Causative homozygous or compound heterozygous mutations in the glucose-6-phosphatase, catalytic subunit 3 gene (G6PC3) have recently been described for a sub-group of patients with severe congenital neutropenia (SCN) (Arostegui et al, 2009; Boztug et al, 2009, 2011; Xia et al, 2009; Germeshausen et al, 2010; Banka et al, 2011a,b; Hayee et al, 2011). In addition to neutropenia and recurrent infections, mutant-positive patients are reported to have varied other congenital abnormalities, such as a prominent superficial venous pattern, structural heart defects, urogenital malformations, skeletal abnormalities, primary pulmonary hypertension, growth and developmental delay (Boztug et al, 2011). Several reports have also indicated considerable variation in bone marrow (BM) morphology, including a lack of the myeloid maturation arrest characteristic of SCN (McDermott et al, 2010; Banka et al, 2011b). We screened 108 CN patients or kindreds, known wild-type for the ELANE, HAX1 and WAS genes, for G6PC3 mutations and identified further phenotypic heterogeneity, in that two mutant-positive patients had neutropenia alone. Another presented with isolated neutropenia and was found to have clinically asymptomatic cardiac abnormalities.
The study had Local Regional Ethical Committee approval. Informed consent was obtained from patients and/or their parents. Polymerase chain reaction (PCR) amplicons of the six G6PC3 exons were screened using denaturing high performance liquid chromatography (Transgenomic Limited, Glasgow, UK) (see Table S1). Samples with abnormal chromatograms were sequenced. Mutations were confirmed by PCR with mismatch primers and restriction enzyme digestion (Table S2). Three different homozygous G6PC3 mutations were detected in four of the 108 patients/kindreds studied. Parents of affected individuals were all heterozygous for the mutation.
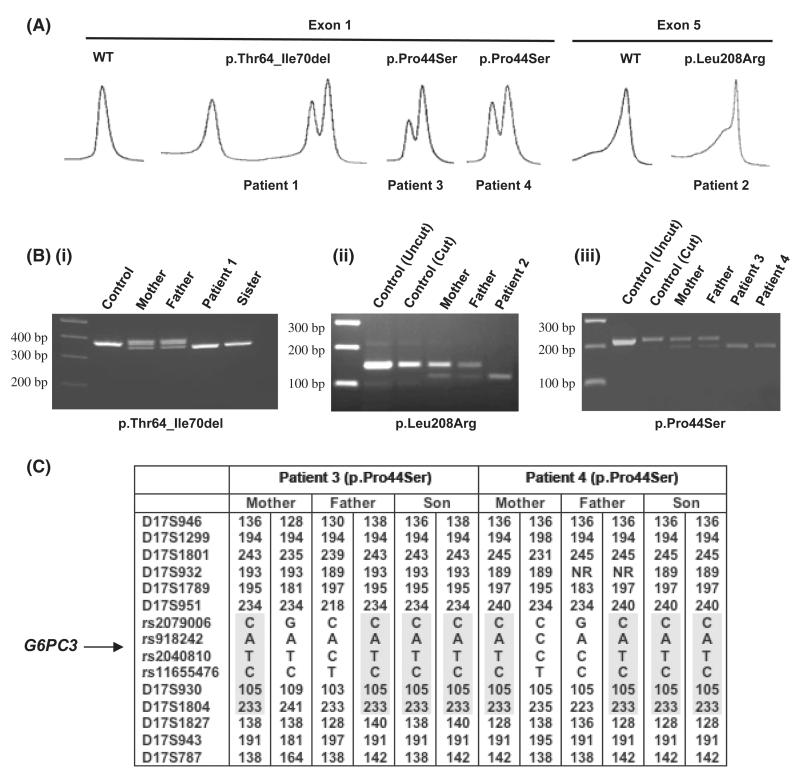
Patient 1 (born 1982, from Pakistan, parents first cousins) had a homozygous 21 bp deletion in exon 1 (c.190_210del) resulting in an in-frame deletion of seven amino acids in the second trans-membrane domain (p.Thr64_Ile70del) (Fig 1A, B). The patients’ phenotype was similar to that of the G6PC3-mutated patients described by Boztug et al (2009). In addition to neutropenia (absolute neutrophil count [ANC] 0·1 × 109/l), first noted aged 9 years, he suffered from recurrent oral ulcers, aspergillus infection and recurrent episodic bacterial pneumonia, but responded to standard antibiotic therapy and now receives regular granulocyte colony-stimulating factor (G-CSF). He was born with an atrial septal defect and has granulomatous inflammatory bowel disease, splenomegaly, digital clubbing and short stature. His affected sister is also homozygous G6PC3 mutant and has a similar phenotype but without the atrial septal defect and opportunistic infection.
Fig 1.
WAVE chromatograms, restriction enzyme digests and haplotype study of G6PC3 mutant-positive kindreds. (A) WAVE chromatograms of exon 1 and exon 5 polymerase chain reaction (PCR) products from mutation-positive patients mixed with known wild-type (WT) PCR products. (B) (i) PCR products of Patient 1, his affected sister and parents carrying the c.190_210del mutation. (ii) TaqI digestion confirming the c.623T>G mutation in Patient 2 and her parents. (iii) DdeI digestion of PCR products confirming the c.130C>T mutation in Patients 3 and 4. Parental results (lanes 3 and 4) were the same for both patients. (C) Microsatellite and SNP analysis of the G6PC3 locus in kindreds 3 and 4 with the c.130C>T mutation. The shared haplotype flanked by markers D17S951 and D17S1827 is highlighted.
Patient 2 (born 2001, from Turkey, consanguineous parents) has a novel c.623T>G mutation in exon 5 resulting in p.Leu208Arg (Fig 1A, B). She presented in infancy with severe neutropenia (ANC <0·1 × 109/l), recurrent otitis media, chronic gingivitis and periodontitis. Initially responsive to 5 μg/kg G-CSF, subsequent doses in excess of 10–15 μg/kg have failed to control her infections. Although otherwise clinically asymptomatic and with no obvious congenital anomalies, echocardiography post-G6PC3 mutation detection showed a patent foramen ovale and tricuspid insufficiency.
Patients 3 and 4, both from Pakistan, are unrelated but have the same homozygous missense mutation in exon 1, c.130C>T resulting in p.Pro44Ser (Fig 1A, B). Both have normal echocardiograms, lack other congenital abnormalities and have had normal growth and development. Patient 3 (born 1989, non-consanguineous parents) presented with neutropenia at age 13 years with intermittent mouth ulcers (ANC 0·4 × 109/l) and has suffered a single bout of myositis. His only treatment has been intermittent G-CSF for recurring severe mouth ulcers. Patient 4 (born 2002, parents first cousins) presented at 3 years of age with recurrent upper respiratory and chest infections, and has had two instances of gluteal abscesses. He has otherwise been well, with a recent ANC of 0·45 × 109/l, and has not received G-CSF. Of note, both patients have normocellular BM morphology with normal neutrophils, although slightly left-shifted myelopoiesis was reported for Patient 3. The mutation was not detected in samples from 54 haematologically normal individuals of Pakistani origin but was reported recently in an SCN patient, also of Pakistani origin, who similarly had normal BM cellularity without maturation arrest, although further information on the patient was not available (Banka et al, 2011b). Pro44 and Leu208 are both conserved amongst species (Fig S1) and, due to their trans-membranous location, the mutations are predicted to be disruptive to normal G6PC3 function.
A possible ancestral founder for the p.Pro44Ser mutation was identified from a common haplotype consisting of two highly polymorphic micro-satellite markers and four single nucleotide polymorphisms spanning a maximum of 4·8Mb in the two unrelated p.Pro44Ser-mutated patients (Fig 1C) (PCR primers and conditions in Table S3). None of 20 Pakistani control samples carried the entire six-marker haplotype, and the probability of this conserved haplotype homozygously presenting in the Pakistani population would be very low.
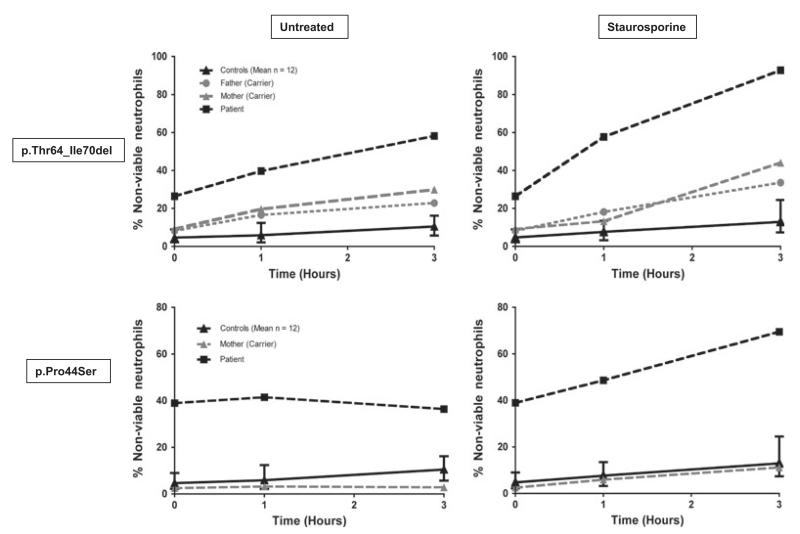
A functional impact on apoptosis consistent with a pathogenic mutation was demonstrated for purified neutrophils from Patients 1 and 4 using previously published assays (Smith et al, 2009). Both patients had a high level of spontaneous apoptosis (26·4%, 38·9% non-viable cells respectively at time zero compared to 3·6 ± 4·7% [mean±2SD] for 12 normal controls) (Fig 2). After 3 h incubation this level had significantly increased in Patient 1 compared to controls (58·2% versus 10·5 ± 7·1%) and was further enhanced in the presence of staurosporine (92·8% versus 12·9% ± 9·5% at 3 h). For Patient 4, the level of apoptosis only increased in the presence of staurosporine (36·4% without, 69·5% with staurosporine at 3 h). In addition, severe neutrophil glycosylation defects have been reported for Patients 1 and 3 (Hayee et al, 2011).
Fig 2.
Percentage of non-viable neutrophils in patients with the p.Thr64_Ile70del and p.Pro44Ser mutations, their carrier parents and controls. Graphical representation of the percentage of non-viable neutrophils (Annexin V-positive, propidium iodide-negative plus Annexin V-positive propidium iodide-positive cells) from patients, parents and controls after incubation for up to 3 h either untreated or in the presence of 5 μmol/l staurosporine.
Overall, the frequency of G6PC3 mutations in our SCN cohort was 3% (4 of 155 patients/kindreds), although this incidence is likely to depend on the racial composition of the patient population investigated. No evidence has been found for a G6PC3 genotype-phenotype correlation (Banka et al, 2011b; Boztug et al, 2011), and it has been suggested that different founder mutations may exist according to racial groups (Banka et al, 2011b). Whether such variable genetic inheritance patterns has a wider impact on phenotype will require analysis of many cases, and the underlying reasons for the phenotypic heterogeneity associated with G6PC3 mutations are therefore currently unclear. Nevertheless, based on our observations of milder disease, isolated neutropenia and normal BM morphology but evident functional abnormalities in our two p.Pro44Ser-mutated cases, we would recommend G6PC3 screening as part of the work-up of all CN/SCN patients lacking other mutations, irrespective of the presence of additional congenital abnormalities.
Supplementary Material
Protein sequence alignment of G6PC3 in different species.
G6PC3 PCR primer sequences, PCR annealing temperatures and WAVE melting analysis temperatures.
Mismatch PCR primer sequences and restriction enzyme digests used to confirm the p.Pro44Ser and p.Leu208Arg mutations.
Micro-satellite markers and SNP PCR primer sequences spanning the G6PC3 locus used in the haplotype study of two Pakistani kindreds carrying the c.130C>T (p.Pro44Ser) mutation.
Acknowledgements
This work was supported by Leukaemia and Lymphoma Research, UK. The work was undertaken at University College London Hospital/University College London, which received a proportion of funding from the Department of Health’s National Institute for Health Research Biomedical Research Centres funding scheme.
Footnotes
Conflict of interest
The authors declare no competing financial interests.
Additional Supporting Information may be found in the online version of this article:
Publisher's Disclaimer: Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Arostegui JI, de Toledo JS, Pascal M, Garcia C, Yague J, Diaz de Heredia C. A novel G6PC3 homozygous 1-bp deletion as a cause of severe congenital neutropenia. Blood. 2009;114:1718–1719. doi: 10.1182/blood-2009-04-219451. [DOI] [PubMed] [Google Scholar]
- Banka S, Chervinsky E, Newman WG, Crow YJ, Yeganeh S, Yacobovich J, Donnai D, Shalev S. Further delineation of the phenotype of severe congenital neutropenia type 4 due to mutations in G6PC3. European Journal of Human Genetics. 2011a;19:18–22. doi: 10.1038/ejhg.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banka S, Wynn R, Newman WG. Variability of bone marrow morphology in G6PC3 mutations: is there a genotype-phenotype correlation or age-dependent relationship? American Journal of Hematology. 2011b;86:235–237. doi: 10.1002/ajh.21930. [DOI] [PubMed] [Google Scholar]
- Boztug K, Appaswamy G, Ashikov A, Schaffer AA, Salzer U, Diestelhorst J, Germeshausen M, Brandes G, Lee-Gossler J, Noyan F, Gatzke AK, Minkov M, Greil J, Kratz C, Petropoulou T, Pellier I, Bellanne-Chantelot C, Rezaei N, Monkemoller K, Irani-Hakimeh N, Bakker H, Gerardy-Schahn R, Zeidler C, Grimbacher B, Welte K, Klein C. A syndrome with congenital neutropenia and mutations in G6PC3. New England Journal of Medicine. 2009;360:32–43. doi: 10.1056/NEJMoa0805051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boztug K, Rosenberg PS, Dorda M, Banka S, Moulton T, Curtin J, Rezaei N, Corns J, Innis JW, Avci Z, Tran HC, Pellier I, Pierani P, Fruge R, Parvaneh N, Mamishi S, Mody R, Darbyshire P, Motwani J, Murray J, Buchanan GR, Newman WG, Alter BP, Boxer LA, Donadieu J, Welte K, Klein C. Extended Spectrum of Human Glucose-6-Phosphatase Catalytic Subunit 3 Deficiency: Novel Genotypes and Phenotypic Variability in Severe Congenital Neutropenia. Journal of Pediatrics. 2012;12:679–683. e2. doi: 10.1016/j.jpeds.2011.09.019. [DOI] [PubMed] [Google Scholar]
- Germeshausen M, Zeidler C, Stuhrmann M, Lanciotti M, Ballmaier M, Welte K. Digenic mutations in severe congenital neutropenia. Haematologica. 2010;95:1207–1210. doi: 10.3324/haematol.2009.017665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayee B, Antonopoulos A, Murphy EJ, Rahman FZ, Sewell G, Smith BN, McCartney S, Furman M, Hall G, Bloom SL, Haslam SM, Morris HR, Boztug K, Klein C, Winchester B, Pick E, Linch DC, Gale RE, Smith AM, Dell A, Segal AW. G6PC3 mutations are associated with a major defect of glycosylation: a novel mechanism for neutrophil dysfunction. Glycobiology. 2011;21:914–924. doi: 10.1093/glycob/cwr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott DH, De Ravin SS, Jun HS, Liu Q, Priel DA, Noel P, Takemoto CM, Ojode T, Paul SM, Dunsmore KP, Hilligoss D, Marquesen M, Ulrick J, Kuhns DB, Chou JY, Malech HL, Murphy PM. Severe congenital neutropenia resulting from G6PC3 deficiency with increased neutrophil CXCR4 expression and myelokathexis. Blood. 2010;116:2793–2802. doi: 10.1182/blood-2010-01-265942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BN, Ancliff PJ, Pizzey A, Khwaja A, Linch DC, Gale RE. Homozygous HAX1 mutations in severe congenital neutropenia patients with sporadic disease: a novel mutation in two unrelated British kindreds. British Journal of Haematology. 2009;144:762–770. doi: 10.1111/j.1365-2141.2008.07493.x. [DOI] [PubMed] [Google Scholar]
- Xia J, Bolyard AA, Rodger E, Stein S, Aprikyan AA, Dale DC, Link DC. Prevalence of mutations in ELANE, GFI1, HAX1, SBDS, WAS and G6PC3 in patients with severe congenital neutropenia. British Journal of Haematology. 2009;147:535–542. doi: 10.1111/j.1365-2141.2009.07888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein sequence alignment of G6PC3 in different species.
G6PC3 PCR primer sequences, PCR annealing temperatures and WAVE melting analysis temperatures.
Mismatch PCR primer sequences and restriction enzyme digests used to confirm the p.Pro44Ser and p.Leu208Arg mutations.
Micro-satellite markers and SNP PCR primer sequences spanning the G6PC3 locus used in the haplotype study of two Pakistani kindreds carrying the c.130C>T (p.Pro44Ser) mutation.