoma), PC3 (prostate adenocarcinoma).
For experiments measuring oxygen consumption and for untargeted metabolite profiling experiments, RPMI was used, which does not contain the PKM2 product pyruvate.
Subcutaneous xenograft experiments
For regular subcutaneous xenograft studies, LN229 cells were transduced to stably express shRNAs (shGLDCdox, shGFP via puromycin selection for 3 days) and then cDNAs (empty vector or RNAi-resistant GLDC cDNA via blastocidin selection for 3 days) then further amplified. Xenografts were initiated with 3 million cells injected subcutaneously per site, with 30% matrigel, 100 µl injection volume in the left and right flanks of female, 6–8 week old NCr nude mice (Taconic). Tumors were allowed to form for two weeks, and at this point the first caliper measurements were taken, and induction started by addition of doxycycline at 2 g/L to drinking water. Tumor volume was calculated using the modified ellipsoid formula ½ (Length × Width2) and expressed as relative fold change to the initial volume of each tumor at the start of doxycycline induction.
For rapid tumor xenograft studies to form ischemic tumor cores, LN229 cells were transduced to stably express both cDNAs (empty vector, RNAi-resistant SHMT2 cDNA, or PKM2 cDNA via blastocidin selection) and shRNAs (via puromycin selection). Xenografts were initiated with 8 million cells injected subcutaneously per site in the left and right flanks of female, 6–8 week old NCr nude mice (Taconic). Tumors were removed at 48 hours post-injection and fixed in 10% formalin.
For the quantification of viable and nonviable regions in the ischemic region, fixed tumors were embedded and sections prepared. Sections were immunostained for SHMT2 and cleaved PARP, and images of the central tumor regions were obtained using a Zeiss Axiovert 200M inverted fluorescent microscope and AxioVision Software. All images were acquired and processed under the same parameters across the entire set. The image labels were scrambled so that analyses could be carried out in a blinded manner, and the Red channel (cPARP) and Blue channel (Hoechst 33342) was analyzed. Using Adobe Photoshop, the entire central necrotic region, labeled by cleaved PARP, was manually outlined with the Lasso tool. Within this tumor region, the total area counts (in pixels) of the dead (cPARP positive and Hoechst positive) and viable (cPARP negative and Hoechst positive) regions were obtained to calculate the percentage of viable region within the central necrotic zone.
Analyses of oncogenomic and other microarray data
We had previously classified a set of 2,752 metabolic enzymes and transporters32. To obtain a list of metabolic genes and transporters that have increased expression in gliomas, we analyzed the 9 expression studies deposited in Oncomine33 that profiled gene expression normal brain tissue and gliomas. For each dataset, the top 10% of genes overexpressed in the glioma relative to normal brain was obtained, and cross referenced with our gene set, which yielded a list of 367 genes which placed within the top 10% of overexpressed genes in at least two separate studies.
To determine the expression of selected metabolic genes in the context of neural stem cells, we analyzed a set of 5 microarray datasets deposited in Gene Expression Omnibus in which neural stem cells are compared with differentiated controls (GSE36484, GSE10721, GSE15209, and two comparison groups in GSE11508). A summary of the datasets and fold change in expression of each gene in each study is provided in Supplemental Table 3.
shRNA expressing lentivirus generation and sequences
For each gene of interest (GLDC, SHMT2, GCAT, GCSH), 5 lentiviral shRNA constructs were obtained from The RNAi Consortium (TRC) and recombinant lentivirus containing supernatant was produced using a transient transfection protocol34. Each lentivirus was separately transduced into LN229 by overnight incubation of virus in trypsin dissociated cells (20,000 cells per ml, 2 ml into each well of a 6-well plate) in the presence of polybrene. Lentiviral expression of shGFP and shLacZ served as negative controls for gene knockdown, and noninfected cells served as negative controls for transduction. Cells were selected with puromycin for 3 days to ensure transduction, and for each gene, the two (or three) most effective shRNAs, in terms of knockdown of protein expression by western blot, were chosen for use in our experiments.
The following shRNA sequences were used:
shGFP: TRCN0000072186, target sequence: TGCCCGACAACCACTACCTGA
shLacZ: TRCN0000072235, target sequence: CCGTCATAGCGATAACGAGTT
shGLDC_1: TRCN0000036599, target sequence: CGAGCCTACTTAAACCAGAAA
shGLDC_2: TRCN0000036603, target sequence: GAAGTTTATGAGTCTCCATTT
shGLDCdox: target sequence same as shGLDC_2, cloned into dox-inducible vector (pLKO_GC11)
shSHMT2_1: TRCN0000238795, target sequence: CGGAGAGTTGTGGACTTTATA
shSHMT2_2: TRCN0000034804, target sequence: CCGGAGAGTTGTGGACTTTAT
shSHMT2_3: TRCN0000234657, target sequence: GTCTGACGTCAAGCGGATATC
shGCSH_1: TRCN0000083395, target sequence: GTGAACTCTATTCTCCTTTAT
shGCSH_2: TRCN0000428788, target sequence: TGAGGAACACCACTATCTTAA
shGCAT_1: TRCN0000034579, target sequence: CCTTAACTTCTGTGCCAACAA
shGCAT_2: TRCN0000034580, target sequence: CCAGAGGTTCCGTAGTAAGAT
shNOTCH2_1: TRCN0000004896, target sequence: CCAGGATGAATGATGGTACTA
shNOTCH2_2: TRCN0000004897, target sequence: CCACACAACAACATGCAGGTT
Cell viability assays with shRNA transduction
For cell viability experiments involving transduction of a single shRNA (e.g. shGLDCs), cell lines (neurosphere forming cell lines, LN229, ACHN, A2058, U251, T47D, MCF7, HMC-1-8, U87, DoTc2-4510, and PC3) were seeded in 96-wells at 3,500 to 5,000 cells per well. The next day, neurosphere forming lines were infected with lentivirus and polybrene via 30-minute spin at 2,250 rpm followed by incubation for 1 hour prior to a media change (due to neurosphere forming cell line sensitivity to prolonged incubation with virus and polybrene), while all the non neurosphere cell lines were infected via overnight incubation of virus and polybrene prior to a media change. For all non-neurosphere cell lines, puromycin selection was started 24h after infection, while for the neurosphere lines it was started 48h after infection (because of their sensitive nature). Cells were incubated for 4–6 additional days as indicated, and overall cell viability was quantified using the Cell Titer Glo (CTG) reagent (Promega) and measuring luminescence. As doubling times and luminescence values per viable cell differ between different cell lines, values are normalized to the same cells transduced in parallel with innocuous shGFP hairpins as indicated.
When comparing sensitivity to GLDC (or GCSH, SHMT2) knockdown across different cell lines, two identical sets of experiments, one which receives puromycin selection and one which does not, were carried out in parallel. Comparing the two ensures that the toxicity observed in the ‘sensitive’ cell lines is due to GLDC knockdown and not due to selection of nontransduced cells, because identical toxicity is also seen in the nonselected plate. Conversely, we can ensure that low toxicity observed in the ‘insensitive’ cell lines is not an artifact of poor transduction because if they had been poorly transduced, then toxicity would be observed in the puromycin selected plate. In this manner, we verified full transduction of cells that we have examined for GLDC effects on viability.
For some experiments, cells are transduced with more than one shRNA and this was carried out in a sequential manner. Cells were infected with the first lentivirus expressing an shRNA (shGFP, shSHMT2_1, or shSHMT2_2, shGCAT_1, shGCAT_2) as described, then selected in puromycin for 3 days, and expanded for 2–5 more days. Equal numbers of each stable cell line were infected with the second lentivirus (shGFP, shGLDC_1, or shGLDC_2), seeded in 96-well plates, and at 5 days following infection, cell viability was measured. Because in some cases (e.g. shGCAT hairpins) the primary transduction itself moderately impairs cell proliferation, viability values for the cells secondarily transduced with shGLDCs are always expressed as relative to the same primary transduced cells, processed in parallel, which are secondarily infected with a control hairpin (shGFP). Because the secondary transduction cannot be selected for (since the cells are already puromycin resistant from the first round of transduction), effective knockdown of the second gene was verified by Western blot.
For cell proliferation experiments, cell counts were determined using a Coulter Counter (Beckman).
CRISPR /Cas9 mediated gene knockdown
In some of our experiments, effective gene knockdown was achieved via CRISPR/Cas-9 mediated genome editing. We utilized pLENTICRISPR, in which both single guide RNA, directed against a target of interest, and the Cas9 endonuclease are both delivered to cells via lentivirus35 in an analogous manner to the TRC shRNA experiments. Three target site sequences, selected based on best scores as previously calculated for all genes36, were cloned into pLENTICRISPR. As described for TRC shRNA transduction, lentiviruses were produced and transduced into trypsin dissociated LN229 cells, via overnight incubation with polybrene. Following media change and puromycin selection, cells were harvested 7 days following infection, and gene knockdown determined by Western blotting. The two most effective target guide sequences, in terms of knockdown of protein expression by western blot, were chosen for use in our experiments.
The following target site sequences, transduced via pLENTICRISPR, were used:
sgGFP: TGAACCGCATCGAGCTGAAG (plus strand)
sgGLDC_1: CGGGACAGCAGCAGTGGCGG (minus strand)
sgGLDC_2: ATTTGGGGTAGACATCGCCC (minus strand)
sgGCAT_1: CCAGCGCTGACTGTGCGCGG (minus strand)
sgGCAT_2: GAAGCATCGGCTGCGCCTGG (plus strand)
Pooled shRNA screening
pLKO.1 lentiviral plasmids encoding shRNAs targeting glycine metabolizing enzymes, metabolic enzymes for other amino acids, as well as nontargeting controls were obtained and combined to form a pool as described in Extended Figure 2. This pool was used to generate a pool or lentiviruses as described34. LN229 cells were infected with the pooled virus at a low titer (multiplicity of infection of 0.7) to ensure that each cell contained only one viral integrant. After cells were selected for 3 days with puromycin, pooled cells were dissociated, divided, and subjected to a secondary infection with either shGFP, shGLDC_1, or shGLDC_2. After 6 days following the secondary infection, a time point corresponding to moderate toxicity as determined by decreased proliferation and moderate changes cell morphology compared to shGFP infected cells, cells were collected to obtain genomic DNA. As previously described32, the shRNAs encoded in the genomic DNA were amplified and analyzed by high throughput sequencing (Illumina) using the following primers:
-
Barcoded Forward Primer (‘N’s indicate location of sample-specific barcode sequence):
AATGATACGGCGACCACCGAGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTG
GAANNGACGAAAC
-
Common Reverse Primer:
CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTTGTGGATGAATACTGCCATTTG
TCTCGAGGTC
Illumina Sequencing Primer: AGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAA
Sequencing reads were deconvoluted using GNU Octave software as described. For each shRNA, Abundance was defined as (number of reads / total number of all reads of pooled cells in either shGFP or shGLDC). Enrichment was defined as (abundance in shGLDC / abundance in shGFP), thus an enrichment score of 2.0 would indicate that an shRNA is twice as abundant in the shGLDC infected pool as it was in the shGFP infected pool. “Fold change” is defined as the enrichment score of an shRNA relative to the mean enrichment score of all 11 nontargeting control shRNAs. For a given gene, the mean fold change (shGLDC_1/shGFP and shGLDC_2/shGFP) was calculated from all shRNAs targeting that gene.
Clonogenic neurosphere formation assay
0308 cells were stably transduced with shRNAs as indicated, and seeded at single cell-per-well density in Poly-D-Lysine coated 384-well plates (Becton Dickinson). Wells containing a single cell were marked and two weeks later, the marked wells containing spheres were counted.
Histology and immunohistochemistry
Immunohistochemical analyses were performed on discarded archival biopsy (7) and autopsy (7) specimens of glioblastoma, World Health Organization Grade IV, seen at the Departments of Pathology, Massachusetts General Hospital and NYU Langone Medical Center, from 2010 to 2013. Approval from respective Institutional Review Board was obtained, and because we used discarded tissue only, a waiver of informed consent was received. Formalin-fixed, paraffin-embedded brain biopsy tissues were stained with routine hematoxylin and eosin stain (H&E), and cases were reviewed by a neuropathologist (M.S.) to select the most representative block/s for immunohistochemical analysis. Paraffin sections of GBM tumors and normal brains, fixed in 10% formalin, were subjected to deparaffinization and antigen retrieval with Borg Decloaker RTU solution pressurized Decloaking Chamber (Biocare Medical). Antibodies were diluted in in 4% horse serum and 0.1% tween in PBS, which was also used for blocking. Vectastain ABC immunoperoxidase detection kit (Vector Labs) and DAB+ substrate kit (Dako) was used for chromogenic labeling. It was noted that antigen presentation for SHMT2 was much weaker in autopsy sections compared to tumor biopsy sections, likely a result of postmortem interval, and thus a more concentrated primary antibody incubation and longer chromogenic development was required for these sections to get comparable signal to the biopsy sections.
Images were acquired using an Olympus BX41microscope and CellSensR software. For immunofluorescence staining of GBM tumors and normal brains, as well as rapid tumor xenografts, fixed in 10% formalin, the same deparaffinization, antigen retrieval, and blocking/antibody incubation steps were used as above. Immunoreactivity was detected using Alexa-fluor 488 and 568 antibodies and nuclei labeled with Hoechst 33352 (Life Technologies), and ProlongR Gold antifade reagent (Life Technologies) was used as mounting medium. Images were acquired using a Zeiss Axiovert 200M inverted fluorescent microscope and AxioVision Software. For all image-based data, acquisition and processing steps were carried out using the same parameters across the entire set, aside from the increased antibody concentration and longer chromogenic development for the set of autopsy sections for SHMT2 immunostaining as described.
Amino acid analyses
Intracellular amino acids were extracted by hot water extraction, and proteins were removed with sulfosalicylic acid. The amino acids were separated by high-resolution ion-exchange chromatography and derivatized with ninhydrin, and analyzed on a Hitachi L-8800 amino acid analyzer37. Amino acids were normalized by wet pellet weight of the cells prior to extraction.
Quantitative CE-MS based metabolite profiling
Capillary electrophoresis mass spectrometry-based targeted quantitative analysis was performed on stably transduced LN229 cells, as previously described38. A total of 116 metabolites involved in glycolysis, pentose phosphate pathway, tricarboxylic acid (TCA) cycle, urea cycle, and polyamine, creatine, purine, glutathione, nicotinamide, choline, and amino acid metabolism were analyzed and listed in Table 3.
Metabolite extraction and LC-MS analysis
Untargeted metabolite profiling, flux experiments, and amino acetone measurements were performed on a Dionex UltiMate 3000 ultra-high performance liquid chromatography system coupled to a Q Exactive benchtop Orbitrap mass spectrometer, which was equipped with an Ion Max source and a HESI II probe (Thermo Fisher Scientific). External mass calibration was performed every 7 days.
For untargeted metabolite profiling and flux experiments, polar metabolites were extracted from cells growing in a 6-well dish using 400 µL of ice cold 80% methanol with 20 ng/mL valine-d8 as an internal extraction standard. After scraping the cells, 400 µL of chloroform was added before vortexing for 10 min at 4°C, centrifugation for 10 min at 4°C at 16,000×g, and drying 150 µL of the upper methanol/water phase under nitrogen gas. Dried samples were stored at −80°C then resuspended in 40 µL 50% acetonitrile/50% water immediately before analysis. Cells were usually left plated for 24–48 h after a media change before extraction in order to allow for media conditioning. Accordingly, U-13C-glucose labelling of cells was achieved by adding a concentrated stock to a final concentration of 11.1 mM after 24 h of media conditioning. Chromatographic separation was achieved by injecting 10 µL of sample on a SeQuant ZIC-pHILIC Polymeric column (2.1×150 mm 5 uM, EMD Millipore). Flow rate was set to 100 µL/min, column compartment was set to 25°C, and autosampler sample tray was set to 4°C. Mobile Phase A consisted of 20 mM ammonium carbonate, 0.1% ammonium hydroxide. Mobile Phase B was 100% acetonitrile. The mobile phase gradient (%B) was as follows: 0 min 80%, 5 min 80%, 30 min 20%, 31 min 80%, 42 min 80%. All mobile phase was introduced into the ionization source set with the following parameters: sheath gas = 40, auxiliary gas = 15, sweep gas = 1, spray voltage = −3.1kV or +3.0kV, capillary temperature = 275°C, S-lens RF level = 40, probe temperature = 350°C. In experiments to measure steady-state levels, metabolites were monitored using a polarity-switching full-scan method. In experiments utilizing U-13C-glucose tracing, metabolites were monitored using a targeted selected ion monitoring (tSIM) method in negative mode with the quadrupole centered on the M-H ion m+1.5, m+2.5, or m+3.5 mass with a 8 amu isolation window, depending on the number of carbons in the target metabolite. Resolution was set to 70,000, full-scan AGC target was set to 106 ions, and tSIM AGC target was set to 105 ions. For tracing experiments, samples were collected at various time points as indicated. Labeling rate was calculated from counts at 6 minutes, and detailed methods for determining the labeling rate and overall flux are provided in the first three worksheets of Supplemental Table 8. Data were acquired and analyzed using Xcalibur v2.2 software (Thermo Fisher Scientific).Full-scan untargeted data was analyzed using Progenesis CoMet v2.0 software (Nonlinear Dynamics) to identify differential peaks (Supplemental Table 4 and 5) and the identified metabolites with greatest predicted change were further analyzed with Xcalibur. Retention times for selected metabolites appearing in the untargeted analyses (AICAR and SAICAR) were confirmed by running a standard. All standards were obtained commercially, except for SAICAR, which was synthesized enzymatically from AICAR and purified by ion-exchange chromatography as described23.
Derivatization and LC-MS detection of aminoacetone
The protocol for aminoacetone derivatization with fluorenylmethyl chloroformate (FMOC-Cl) and subsequent detection via LC-MS was adapted from previous studies39,40. LN229 cells grown in 6cm plates were quickly washed in cold PBS, then extracted with scraping in 500 µl acetonitrile containing 1 µM ethylamine as an internal control for sample recovery and derivatization efficiency. Following vortexing, centrifugation, and transfer of supernatant to eliminate insoluble material, potassium borate buffer at pH 10.4 (final concentration 33 µM) and FMOC-Cl (final concentration 400 µg/ml) were added. Samples were completely dried, 100 µl of water added followed by addition of 800 µl hexane. Following vortexing and centrifugation, the upper phase was transferred to a new tube, dried, and the pellet extracted in acetonitrile.
For LC separation, 10 µL of each biological sample was injected onto an Ascentis Express C18 2.1×150 mm (2.7 µm particle size) column (Sigma-Aldrich). Mobile phase A was 0.1% formic acid and mobile phase B was 0.1% formic acid in acetonitrile. The chromatographic gradient was as follows, all at a flow rate of 0.25 mL/min.: 0–2 min.: hold at 5% B; 2–20 min.: increase linearly to 75% B; 20–20.1 min.: increase linearly to 95% B; 20.1–24 min.: hold at 95% B; 24–24.1 min.: decrease linearly to 5% B; 24.1–28 min.: hold at 5% B. The autosampler was held at 4°C and the column compartment was held at 35°C. To minimize carryover, blank injections were performed after every six analytical runs.
All mobile phase was introduced into the ionization source with the spray voltage set to +3.0 kV and the same temperature and gas parameter settings as described in the previous section. The MS data acquisition was performed by tSIM of aminoacetone-FMOC and ethylamine-FMOC (internal standard) with the resolution set at 70,000, the AGC target at 105, the maximum injection time at 150 msec, and the isolation window at 1.0 m/z. The full scan range was 150–2000 m/z. Quantitation of the data was performed with Xcalibur v2.2 using a 5 ppm mass tolerance by a researcher (E.F.) blinded to the identity of the samples.
Peak areas for aminoacetone-FMOC were normalized to peak areas for ethylamine-FMOC from the same sample, and further normalized to total protein (µg) and expressed relative to the control sample.
Oxygen consumption measurements
Oxygen consumption of LN229 cells was measured using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience). 60,000 cells were plated per well the night before the experiments, and RPMI 8226 media (US Biological #9011) containing 2 mM glutamine and 10 mM glucose without serum was used as the assay media. Oxygen consumption measurements were normalized based on protein concentration obtained from the same plate used for the assay.
Lactate dehydrogenase (LDH)-linked pyruvate kinase activity assay
Concentrated (5–10 mg/mL) hypotonic lysate was prepared from cells by swelling on ice for 10min in one equivalent of 1xHypotonic Lysis Buffer (20 mM HEPES pH 7.0, 5 mM KCl, 1 mM MgCl2, 2 mM DTT, 1 tablet/10mL Complete EDTA-free protease inhibitor (Roche)), then passing through a 26 gauge needle 3×, then spinning 10min at 4C at 16,000×g. Concentrated lysate was diluted 1:100 in 1xHypotonic Lysis Buffer and immediately assayed with 500 µM final PEP, 600 µM final ATP, 180 µM final NADH, and 0.16mg/mL LDH in 1xReaction Buffer (50 mM Tris pH 7.5, 50 mM KCl, 1 mM DTT) in 100 µL total. Decrease in NADH fluorescence was followed in a Tecan plate reader and a regression on the slope of the decrease was taken as the activity. Bradford assay was performed on the concentrated lysate and activities were normalized to total protein.
Mitochondrial isolation and glycine cleavage assay
Intact mitochondria were isolated from mechanically lysed cells using differential centrifugation as described41, and the intact state of mitochondria verified using the JC-1 dye. Equal quantities of isolated mitochondria were resuspended in a buffer to support glycine cleavage activity as described42 (100 mM KCl, 50 mM mannitol, 20 mM sucrose, 10 mM KH2PO4, 0.1 mM EGTA, 1 mM MgCl2, 0.175 mM pyridoxal phosphate, 1 mM ADP, 25 mM HEPES, pH 7.4) with the addition of 1 µM NAD+, 2 µM tetrahydrofolate, and 10 µM beta-mercaptoethanol. Upon addition of U-14C serine, the reaction mixture containing mitochondria were incubated for 40 min at 37°C, and CO2 produced by the reactions were collected in phenylethylamine-coated paper overnight at 30°C, and the 14C content was measured using a scintillation counter.
Statistics and animal models
All experiments reported in Figs 1,2,3,4 were repeated at least three times, except 1f, 1k, 2b, and 4c, which were repeated twice; 1c, 2a, 2f, and 4a were performed once. In addition, histological analyses experiments (3c) and xenograft based experiments (1h–j, 3g, 4g) were performed once, with n’s indicating the number of individual patient-based tumors or xenograft tumors. All center values shown in graphs refer to the mean. t-tests were heteroscedastic to allow for unequal variance and distributions assumed to follow a Student’s t distribution, and these assumptions are not contradicted by the data. No samples or animals were excluded from analysis, and sample size estimates were not used. Animals were randomly assigned to groups. Studies were not conducted blind with the exception of Fig 3g and 4g. All experiments involving mice were carried out with approval from the Committee for Animal Care at MIT and under supervision of the Department of Comparative Medicine at MIT.
Extended Data
Extended Data Figure 1. GLDC and SHMT2 expression and function in neurosphere forming cells.
a, Micrographs of cells (0308 cell line) cultured under neurosphere forming conditions (top panel) or differentiated into their non-tumorigenic counterparts (bottom panel). b, Immunoblots from neurosphere forming cells maintained in the neurosphere state or differentiated by serum treatment for 1 week, with SOX-2 as a marker for the neural stem cell-like state. c, Immunoblots from neurosphere forming cells transduced with control shRNA (shGFP), or NOTCH2 shRNAs which induce differentiation, for 1 week. d, Immunoblot showing suppression of GLDC expression in BT145 cells transduced with the indicated shRNAs for 5 days. e, Micrographs from undifferentiated or serum-differentiated BT145 cells expressing shGFP or GLDC shRNAs for 6 days.
f. Viability of LN229 cells overexpressing blank vector or mouse GLDC, either untreated or treated with 1 mM esterified glycine for 3.5 days as indicated. Values are relative to that of the same cells left untreated. g, Immunoblot showing suppression of SHMT2 expression in BT145 cells transduced with the indicated shRNAs for 5 days. h, Immunoblots for SHMT2 expression in cells maintained in the neurosphere forming state or induced to differentiate by serum treatment, which are from the same blot as shown in Fig. 1b. i, Immunoblots for SHMT2 expression in neurosphere forming cells transduced with control (shGFP) or NOTCH2 shRNAs, which induce differentiation, for 1 week, and are from the same blot as shown in Fig. 1c. j, Micrographs showing morphology of BT145 cells transduced with the indicated shRNAs for 6 days. k, Cell viability of 0308 cells transduced with the indicated shRNAs for 6 days. Values are normalized to the viability of shGFP transduced cells. l, Clonogenic sphere formation in 0308 cells transduced with the indicated shRNAs and then plated as single cells. The proportion of wells containing single cells that were able to form spheres are shown as values relative to shGFP- transduced cells. For l, n=2 independent biological replicates. For f and k, n=3 independent biological replicates; error bars are SD. *P<0.05 (student’s t test).
Extended Data Figure 2. Identification of GCAT as a mediator of toxicity caused by GLDC suppression.
a, Schematic presentation of pooled shRNA screen carried out in LN229 cells. Detailed procedures are provided in Methods. b, Table of a subset of genes examined in the pooled screen, the average fold change increase in relative abundance of all shRNAs for each gene in GLDC-suppressed conditions (shGLDC_1 and shGLDC_2) compared to the set of nontargeting control shRNAs, as described in Supplemental methods. Genes are sorted by ascending p value; the top 15 out of the 25 genes are shown. Asterisks indicate metabolic genes which do not function in glycine metabolism, included as additional controls. All shRNAs used and their abundance in each condition are shown in Supplemental Table 4. c, Relative representation, in the shGLDC infected pool, of each shRNA against non-targeting controls, SHMT2, and GCAT. A value of 1.0 indicates the average for all hairpins in the screen. Representation in both shGLDC_1 and shGLDC_2 are shown, so each hairpin is represented twice in the plot. Bars are mean±SEM. *P<0.05 (student’s t test). d, Immunoblots of LN229 cells transduced with shRNAs against GCAT as indicated. e, Extracted ion chromatogram showing peaks from FMOC derivatized aminoacetone and ethylamine (an internal standard spiked into each sample as a control for efficiency of derivatization, and recovery and detection) from pure standards (lower graph) and from a representative LN229 xenograft tumor sample (upper graph), showing a match between predicted and observed m/z values and retention times. f, Aminoacetone levels in control (no shRNA) cells, cells transduced with shGLDCdox, and cells with shGLDCdox plus shRNA-resistant mouse GLDC, which were all induced with dox for 5 days; n=3 independent biological replicates; error bars are SD. *P<0.05 (student’s t test). g, Methylglyoxal levels (argpyrimidine antibody) in LN229 cells transduced with Cas9 and single guide RNAs against GLDC for 7 days. h, Expression of GCAT in cells transduced with Cas9 and single guide RNAs against GCAT for 7 days. i, Methylglyoxal levels (argpyrimidine antibody) in LN229 cells transduced with Cas9 and the indicated single guide RNAs for 7 days, then secondarily transduced with shGLDC_2 for 5 days.
Extended Data Figure 3. Effects of glycine cleavage system inhibition on cells with high or low SHMT2 expression levels.
a, Overview of the serine hydroxymethyltransferase and glycine cleavage reactions mediated by SHMT2 and GLDC, respectively. Asterisks indicate metabolites that are labeled with 13C during mitochondrial metabolism of U-13C serine. Only upon completion of both reactions will 13C-labeled CO2 be formed, which is captured and detected by scintillation as described in Methods. b, Measurement of 13CO2 production, a readout of sequential SHMT2 and GLDC activity on [U-13C]-serine, in intact mitochondria isolated from LN229s expressing shGFP or shSHMT2_1, as described in Methods. c, Table indicating cell viability in various cell lines following transduction of GLDC shRNAs for 6–7 days. Values are relative to the CTG signal of the same cells secondarily transduced with shGFP and grown in parallel. d, SHMT2 and GLDC expression in LN229 cells stably transduced with shSHMT2_1 or shGFP, then secondarily transduced shGLDC or shGFP as indicated. e, Viability of LN229 cells first transduced with control or SHMT2 shRNAs, then transduced with shGFP or GLDC shRNAs for 5 days. Values are relative to that of cells secondarily transduced with shGFP. f, Viability of U251 cells first transduced with control or SHMT2 shRNAs, then transduced with shGFP or GLDC shRNAs for 7 days. Values are relative to that of cells secondarily transduced with shGFP. g,h,i, Viability of various cell lines transduced with shRNAs targeting (g) GLDC, (h) GCSH (glycine cleavage system protein H, another integral component of the glycine cleavage system), or (i) SHMT2. Values are relative to those of the cells expressing shGFP, which were grown in parallel. For b,c, and e–i, n=3 independent biological replicates; error bars are SD. *P<0.05 (student’s t test).
Extended Data Figure 4. GLDC and SHMT2 expression in GBM tumors.
a, SHMT2 and GLDC expression across GBM and normal brain regions as examined in autopsy sections. A whole coronal section is shown, with the GBM bulk tumor outlined in white. Insets indicate magnified micrographs from the regions, indicated by the small red squares, from the same brain. For the bulk tumor insets, cells around the pseudopalisading necroses are shown. Additional GBM and normal brain are shown in Extended Data Figure 4. Scale bar for whole coronal section = 1 cm, and for insets = 100 µm. b, SHMT2 expression at the GBM-normal brain interface, showing SHMT2-immunoreactivity in migrating cells. Scale bar = 100 µm. c, Magnified image of the boxed region in b. d and e, High magnification micrographs of SHMT2 and GLDC expression, respectively, in 1) perinecrotic GBM tumor, 2) non-ischemic bulk tumor, 3) frontal cortex, 4) temporal white matter, and 5) striatum from autopsy cases. Scale bar = 100 µm. f, Semi-quantitative scoring of GLDC staining intensity by neuropathologist (M.S.) on 7 tumor biopsy cases (left) and 7 autopsy cases (right). g, Semi-quantitative scoring of SHMT2 staining intensity by neuropathologist (M.S.) on 7 tumor biopsy cases (left) and 7 autopsy cases (right). h, Viability of LN229 cells expressing an empty vector control or mouse GLDC cDNA and cultured in 0.5% hypoxia for 8 days. Values are relative to that of the same cells cultured in parallel in normoxia. i, Cell number counts from LN229 cells expressing an empty vector control or SHMT2 cDNA and cultured in 0.5% hypoxia for 8 days. Values are relative to the counts of the same cells cultured in parallel in normoxia. j. Viability of U251 cells expressing shRNA or cDNAs as indicated and cultured in 0.5% hypoxia for 6 days. Values are relative to that of the same cells cultured in parallel in normoxia. For f and g, bars indicate the mean. For h,i, and j, n=3 independent biological replicates; error bars are SD. *P<0.05 (student’s t test).
Extended Data Figure 5. Effects of SHMT2 expression on PKM2 activity and cell metabolism.
a, Absolute quantitative CE-MS measurement of intracellular metabolites from LN229 cells stably expressing indicated shRNAs without media change for 84 hours. The metabolites with the greatest fold change are listed, in units of pmol/106 cells. All metabolites listed are changed in a statistically significant manner (student’s t test, P<0.05). Data for all 116 metabolites in the analysis are in Supplementary Table 4. b, Oxygen consumption in LN229 cells (in RPMI media) transduced with indicated shRNAs and cDNAs. Error bars are SD (n=4 technical replicates). c, Pyruvate kinase activity assay from lysates of U251 cells transduced with indicated shRNAs or cDNAs. d, Proposed mechanism by which SHMT2 inhibition could lead to an increase in AICAR and SAICAR. In cells with high SHMT2 expression, 10-formyl-THF, which is a downstream product of SHMT2 activity, serves as a cofactor for cytosolic 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase (ATIC) in the conversion of AICAR to FAICAR during purine biosynthesis. In the absence of SHMT2, a lack 10-formyl-THF production could lead to accumulation of AICAR and SAICAR. As indicated by the asterisk, the contribution of SHMT2 to cytosolic 10-formyl-THF formation may be direct, occur via export of formate from the mitochondria, or occur indirectly by changing serine/glycine levels and thereby altering SHMT1 activity. e, 13C labeling rates of pyruvate and its downstream metabolites in cells from e. Labeling rates are relative and expressed as fold changes in cells transduced with shSHMT2_1 versus cells transduced with shGFP, with or without RNAi-resistant SHMT2 cDNA. Estimated PK flux is the calculated total flux to the four product species in moles of 13C per unit time. Plots for individual labeled species over time are shown in Extended Data Figure 5d and Supplementary Table 9. f, Scheme for calculating total PK flux by measuring the net molar labeling of [U-13C]-glucose derived, PK product species. Estimated PK flux is the calculated total flux to the four product species in moles of 13C per unit time. g, Plots of glucose-derived labeled species abundance over time for shGFP, shSHMT2, and shGFP plus PKM2 cDNA expressing LN229 cells shown in Fig. 4e–g. Raw data, calculations and plots for all stable cell lines are shown in Supplemental data tables 8 and 9. h, SHMT2 and PKM2 expression in LN229 cells transduced with shRNAs and cDNAs as indicated. Asterisk indicates the overexpressed PKM2, which shows higher migration due to Flag tag. i, Oxygen consumption in LN229 cells expressing shGFP or shSHMT2_1 in RPMI media with or without 1 mM pyruvate. Error bars are SD (n=4 technical replicates). j, Viability of LN229 cells transduced with shRNAs or cDNAs as indicated, and also treated with vehicle or 50 uM of TEPP-46 or DASA-58 as indicated, then subjected to hypoxia for 6 days. Values are relative to the same cells grown in parallel in normoxia. k, Overview of effects of SHMT2 expression on cell metabolism, tumor cell survival, and liability to toxic glycine accumulation. Red arrow indicates upregulation, and blue arrow indicates downregulation. Gray bar indicates the inhibitory effect of SHMT2 activity on PKM2 activity. Depiction of pseudopalisading necrosis is adapted from a previous review (Brat and Meir, Laboratory Investigation 84(4):397–405). Illustration by Mica Duran. For a,c,e,g, and j, n=3 independent biological replicates; error bars are SD. *P<0.05 (student’s t test).
Supplementary Material
Acknowledgements
We thank members of the Sabatini lab for assistance and feedback, in particular Yoav Shaul, Tim Wang, Shuyu Wang, and Omer Yilmaz. Authors would like to thank Dr. Jennie Taylor (Massachusetts General Hospital) with GBM sample collection, and Tom DiCesare with illustrations. This work was supported by a Basic Research Fellowship from the American Brain Tumor Association to D.K.; MIT School of Science Fellowship in Cancer Research and NIH T32GM007287 to B.P.F., fellowships from the Jane Coffin Childs Memorial Fund and Leukemia and Lymphoma Society to K.B.; a grant from the NIH (K99 CA168940) to R.P.; an American Cancer Society fellowship and an American Brain Tumor Association Discovery Grant to Y.C.; a fellowship from the US National Institute of Aging to W.W.C.; NIH (K08-NS087118) to S.H.R.; support from NIH (R01CA168653, 5P30CA14051), the Smith Family Foundation, the Burroughs Wellcome Fund, the Damon Runyon Cancer Research Foundation, and the Stern family to M.G.V.H.; DOD CDMRP Discovery Award, grants from the David H. Koch Institute for Integrative Cancer Research at MIT, The Alexander and Margaret Stewart Trust Fund, and NIH (CA103866, CA129105, and AI07389) to D.M.S. D.M.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information
The following supplementary files are provided:
Supplementary Methods
Supplemental Table 1. List of genes whose loss causes toxicity in the developing brain, and the associated disorder, targeted organs, and other relevant information.
Supplemental Table 2. Data from oncomine-based gene expression studies comparing glioma vs. normal brain, and list of metabolic genes that are highly overexpressed in gliomas in each study.
Supplemental Table 3. Data summary for gene expression omnibus microarray-based studies comparing expression of select genes in neural stem cells versus differentiated controls.
Supplemental Table 4. List of individual shRNAs used and abundance scores for each shRNA under each pool condition.
Supplemental Table 5. Data for all metabolites in LN229 cells expressing shGFP or shSHMT2_1 as measured using CE-MS based, quantitative metabolite profiling.
Supplemental Table 6. List of metabolites identified through LC-MS based, untargeted discovery using Progenesis software, in positive mode.
Supplemental Table 7. List of metabolites identified through LC-MS based, untargeted discovery using Progenesis software, in negative mode.
Supplemental Table 8. Methods and data for 13-C labeled species in the labeling rate analyses shown in Fig. 4e–g and Extended Fig. 5c–d.
Supplemental Table 9. Plots for 13-C labeled species abundance over time for the labeling rate analyses shown in Fig. 4e–g and Extended Fig. 5c–d.
Author contributions
D.K. and D.M.S. conceived the study and designed most of the experiments. D.K. performed most of the experiments (cell viability and proliferation, western blotting, immunohistochemistry, xenografts) with assistance from K.B., R.L.P., Y.C., W.W.C., S.K., and M.K. B.P.F. and M.G.V.H. designed, carried out, and analyzed pyruvate kinase activity and LC-MS based experiments with input and assistance from E.F., M.E.P., and D.Y.G. E.F., D.K., and J.C. designed and carried out LC-MS based derivatization experiments measuring aminoacetone levels. K.K. and L.M.S. conducted and analyzed CE-MS metabolite profiling. M.S. provided GBM sections and conducted analyses and imaging of IHC. S.H.R. and K.L.R. assisted with neurosphere forming cell characterizations. D.K. and D.M.S. wrote and all authors edited the manuscript.
REFERENCES
- 1.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tennant DA, Duran RV, Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattson MP, Shea TB. Folate and homocysteine metabolism in neural plasticity and neurodegenerative disorders. Trends Neurosci. 2003;26:137–146. doi: 10.1016/S0166-2236(03)00032-8. doi:S0166-2236(03)00032-8 [pii] 10.1016/S0166-2236(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 5.Saudubray JM, Van den Berghe G, Walter J. Inborn metabolic diseases : diagnosis and treatment. 5th edn. Springer; 2012. [Google Scholar]
- 6.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.Zhang WC, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 8.Tibbetts AS, Appling DR. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. doi:S1535-6108(06)00117-6 [pii] 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Shikano N, et al. Stimulation of 125I-3-iodo-alpha-methyl-L-tyrosine uptake in Chinese hamster ovary (CHO-K1) cells by tyrosine esters. Nucl Med Biol. 2010;37:189–196. doi: 10.1016/j.nucmedbio.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Dale RA. Catabolism of threonine in mammals by coupling of L-threonine 3-dehydrogenase with 2-amino-3-oxobutyrate-CoA ligase. Biochim Biophys Acta. 1978;544:496–503. doi: 10.1016/0304-4165(78)90324-0. [DOI] [PubMed] [Google Scholar]
- 13.Tressel T, Thompson R, Zieske LR, Menendez MI, Davis L. Interaction between L-threonine dehydrogenase and aminoacetone synthetase and mechanism of aminoacetone production. J Biol Chem. 1986;261:16428–16437. [PubMed] [Google Scholar]
- 14.Sartori A, et al. Aminoacetone, a putative endogenous source of methylglyoxal, causes oxidative stress and death to insulin-producing RINm5f cells. Chem Res Toxicol. 2008;21:1841–1850. doi: 10.1021/tx8001753. [DOI] [PubMed] [Google Scholar]
- 15.Kalapos MP. Methylglyoxal in living organisms: chemistry, biochemistry, toxicology and biological implications. Toxicol Lett. 1999;110:145–175. doi: 10.1016/s0378-4274(99)00160-5. doi:S0378427499001605 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 17.Jain M, et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narkewicz MR, Sauls SD, Tjoa SS, Teng C, Fennessey PV. Evidence for intracellular partitioning of serine and glycine metabolism in Chinese hamster ovary cells. Biochem J. 1996;313(Pt 3):991–996. doi: 10.1042/bj3130991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rong Y, Durden DL, Van Meir EG, Brat DJ. 'Pseudopalisading' necrosis in glioblastoma: a familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J Neuropathol Exp Neurol. 2006;65:529–539. doi: 10.1097/00005072-200606000-00001. doi:00005072-200606000-00001 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Nelson DA, et al. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes Dev. 2004;18:2095–2107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaneton B, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gui DY, Lewis CA, Vander Heiden MG. Allosteric regulation of PKM2 allows cellular adaptation to different physiological states. Sci Signal. 2013;6:pe7. doi: 10.1126/scisignal.2003925. [DOI] [PubMed] [Google Scholar]
- 23.Keller KE, Tan IS, Lee YS. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anastasiou D, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 26.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. doi:nchembio.1060 [pii] 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 28.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. doi:S1550-4131(06)00060-X [pii] 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Brown JM, Wilson WR. Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 30.Mehta S, et al. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359–371. doi: 10.1016/j.ccr.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chudnovsky Y, et al. ZFHX4 Interacts with the NuRD Core Member CHD4 and Regulates the Glioblastoma Tumor-Initiating Cell State. Cell Rep. 2014;6:313–324. doi: 10.1016/j.celrep.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ptolemy AS, et al. A 9-month-old boy with seizures and discrepant urine tryptophan concentrations. Clin Chem. 2011;57:545–548. doi: 10.1373/clinchem.2010.144899. [DOI] [PubMed] [Google Scholar]
- 38.Kami K, et al. Metabolomic profiling of lung and prostate tumor tissues by capillary electrophoresis time-of-flight mass spectrometry. Metabolomics. 2013;9:444–453. doi: 10.1007/s11306-012-0452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao S, Yu PH. A fluorometric high-performance liquid chromatography procedure for simultaneous determination of methylamine and aminoacetone in blood and tissues. Anal Biochem. 2009;384:20–26. doi: 10.1016/j.ab.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 40.Kazachkov M, Yu PH. A novel HPLC procedure for detection and quantification of aminoacetone, a precursor of methylglyoxal, in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;824:116–122. doi: 10.1016/j.jchromb.2005.07.006. doi:S1570-0232(05)00465-4 [pii] 10.1016/j.jchromb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Rotem R, et al. Jasmonates: novel anticancer agents acting directly and selectively on human cancer cell mitochondria. Cancer Res. 2005;65:1984–1993. doi: 10.1158/0008-5472.CAN-04-3091. doi:65/5/1984 [pii] 10.1158/0008-5472.CAN-04-3091. [DOI] [PubMed] [Google Scholar]
- 42.Jois M, Hall B, Fewer K, Brosnan JT. Regulation of hepatic glycine catabolism by glucagon. J Biol Chem. 1989;264:3347–3351. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.