Abstract
Progesterone receptor membrane component 1 (PGRMC1) is a 25 kDa protein with an N-terminal transmembrane domain and a putative C-terminal cytochrome b5 domain. Heme-binding activity of PGRMC1 has been shown in various homologues of PGRMC1. Although the general definition of PGRMC1 is as a progesterone receptor, progesterone-binding activity has not been directly demonstrated in any of the purified PGRMC1 proteins fully loaded with heme. Here, we show that the human homologue of PGRMC1 (hPGRMC1) binds heme in a five-coordinate (5C) high-spin (HS) configuration, with an axial tyrosinate ligand, likely Y95. The negatively charged tyrosinate ligand leads to a relatively low redox potential of approximately −331 mV. The Y95C or Y95F mutation dramatically reduces the ability of the protein to bind heme, supporting the assignment of the axial heme ligand to Y95. On the other hand, the Y95H mutation retains ~90% of the heme-binding activity. The heme in Y95H is also 5CHS, but it has a hydroxide axial ligand, conceivably stabilized by the engineered-in H95 via an H-bond; CO binding to the distal ligand-binding site leads to an exchange of the axial ligand to a histidine, possibly H95. We show that progesterone binds to hPGRMC1 and introduces spectral changes that manifest conformational changes to the heme. Our data offer the first direct evidence supporting progesterone-binding activity of PGRMC1.
Progesterone receptor membrane component 1 (PGRMC1) is a member of the membrane-associated progesterone receptor (MAPR) protein family. It is a small heme-binding protein (~25 kDa) with an N-terminal transmembrane domain and a C-terminal putative cytoplasmic cytochrome b5 (Cyt b5) domain. PGRMC1 is expressed in a variety of animal tissues, in particular those with high P450 activity, such as liver, kidney, and adrenals.1–4 It is also strongly expressed in a range of cancers, such as lung, breast, and ovarian cancer, in which it promotes cancer cell survival and stimulates insensitivity of cancer cells to chemotherapy,5–7 making it a potential molecular target for cancer therapy and prevention.
Although PGRMC1 belongs to the MAPR protein family, it shares no homology with the nuclear or membrane-bound steroid receptors.8 Instead, it resembles Cyt b5 proteins, despite the fact that neither of the two histidine residues responsible for heme coordination is present in any of the PGRMC1 proteins. So far, the progesterone-binding activity of PGRMC1 has been demonstrated only in cellular fractions or partially purified protein samples, in which the molecular target was not well-defined.9,10 Consequently, it remains unclear if PGRMC1 binds progesterone.6 On the other hand, it is well-established that PGRMC1 binds multiple P450 enzymes and regulates their activities in a heme-dependent manner.1,11–14 Hughes et al. showed that PGRMC1 binds CYP51A1, a P450 involved in the cholesterol synthetic pathway, thereby regulating its activity; in addition, the loss of PGRMC1 blocks cholesterol synthesis, thereby leading to accumulation of toxic sterol intermediates.11 It was demonstrated that, in addition to CYP51A1, PGRMC1 also binds other P450s, such as CYP3A4 (a major drug metabolizing enzyme), CYP7A1 (an important enzyme involved in bile acid synthesis), and CYP21A2 (a progesterone 21-hydroxylase).11 Later studies confirmed that PGRMC1 binds a wide range of P450s and regulates their activities.12,13 Szczesna-Skorupa et al.12 indicated that PGRMC1 binds P450 reductase and modestly inhibits the activity of several drug metabolizing P450s, including CYP2C2, CYP2C8, and CYP3A4, suggesting that PGRMC1 modulates P450 activity in an isozyme-specific manner, adding a layer of complexity to the function of PGRMC1. Until now, there has been no general consensus on the mechanism by which PGRMC1 regulates the activity of P450s. In addition to P450s, several other binding partners of PGRMC1 have been identified, such as cholesterol regulators, Insig and SCAP, and a membrane-associated protein involved in antiapoptotic action of progesterone, PAIR-BP1.4,15
In 2007, Hughes et al. showed that a yeast homologue of PGRMC1, Dap1 (damage resistance protein 1), also binds heme and positively regulates two P450s, CYP51A1 and CYP61A1, both of which are required for sterol biosynthesis.11 In vitro characterization of Dap1 showed that heme is noncovalently attached to the protein.11 Spectroscopic characterization with UV–vis, EPR, and MCD indicated that the heme in the ferric state is five-coordinate (5C) high-spin (HS), possibly with a conserved tyrosine, Y138, coordinated to the heme as the axial ligand,14 in contrast to the six-coordinate (6C) low-spin (LS) heme of Cyt b5.
In addition to animals and yeast, PGRMC1 has also been identified in Arabidopis. The three-dimensional structure of the apo form of the Arabidopis homologue of PGRMC1, At2g24940 (with >60% sequence homology with Dap1), has been solved by NMR independently by Song et al.16 (PDB code: 1T0G) and Suzuki et al.17 (PDB code: 1J03), which serve as the only structures available for the MAPR family of proteins.4,6,16
In this work, we expressed and purified a truncated version of human PGRMC1 (hPGRMC1), in which the N-terminal transmembrane domain was deleted. We have characterized its molecular properties and evaluated its progesterone-binding activity by UV–vis and resonance Raman (RR) spectroscopies. To examine the potential role of Y95 (equivalent to Y138 in Dap1) as the proximal axial heme ligand, we carried out comparative studies of the wild-type (WT) protein and three Y95 mutants: Y95F, Y95C, and Y95H.
■ MATERIALS AND METHODS
Cloning and Sequence
Escherichia coli codon-optimized gene encoding the heme domain of hPGRMC1 was subcloned into pET 28a (+) from Novagen with an N-terminal cleavable 6×His tag between sites NdeI and BamHI. The first 70 amino acids were deleted from the full-length protein. The sequence cloned is shown below.
DFTPAELRR FDGVQDPRIL MAINGKVFDV TKGRKFYGPE GPYGVFAGRD ASRGLATFCL DKEALKDEYD DLSDLTAAQQ ETLSDWESQF TFKYHHVGKL LKEGEEPTVY SDEEEPKDES ARKND
Y95 mutants (Y95F, Y95C, and Y95H) were generated by standard site-directed mutagenesis procedures. All of the sequences were verified by DNA sequencing.
Expression and Purification
Expression of the wild-type (WT) and Y95 mutants of hPGRMC1 proteins was achieved by growing BL21 DE3 cells (harboring the respective protein gene) at 37 °C in 1 L flasks of Luria–Bertani broth (LB) medium containing the appropriate antibiotic until the optical density at 600 nm reached ~0.6–0.8. Recombinant protein production was induced with 1 mM isopropyl 1-thio-d-galactopyranoside and supplemented with 5 µM hemin chloride. The culture was incubated overnight at 25 °C in a shaking incubator at 100 rpm. The cells were harvested by centrifugation for further purification.
Cell pellets were resuspended in 50 mM potassium phosphate (pH 7.4) containing 5% glycerol, 0.1% Triton X-100, and 150 mM NaCl and were then lysed by sonication at 4 °C. Immediately prior to lysis, hemin was added to the resuspended cell pellet at a final concentration of ~20 µM to ensure maximum saturation of the protein with heme. The crude extracts were centrifuged at 16 000 rpm for 1 h. The supernatant was then collected and loaded onto a Ni- IMAC column (Bio-Rad), prewashed with lysis buffer containing 10 mM imidazole.
The target protein was eluted with an elution buffer, 50 mM potassium phosphate (pH 7.4) and 150 mM NaCl containing 200 mM imidazole. The eluted protein was pooled, buffer-exchanged into an imidazole-free elution buffer, and concentrated. The protein then was incubated with high-purity bovine thrombin (approximately 15 units of thrombin per milligram of protein) at room temperature for 12 h to cleave the His tag. The protein mixture was reloaded onto a Ni-IMAC column. The flow-through was collected, concentrated, and further purified by a Superdex-75 column (GE Healthcare) using the imidazole-free elution buffer containing 1 mM DTT. The purity of the protein was checked with SDS-PAGE analysis. The identity of the WT protein and presence of heme (protoporphyrin IX) were confirmed by mass spectrometry.
UV–Vis Spectroscopic Measurements
UV–vis spectra were obtained at room temperature using a UV2100 spectrophotometer (Shimadzu Scientific Instruments, Inc., Columbia, MD). The proteins were buffered with 100 mM potassium phosphate buffer (pH of 7.4) containing 150 mM NaCl and 1 mM DTT. The deoxy proteins were prepared by injecting a minimum amount of sodium dithionite into the protein samples prepurged with nitrogen gas with a gastight syringe. The 13C18O adducts were prepared by injecting 200 µL of 13C18O gas into freshly prepared deoxy protein samples, whereas the 12C16O adducts were generated by adding a minimum amount of sodium dithionite to 12C16O-purged protein samples. For progesterone-binding studies, a small aliquot of a progesterone stock in methanol was added to each protein sample at room temperature such that the final protein and progesterone concentrations were 5 and 80 µM, respectively. The final methanol concentration was kept at ~1% (v/v). Control experiments confirmed that 1% methanol introduced only negligible changes to the UV–vis and resonance Raman (RR) spectra.
RR Spectroscopic Measurements
RR measurements with a spectral resolution of 1 cm−1 were performed as described in earlier work.18,19 Briefly, the 413.1 nm output from a Kr ion laser (Spectra Physics, Mountain View, CA) was focused to a ~30 µm spot on a spinning quartz cell (~6000 rpm). The scattered light was collected at a 90° geometry and focused on the 100 µm wide slit of a 1.25 m Spex spectrometer equipped with a 1200 grooves/mm grating (Horiba Jobin Yvon, Edison, NJ), where it was dispersed and detected by a CCD detector (Princeton Instruments, Trenton, NJ). Rayleigh scattering was removed by a holographic notch filter (Kaiser, Ann Arbor, MI). The Raman shifts were calibrated by using indene and an acetone/ferricyanide mixture for the 200–1700 and 1700–2300 cm−1 spectral windows, respectively. The laser power was kept at ~5 mW for ferric and ferrous samples and ~250 µW for CO-bound samples (to prevent photodissociation of heme iron-bound CO).
Reduction Potential Measurements
The redox potentials of the WT and Y95H mutant of hPGRMC1 were first determined by potentiometric titration method as reported elsewhere.20 Briefly, the ferric or deoxy ferrous protein sample (~5 µM), containing a cocktail of electron mediators (Safranin T and Nile blue) to mediate electron transfer in the −116 to −289 mV window, was prepurged with nitrogen gas. It was then titrated with sodium dithionite (as a reductant) or potassium ferricyanide (as an oxidant) under anaerobic conditions. The ratio of the reduced and oxidized protein was determined by UV–vis measurements. Our data indicated that small amount of heme was released out of the protein during the measurements, possibly due to nonspecific interactions between the electron mediators and the protein, which complicated the analysis of the data. As an alternative approach, we sought to estimate the redox potential of hPGRMC1 by monitoring the electron distribution between the protein and a dye, safranin T, with a known potential (−289 mV vs SHE), as a function of [dithionite] at 25 °C under anaerobic conditions. The reaction mixture, containing 4 µM protein and 4 µM safranin T, was loaded into an aerobic cuvette. It was purged with nitrogen gas and then titrated with dithionite under anaerobic conditions. The reduction reaction following each addition of dithionite was followed every 1 min until reduction of the protein and dye reached equilibrium (typically, ~5 min) using UV–vis. The ratio of the oxidized and reduced protein was determined by absorbance at 399 nm (the isosbestic point of the dye), whereas that of the oxidized and reduced dye was monitored at 538 nm (the isosbestic point of the protein). Linear Nernst plots for one-electron reduction of the heme in hPGMC1 and two-electron reduction of the dye were used to estimate the redox potential of the protein. All of the potentials cited in this work are referenced to the standard hydrogen electrode (SHE).
■ RESULTS AND DISCUSSION
The WT and mutants of hPGRMC1 were expressed and purified with a cleavable N-terminal 6×His tag. To evaluate if the 6×His tag perturbs the molecular properties of the proteins, the samples with and without the tag were compared with UV–vis and RR spectroscopies. Our data show that the presence of the 6×His tag significantly modulates the spectral properties of the proteins (data not shown). Thus, the data presented herein are based on the tag-free proteins.
To evaluate if Y95 (equivalent to Y138 in Dap1) is the axial heme ligand in hPGRMC1, we first examined two mutants of Y95, Y95C and Y95F, with respect to the WT protein with UV–vis spectroscopy. Our data indicate that the mutations significantly diminish heme-binding activity of the protein. Specifically, no heme binding was observed in the Y95C mutant, whereas heme binding was ~15% of that of WT in the Y95F mutant (Table 1). The residual heme-binding activity of Y95F suggests that the heme might be partially stabilized by π−π interactions between the phenyl ring of the engineered-in F95 and the porphyrin macrocycle. In any case, the data support the view that Y95 is the axial heme ligand in hPGRMC1. To further examine the heme-binding properties of hPGRMC1, we sought to construct and examine an additional mutant, Y95H. We found that Y95H mutation retains ~90% heme binding. The heme coordination states of the WT and Y95H mutant of hPGRMC1 will be further discussed below.
Table 1.
Soret Maximum and R/Z Ratio of the WT and Three Y95 Mutants (Y95H, Y95F, and Y95C) of hPGRMC1a
| PGRMC1 | Soret maximum (nm) | R/Z ratio |
|---|---|---|
| WT | 400 | 1 |
| Y95H | 396 | 0.90 |
| Y95F | 402 | 0.15 |
| Y95C | ND | ND |
ND, not determined due to the incompetence the protein to bind heme.
Redox Potential of hPGRMC1
The redox potential of a heme is closely correlated with the electronic properties of its axial ligands; typically, a stronger electron-donating ligand gives rise to a more negative redox potential. Consequently, the redox potential of a heme protein with a negatively charged ligand (such as a thiolate21 or a tyrosinate22) is lower than that with a neutral ligand (such as a histidine23).
Our first attempt at redox potential measurements with a potentiometric titration method was not successful due to heme release triggered by interactions between the protein and the electron mediators (data not shown). As an alternative approach, we sought to determine the redox potential by measuring electron distribution between the protein and a dye, safranin T, with a known potential (−289 mV vs SHE), as a function of reduction by dithionite. Our data show that, with this method, heme release is dramatically reduced, although the isosbestic point of the Soret band of the heme during the redox transition is slightly blurred (Figure S1). Nonetheless, on the basis of the Nernst plot shown in Figure 1A, we estimated the redox potential of the WT protein to be −331 mV, which is similar to that reported for Dap1 (−307 mV).24 The relatively low redox potentials of hPGRMC1 are consistent with the assignment of its proximal heme ligand to a negatively charged ligand, a tyrosinate. Intriguingly, the Y95H mutation only leads to ~+50 mV increase in the redox potential (−331 → −284 mV; Figure 1B), suggesting an axial ligand with a negative charge, possibly a hydroxide ion (vide inf ra).
Figure 1.
Linear Nernst plots based on one-electron reduction of WT (A) and Y95H mutant (B) of hPGRMC1 and two-electron reduction of a dye, safranin T. Pox/Pred and Dox/Dred stand for the ratio of oxidized protein vs reduced protein and the ratio of oxidized dye vs reduced dye, respectively. The solid lines are the best fit lines with the slopes fixed to be 1. The redox potentials of the WT and Y95H mutant of hPGRMC are estimated based on the intercepts of the lines to be ~−331 and −284 mV, respectively, with the assumption that E° (safranin T) = −289 mV (vs SHE).
Spectral Properties of the WT hPGRMC1
Figure 2A shows the UV–vis spectra of the WT protein. The ferric protein exhibits a Soret band at 400 nm and β/α bands at 500/533 nm as well as a charge-transfer band at 618 nm. The λmax of the Soret band is relatively low, suggesting a 5CHS heme.25 The reduction of the protein from the ferric to ferrous state leads to red shift of the Soret band to 416 nm and β/α bands to 533/567 nm. The λmax of the Soret band is unusually low, but it is consistent with a tyrosinate-coordinated 5CHS heme (Table 2). The presence of a small shoulder at ~400 nm suggests incomplete reduction of the heme iron, possibly due to its low redox potential. In general, the UV–vis spectra of hPGRMC1 are similar to those of the yeast analogue, Dap1.14,24 More intriguingly, they are also analogous to a group of recently discovered heme-transport proteins with a tyrosinate as the sole axial heme ligand, such as ShuT.14,26
Figure 2.
UV–vis (A) and high-frequency RR (B) spectra of WT hPGRMC1. The UV and RR spectra were obtained with ~5 and 80 µM protein, respectively, in 100 mM pH 7.4 potassium phosphate buffer.
Table 2.
Absorption Maxima of WT and Y95H Mutant of hPGRMC1 Determined in This Work with Respect to That of Other Heme Proteinsa
| hPGRMC1 (WT) | hPGRMC1 (Y95H) | Dap114 | ShuT26 | CYP2B4 (C436S)28 | swMb31 | |
|---|---|---|---|---|---|---|
| FeIII | 400, 500/533, 618b | 396, 500/538, 621b | 398, 500/530, 620b | 400, 500/521, 617b | 404, 500/530, 620b | 408, 502, 632b |
| FeII | 416, 533/567 | 418, 558 | 418, 538/562 | ND | 422, 530c/561 | 432, 554 |
| CO | 410, 533/565 | 419, 537/567 | ND | ND | 413, 534/568 | 422, 542/578 |
ND, not determined.
A charge transfer band.
The number was estimated from a small shoulder on the UV side of the 561 nm band.
CO binding to the ferrous protein leads to a Soret band centered at 410 nm and β/α bands at 533/565 nm. The spectral features, in particular the uncommonly low λmax of the Soret maximum, are similar to those reported for CO-bound heme proteins with an oxygen-based proximal ligand, such as the H25Y mutant of HO-127 and the C346S mutant of P450 2B428 or the C400S mutant of P450 BM3 (Table 2), supporting the scenario that Y95 is the proximal ligand of the heme.
Figure 2B shows the RR spectra of the WT protein in the high-frequency (HF) window (1100–1700 cm−1). The totally symmetric in-plane ring breathing modes of the porphyrin macrocycle present in the HF window, in particular, the ν2, ν3, and ν4 modes, are sensitive to the oxidation state, spin state, and the nature of axial ligands of the heme.18,19 The RR spectrum of the ferric hPGRMC1 confirms that the heme is 5CHS, as indicated by the ν2, ν3, and ν4 modes at 1566, 1488, and 1373 cm−1, respectively. The unusually high relative intensity of the ν3 mode with respect to the ν4 mode suggests an oxygen-based proximal ligand, such as a hydroxide or tyrosinate.26,29–34 Comparable spectra obtained in H2 16O, D2 16O, or H2 18O solvents revealed no hydroxide-related vibrational modes (data not shown), suggesting that the axial heme ligand is not a hydroxide.
The ferrous protein displays ν2, ν3, and ν4 modes at 1563, 1471, and 1359 cm−1, respectively, typical of a 5CHS ferrous heme. The small shoulders on the high-frequency sides of the ν3 and ν4 modes are attributed to residual ferric protein due to incomplete reduction, consistent with that suggested by the UV–vis data. The CO adduct showed ν2, ν3, and ν4 modes at 1581, 1500, and 1373 cm−1, respectively, characteristic of a 6CLS CO-bound heme.
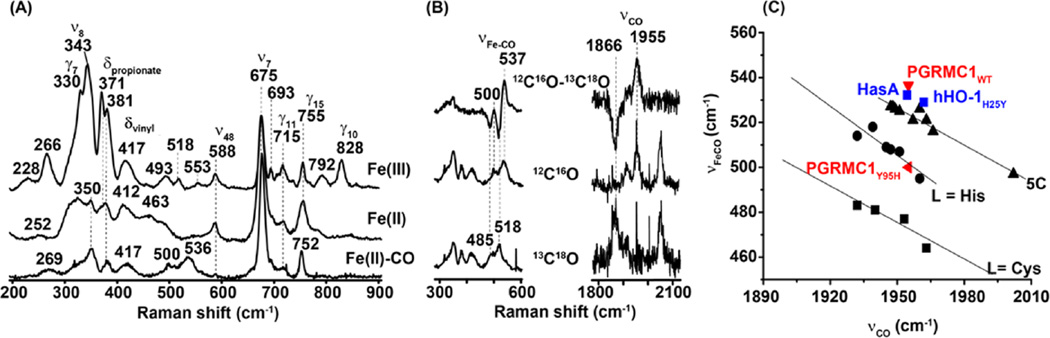
Figure 3A shows the RR spectra of hPGRMC1 in the low-frequency (LF) window (200–900 cm−1). The assignments of the major LF modes are as indicated. The LF spectrum is composed of in-plane and out-of-plane porphyrin skeletal modes (labeled ν and γ, respectively) and the bending modes associated with the propionate and vinyl groups attached to the porphyrin macrocycle (labeled δpropionate and δvinyl, respectively) as well as iron–ligand associated modes. Hence, it offers rich information regarding the nature of the axial heme ligands and the conformation of the porphyrin ring and peripheral groups as well as the surrounding protein environment.18,19,35–39 The presence of the γ modes in the spectrum of the ferric hPGRCM1 (top trace), which are typically silent in the spectrum of a planar heme with D4h symmetry, indicates that the heme is distorted out of plane. The reduction of the protein from the ferric to ferrous state leads to changes in their relative intensities (such as the reduction in intensity of the γ10 and γ12 modes), indicating the alteration in the symmetry type of the heme. On the other hand, the δpropionate modes at 371 and 381 cm−1 change their relative intensities, whereas the δvinyl mode shifts from 417 to 412 cm−1, indicating perturbation in the conformation of the heme peripheral groups, as commonly observed upon reduction of a heme from ferric to ferrous state in other heme protein systems. CO binding to the ferrous protein leads to additional changes, such as the shift of the δvinyl mode from 412 to 417 cm−1 and the appearance of the COassociated modes (vide inf ra), as expected for the conversion of a 5CHS heme to a CO-bound 6CLS heme.
Figure 3.
Low-frequency RR spectra (A) and CO isotope-sensitive νFe–CO/νC–O modes of WT hPGRMC1 (B). νFe–CO vs νC–O inverse correlation plot (C). In (C), the data points associated with P450s (L = Cys), myoglobin variants (l-His), and 5C heme are taken from the literature27,42,44,62,63 and are presented by black squares, circles, and triangles, respectively; those associated with PGRMC1, HasA,44 and H25Y mutant of hHO-127 are as indicated. The data were obtained with ~80 µM protein in 100 mM pH 7.4 potassium phosphate buffer.
Distal Ligand Environment
In the LF RR spectrum of the CO adduct (Figure 3A), the band at 536 cm−1 is assigned to the Fe–CO stretching mode (νFe–CO), as evident by its shift to 518 cm−1 upon 13C18O isotope substitution (Figure 3B). The νFe–CO mode is associated with a C–O stretching mode (νC–O) at 1955 cm−1, which shifts to 1866 cm−1 upon 13C18O isotope substitution.
The νC–O frequency of gaseous CO is 2143 cm−1.40 When CO is coordinated to a ferrous heme iron, it typically shifts to ~1900–1970 cm−1, due to the bonding interaction described by two resonance forms illustrated below
Fe−–C≡O+ (I) ↔ Fe=C=O (II)
As a general rule, a more positive electrostatic environment of CO in the CO adduct of a heme protein with a given proximal ligand destabilizes form (I), leading to a stronger Fe–C bond (associated with a higher νFe–CO frequency) and a weaker C–O bond (associated with a lower νC–O frequency). On this basis, a well-known inverse correlation between νFe–CO and νC–O has been established.41–43
The offset of the inverse correlation line made up by heme proteins with a given proximal ligand is sensitive to the electronic properties of the proximal ligand. As such, the line defined by P450s, with a thiolate proximal ligand (L = Cys), lies below the line defined by globins, with a histidine as the proximal ligand (L = His), which lies below the 5C inverse correlation line made up by CO adducts of model complexes, in which the proximal ligand-binding site is empty (Figure 3C). Recently, it was reported that the data points associated with several heme proteins with a tyrosinate as the proximal ligand, such as HasA44 and H25Y27 mutant of hHO-1, also lie on the 5C inverse correlation line, manifesting the weak nature of the iron–tyrosinate bond.45 Our current studies show that the νFe–CO/νC–O data point of hPGRMC1 (535/1955 cm−1) lies within the cluster of data points associated with the 5C inverse correlation line, consistent with a weak tyrosinate proximal ligand.
It is noted that a weak νFe–CO band is evident at 500 cm−1, which shifts to 485 cm−1 with 13C18O, indicating the presence of an additional minor Fe–C–O conformer. We tentatively assigned it to a conformation with the heme iron coordinated by one of the two histidine residues (H96 and H97) that is adjacent to residue 95, considering the fact that a νFe–CO/νC–O data point at 500/1955 cm−1 would put it on the L = His line. In any case, due to its low population, it will not be further discussed.
Spectral Properties of the Y95H Mutant
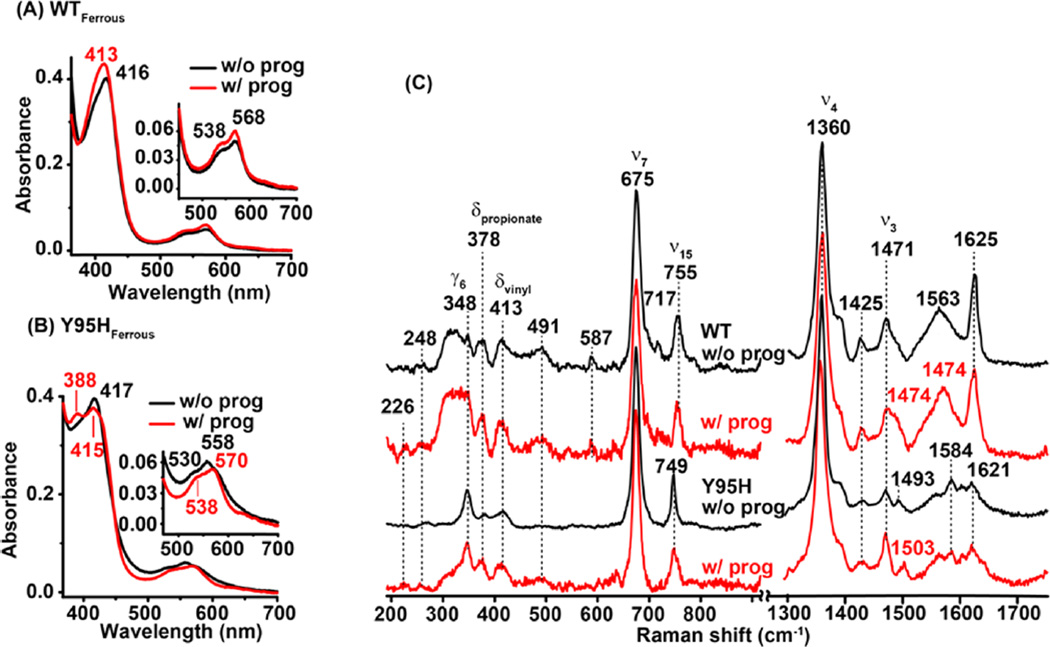
Figure 4A–D shows the UV–vis and RR spectra of the Y95H mutant. The data show that Y95H mutation in the ferric protein leads to the broadening and blue shift (400 → 395 nm) of the Soret band as well as a red shift of the charge transfer band (618 → to 621 nm), suggesting that the heme remains as 5CHS but has a different axial ligand. On the other hand, the HF RR data confirm that the heme has a 5CHS configuration, whereas the LF RR data reveal that the mutation leads to the merging of the two δpropionate modes at 371/381 cm−1 into one band at 381 cm−1, the shift of the ν8 mode from 343 to 349 cm−1, and the reduction in intensity of several out-of-plane heme modes (including γ10, γ11, and γ6), indicating changes in the symmetry of the porphyrin ring and the peripheral groups attached to it.
Figure 4.
UV–vis (A), high-frequency RR (B), low-frequency RR (D) spectra, and νFe–CO and νC–O modes (C) of the Y95H mutant of hPGRMC1. The UV and RR spectra were obtained with ~5 and 80 µM protein, respectively, in 100 mM pH 7.4 potassium phosphate buffer.
The mutation in the ferrous protein gives rise to a slight sharpening and red shift of the Soret band (416 → 418 nm) that is accompanied by significant blue shift of the β-band (567 → 558 nm) as well as a weaker α-band. The HF RR data show that the heme is mostly 5CHS, as indicated by the ν3 and ν4 modes at 1471 and 1359 cm−1, respectively. The UV–vis spectral features of the ferrous Y95H mutant are distinct from those of a typical 5CHS heme with a proximal histidine ligand (see Mb data in Table 2), suggesting that the engineered-in H95 is not able to coordinate to the heme iron. This conclusion is consistent with the fact that the νFe–His mode (typically active in the RR spectrum of a 5CHS heme with a histidine ligand) is not observed in the 200–300 cm−1 spectral window (Figure 4D).
We hypothesize that the imidazole side chain of H95 in the Y95H mutant, rather than coordinating to the heme iron, stacks against the porphyrin macrocycle, with an orientation similar to the phenyl ring of the native Y95 (Scheme 1) as well as those of other heme proteins with tyrosinate as an axial heme ligand.46–53 The imidazole ring of H95 is not in a favorable orientation to coordinate to the heme iron; instead, it positions a hydroxide ion, via an H-bond, to coordinate to the heme iron. This hypothesis is consistent with the observed low redox potential of the Y95H mutant (−284 mV), which is too low to be associated with a heme with an axial histidine ligand.
Scheme 1.

Postulated Coordination States of the WT and Y95H Mutant of hPGRMC1
The mutation in the CO-bound protein, on the other hand, results in shifts of the Soret and β/α bands to 419 and 537/567 nm, respectively, characteristic of a 6CLS heme with CO and histidine as the axial ligands. Consistent with this view, the νFe–CO/νC–O data point of the mutant, determined to be 500/1955 cm−1, lies on the L = His line, instead of the 5C line, in the inverse correlation plot (Figure 3C), demonstrating that the proximal ligand of the heme is exchanged from a hydroxide to a histidine, possibly H95. Although H95 is not able to coordinate to the ferric and deoxy ferrous heme iron, the data suggest that CO binding to the distal site strengthens the proximal iron–histidine bond due to trans effects,54 which provides a thermodynamic driving force to elicit conformational changes required for the coordination of H95 to the heme iron. The observation that the mutation of Y95 to a histidine and a phenylalanine, but not a cysteine, retains the ability to bind heme (Table 1) indicates that the heme is specifically recognized by the surrounding protein matrix, in part by π-stacking interactions between the side chain of residue 95 and the porphyrin macrocycle.
The HF RR data of the CO adduct of the Y95H mutant confirm that the heme has a 6CLS configuration; in addition, the LF RR data show a shift of the ν15 mode from 755 to 747 cm−1 and weaker ν48, δpropionate, and δvinyl modes, manifesting the heme conformational change induced by the mutation.
It is notable that we attempted to detect the νFe–OH mode in the RR spectrum of the ferric enzyme by carrying out H2 16O–D2 16O/H2 18O isotope substitution experiments; unfortunately, we could not detect any isotope-sensitive mode (Figure S2), possibly due to its weak nature. We have also tried detecting the νFe–His mode in the RR spectrum of the CO adduct by photodissociating CO with high laser power; unfortunately, only a negligible amount of CO adduct could be photodissociated with laser power as high as 150 mW.
Progesterone-Binding in the WT hPGRMC1
PGRMC1 has attracted a great deal of attention since Meyer et al. reported indirect evidence supporting its high progesterone-binding activity in porcine liver membranes in 1996.55 On the basis of that report, a high-affinity site with a Kd of 11 nM and a low-affinity site with a Kd of 286 nM were determined, although the molecular target of progesterone was not specifically identified as PGRMC1.55 Similarly, a high-affinity progesterone-binding site with a Kd of ~35 nM was determined in a partially purified GFP–PGRMC1 fusion protein.56 So far, however, there has been no direct evidence demonstrating the progesterone-binding activity in any purified PGRMC1 protein fully reconstituted with heme.
To examine if hPGRMC1 binds progesterone, we repeated the UV–vis and RR studies in the presence of progesterone. As shown in Figure 5A,C, the presence of progesterone only slightly perturbs the UV–vis spectrum of the ferric protein, but it significantly modulates the intensities and positions of a few RR modes, including the merging of the two propionate bending modes at 371 and 381 cm−1 into one band at 371 cm−1 (indicating changes in the H-bonding interactions surrounding the propionate groups), the broadening of the vinyl bending mode at 415 cm−1 (indicating changes in the planarity of the vinyl groups), the shift of the ν8 mode from 343 to 348 cm−1, and a weaker out-of-plane mode (γ6) at 331 cm−1, as well as the change in the intensity ratio of the ν2, ν3, and ν4 modes (1567, 1488, and 1373 cm−1). Likewise, the presence of progesterone in the CO adduct does not significantly disturb the UV–vis spectrum, but it leads to the enhancement and sharpening of the 537 cm−1 band (Figure S3), suggesting a perturbed ligand environment. In contrast, the presence of progesterone in the ferrous protein leads to a shift of the Soret maximum from 416 to 413 nm as well as a slight perturbation to the Q-bands (Figure 6A), whereas the RR spectrum remains almost the same (Figure 6C). In general, the data demonstrate that progesterone binds to hPGRMC1 in both the ferric and deoxy ferrous states and introduces changes in the heme, possibly due to its colocalization with the heme in the putative heme/ligand-binding cleft.
Figure 5.
UV–vis (A, B) and RR (C) spectra of the WT and Y95H mutant of ferric hPGRMC1 with (red) and without (black) progesterone. The concentrations of protein/progesterone are ~5/50 and 80/500 µM (in 100 mM pH 7.4 potassium phosphate buffer) for the UV–vis and RR measurements, respectively.
Figure 6.
UV–vis (A, B) and RR (C) spectra of the WT and Y95H mutant of deoxy ferrous hPGRMC1 with (red) and without (black) progesterone. The concentrations of protein/progesterone are ~5/50 and 80/500 µM (in 100 mM pH 7.4 potassium phosphate buffer) for the UV–vis and RR measurements, respectively.
Taken together, the data provide the first direct evidence supporting progesterone-binding activity in PGRMC1. Although additional work is required to further identify specific progesterone–protein interactions and to determine the progesterone affinity, our data suggest that progesterone binds in the vicinity of the heme.
Progesterone-Binding in the Y95H Mutant of hPGRMC1
The presence of progesterone in the ferric form of Y95H leads to slight shift of the Soret maximum from 395 to 397 nm and the charge transfer band from 621 to 614 nm (Figure 5B) as well as a perturbed intensity ratio of the ν2, ν3, and ν4 modes (Figure 5C), although the LF RR spectrum is only minimally perturbed. On the other hand, the presence of progesterone in the ferrous form of Y95H gives rise to a shift in the Soret maximum from 417 to 415 nm and the β/α bands from 530/558 to 538/570 nm; in addition, it slightly perturbs the RR spectrum, in particular the propionate mode at 378 cm−1, the vinyl bending mode at 413 cm−1, the ν15 mode at 749 cm−1, and the ν3 mode at 1471 cm−1 (Figure 6C). It is also notable that the presence of progesterone leads to a shift in the small ν3 component at 1493 cm−1 to 1503 cm−1, suggesting a change in the core size and/or distortion of the heme macrocycle. Together, the data demonstrate that Y95H, like the WT protein, specifically binds progesterone in the vicinity of the heme.
Intriguingly, progesterone binding in the ferric protein also causes an increase in the intensity of the Soret band of the protein at the expense of that of the free heme (appearing as a shoulder on the UV side of the 395 nm band), indicating that progesterone binding promotes heme binding. Conversely, progesterone binding in the ferrous protein gives rise to partial heme dissociation, as indicated by the intensity decrease in the Soret band at 417 nm and the concurrent intensity increase at 388 nm (Figure 6B). These data are consistent with the view that heme and progesterone share the putative heme/ligand-binding cleft in the protein. They suggest that the heme–progesterone–protein interactions are sensitive to the redox state of the heme.
Comparison with the Yeast Homologue, Dap1, and the Rat Homologue, rPGRMC1
At the molecular level, the yeast homologue, Dap1, is arguably the best characterized member of MARP family of proteins.14,24 The UV–vis and RR spectra of the ferric and deoxy ferrous derivatives of hPGRMC1 reported here closely resemble those of Dap1. Furthermore, the νFe–CO of the CO adduct identified at 537 cm−1 is similar to that of Dap1 at 533 cm−1.24 Likewise, the redox potential of hPGRMC1, −331 mV, is comparable to that of Dap1 (−307 mV).24 The Y138F mutation in Dap1, equivalent to Y95F mutation in hPGRMC1, was first thought to abolish the heme-binding activity of the protein,14 but later studies revealed that it does bind heme, but it does so with 500-fold lower affinity toward the ferric heme (Kd of 200 nM instead of 400 pM) and 5-fold lower affinity toward the ferrous heme (Kd of 10 µM instead of 2 µM).24 Due to the weak affinity of the ferrous heme toward the protein, the redox potential of the Y138F mutant could not be directly determined, but it was estimated to be −155 mV based on the redox potential of the WT protein and the free energy differences in heme binding in the WT and mutant proteins.24 In contrast, we found that the Y95F mutant of hPGRMC1 exhibits only 15% heme-binding ability; although Y95H retains 90% heme binding, its redox potential was identified to be −284 mV, much lower than that of the Y138F mutant of Dap1, suggesting that the redox potential of PGRMC1 is sensitive to the chemical nature of residue 95.
Intriguingly, although hPGRMC1 and the rat homologue (rPGRMC1) share very high sequence identity (>90%),14 the absorption maxima of hPGRMC1 in the deoxy and CO-bound states at 416/567 and 410/533/565 nm, respectively, are significantly different from those of rPGRMC1 reported by Min et al. at 426/559 and 420/538/567 nm, respectively.1 It should be noted that the rPGRMC1 protein used by Min et al. was prepared with an N-terminal 6×His-tag or GST-tag attached to it. Although the authors argued that the spectral properties of rPGRMC1 are not sensitive to the type of tag attached to it, the data listed in their Supplemental Table 2 show that the absorption maxima of the deoxy ferrous protein with a GST-tag, 422/540/570 nm, are, in fact, significantly different from those of the protein with a 6×His tag (426/559 nm). Hence, the unique spectral features of rPGRMC1 reported by Min et al. might be attributed to the structural perturbation introduced by the N-terminal tags. This scenario is consistent with the observation we made that the presence of an N-terminal 6×His tag significantly disturbs the spectral properties of hPGRMC1 (vide supra).
■ CONCLUSIONS
The data reported here demonstrate that hPGRMC1 binds heme in both the ferric and ferrous states. The heme has a 5CHS electronic configuration; it is coordinated by a tyrosinate, Y95, one of the few conserved residues lining a putative heme/ligand-binding cleft in MAPR family of proteins.6 The assignment of the axial ligand to a tyrosinate is confirmed by the relatively low redox potential of the protein (−331 mV; Figure 1A) and the unique location of the νFe–CO/νC–O data point on the 5C line in the inverse correlation plot (Figure 3C). Our data suggest that the Y95H mutation leads to a new 5CHS species, with a hydroxide axial ligand, which is stabilized by the engineered-in H95 via an H-bond (Scheme 1). CO binding to Y95H leads to ligand exchange from a hydroxide to a histidine, possibly H95, due to the trans effect exerted by the CO ligand bound in the distal site.
We propose that in the wild-type protein the heme is anchored in position by the iron–tyrosinate bond that is further stabilized by π stacking between the phenyl ring of Y95 and the porphyrin macrocycle. The π-stacking interaction accounts for the 90 and 15% heme-binding activity retained in the Y95H and Y95F mutants, respectively, in which their imidazole ring or phenyl ring could stack with the porphyrin macrocycle, in contrast to the totally abolished heme-binding activity of the Y95C mutant. The observation that the two histidine residues adjacent to residue 95 are not able to rescue the heme-binding activity of Y95C by coordinating to the heme iron supports the view that heme is specifically recognized by the surrounding protein matrix, including an aromatic side chain of residue 95.
Our data provide the first direct evidence supporting progesterone-binding activity of PGRMC1. We show that progesterone binding introduces changes to the symmetry type of the porphyrin macrocycle and the conformation of the vinyl and propionate groups attached to it, consistent with the view that heme and progesterone co-occupy the putative heme/ligand-binding cleft and that heme and progesterone binding are not mutually exclusive. The observation that progesterone binding does not perturb the νFe–CO or νC–O mode of the CO adduct indicates that progesterone binds to a site remote from the distal CO-binding site.
It is well-accepted that PGRMC1 plays key roles in a number of important biological processes.4,57 However, the exact biological function of PGRMC1 remains elusive. Our studies reveal striking similarities between hPGRMC1 and a group of heme transporters found in bacteria, which comprise a tyrosinate as an axial heme ligand, such as the heme acquisition system protein, HasA,30,44,58,59 and the periplasmic heme transport proteins, ShuT26 and PhuT.60 This observation hints that PGRMC1 might play a role in heme transport in vivo by cycling between the ferric and ferrous states, as the negatively charged tyrosinate ligand might discourage heme binding in the ferrous state, thereby promoting heme unloading. Along a similar line, Ghosh et al.14 and Mallory et al.61 suggested that Dap1 acts as a heme chaperone and donates heme to newly synthesized Erg11p (a P450 involved in ergosterol synthesis), thereby regulating its activity. Likewise, Hughes et al. demonstrated that hPGRMC1 binds and regulates P450 activity in a heme-dependent manner,11 consistent with its function as a heme transporter. Nonetheless, many more studies need to be carried out to evaluate the potential function of PGRMC1 as a heme transporter.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Denis L Rousseau for valuable discussions.
Funding This work was supported by NIH grant nos. GM086482 to S.-R.Y. and GM57353 to T.L.P. and NIH/NCI Institutional Training Grant Fellowship T32CA009054 to D.B.
ABBREVIATIONS
- hPGRMC1
human progesterone receptor membrane component 1
- MAPR
membrane associated progesterone receptor
- Dap1
damage associated protein 1
- RR
resonance Raman spectroscopy
- 5C
five-coordinate
- 6C
six-coordinate
- HS
high spin
- LS
low spin
- SHE
standard hydrogen electrode
- Cyt b5
cytochrome b5
Footnotes
ASSOCIATED CONTENT
UV–vis spectra associated with redox potential measurements; RR spectra of the ferric Y95H mutant of hPGRMC1 in H2 16O, D2 16O, and H2 18O, as well as the associated isotope difference spectra; and UV–vis and RR spectra of CO adducts of hPGRMC1. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Min L, Strushkevich NV, Harnastai IN, Iwamoto H, Gilep AA, Takemori H, Usanov SA, Nonaka Y, Hori H, Vinson GP. Molecular identification of adrenal inner zone antigen as a heme-binding protein. FEBS J. 2005;272:5832–5843. doi: 10.1111/j.1742-4658.2005.04977.x. [DOI] [PubMed] [Google Scholar]
- 2.Raza FS, Takemori H, Tojo H, Okamoto M, Vinson GP. Identification of the rat adrenal zona fasciculata/reticularis specific protein, inner zone antigen (IZAg), as the putative membrane progesterone receptor. Eur. J. Biochem. 2001;268:2141–2147. doi: 10.1046/j.1432-1327.2001.02096.x. [DOI] [PubMed] [Google Scholar]
- 3.Min L, Takemori H, Nonaka Y, Katoh Y, Doi J, Horike N, Osamu H, Raza FS, Vinson GP, Okamoto M. Characterization of the adrenal-specific antigen IZA (inner zone antigen) and its role in the steroidogenesis. Mol. Cell. Endocrinol. 2004;215:143–148. doi: 10.1016/j.mce.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 4.Lösel RM, Besong D, Peluso JJ, Wehling M. Progesterone receptor membrane component 1–many tasks for a versatile protein. Steroids. 2008;73:929–934. doi: 10.1016/j.steroids.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Selmin O, Lucier GW, Clark GC, Tritscher AM, Heuvel JPV, Gastel JA, Walker NJ, Thomas R, Bell DA. Isolation and characterization of a novel gene induced by 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin in rat liver. Carcinogenesis. 1996;17:2609–2615. doi: 10.1093/carcin/17.12.2609. [DOI] [PubMed] [Google Scholar]
- 6.Cahill MA. Progesterone receptor membrane component 1: an integrative review. J. Steroid Biochem. Mol. Biol. 2007;105:16–36. doi: 10.1016/j.jsbmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed IS, Rohe HJ, Twist KE, Mattingly MN, Craven RJ. Progesterone receptor membrane component 1 (Pgrmc1): a heme-1 domain protein that promotes tumorigenesis and is inhibited by a small molecule. J. Pharmacol. Exp. Ther. 2010;333:564–573. doi: 10.1124/jpet.109.164210. [DOI] [PubMed] [Google Scholar]
- 8.Mifsud W, Bateman A. Membrane-bound progesterone receptors contain a cytochrome b5-like ligand-binding domain. Genome Biol. 2002;3:1–5. doi: 10.1186/gb-2002-3-12-research0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peluso JJ, Romak J, Liu X. Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone’s antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations. Endocrinology. 2008;149:534–543. doi: 10.1210/en.2007-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peluso JJ, Liu X, Gawkowska A, Johnston-MacAnanny E. Progesterone activates a progesterone receptor membrane component 1-dependent mechanism that promotes human granulosa/luteal cell survival but not progesterone secretion. J. Clin. Endocrinol. Metab. 2009;94:2644–2649. doi: 10.1210/jc.2009-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes AL, Powell DW, Bard M, Eckstein J, Barbuch R, Link AJ, Espenshade PJ. Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab. 2007;5:143–149. doi: 10.1016/j.cmet.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Szczesna-Skorupa E, Kemper B. Progesterone receptor membrane component 1 inhibits the activity of drug-metabolizing cytochromes P450 and binds to cytochrome P450 reductase. Mol. Pharmacol. 2011;79:340–350. doi: 10.1124/mol.110.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oda S, Nakajima M, Toyoda Y, Fukami T, Yokoi T. Progesterone receptor membrane component 1 modulates human cytochrome P450 activities in an isoform-dependent manner. Drug Metab. Dispos. 2011;39:2057–2065. doi: 10.1124/dmd.111.040907. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh K, Thompson AM, Goldbeck RA, Shi X, Whitman S, Oh E, Zhiwu Z, Vulpe C, Holman TR. Spectroscopic and biochemical characterization of heme binding to yeast Dap1p and mouse PGRMC1p. Biochemistry. 2005;44:16729–16736. doi: 10.1021/bi0511585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suchanek M, Radzikowska A, Thiele C. Photoleucine and photo-methionine allow identification of protein–protein interactions in living cells. Nat. Methods. 2005;2:261–268. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 16.Song J, Vinarov DA, Tyler EM, Shahan MN, Tyler RC, Markley JL. Letter to the Editor: Hypothetical protein At2g24940. 1 from Arabidopsis thaliana has a cytochrome b5 like fold. J. Biomol. NMR. 2004;30:215–218. doi: 10.1023/b:jnmr.0000048943.34504.29. [DOI] [PubMed] [Google Scholar]
- 17.Yoshitani N, Satou K, Saito K, Suzuki S, Hatanaka H, Seki M, Shinozaki K, Hirota H, Yokoyama S. A structure-based strategy for discovery of small ligands binding to functionally unknown proteins: combination of in silico screening and surface plasmon resonance measurements. Proteomics. 2005;5:1472–1480. doi: 10.1002/pmic.200401032. [DOI] [PubMed] [Google Scholar]
- 18.Egawa T, Yeh S-R. Structural and functional properties of hemoglobins from unicellular organisms as revealed by resonance Raman spectroscopy. J. Inorg. Biochem. 2005;99:72–96. doi: 10.1016/j.jinorgbio.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Lu C, Zhao X, Lu Y, Rousseau DL, Yeh S-R. Role of copper ion in regulating ligand binding in a myoglobin-based cytochrome c oxidase model. J. Am. Chem. Soc. 2010;132:1598–1605. doi: 10.1021/ja907777f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leslie Dutton P. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 21.Munro AW, Noble MA, Robledo L, Daff SN, Chapman SK. Determination of the redox properties of human NADPH-cytochrome P450 reductase. Biochemistry. 2001;40:1956–1963. doi: 10.1021/bi001718u. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Wang J, Zhou F. Direct electrochemistry of catalase at a gold electrode modified with single-wall carbon nanotubes. Electroanalysis. 2004;16:627–632. [Google Scholar]
- 23.Antonini E, Brunori M. Hemoglobin and Myoglobin in Their Reactions with Ligands. Amsterdam: North-Holland Publishing Company; 1971. [Google Scholar]
- 24.Thompson AM, Reddi AR, Shi X, Goldbeck RA, Moënne-Loccoz P, Gibney BR, Holman TR. Measurement of the heme affinity for yeast dap1p, and its importance in cellular function. Biochemistry. 2007;46:14629–14637. doi: 10.1021/bi7013739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smulevich G, Neri F, Willemsen O, Choudhury K, Marzocchi MP, Poulos TL. Effect of the His175 .fwdarw. Glu mutation on the heme pocket architecture of cytochrome c peroxidase. Biochemistry. 1995;34:13485–13490. doi: 10.1021/bi00041a028. [DOI] [PubMed] [Google Scholar]
- 26.Eakanunkul S, Lukat-Rodgers GS, Sumithran S, Ghosh A, Rodgers KR, Dawson JH, Wilks A. Characterization of the periplasmic heme-binding protein shut from the heme uptake system of Shigella dysenteriae . Biochemistry. 2005;44:13179–13191. doi: 10.1021/bi050422r. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Moënne-Loccoz P, Hildebrand DP, Wilks A, Loehr TM, Mauk AG, Ortiz de Montellano PR. Replacement of the proximal histidine iron ligand by a cysteine or tyrosine converts heme oxygenase to an oxidase. Biochemistry. 1999;38:3733–3743. doi: 10.1021/bi982707s. [DOI] [PubMed] [Google Scholar]
- 28.Vatsis KP, Peng H-M, Coon MJ. Replacement of active-site cysteine-436 by serine converts cytochrome P450 2B4 into an NADPH oxidase with negligible monooxygenase activity. J. Inorg. Biochem. 2002;91:542–553. doi: 10.1016/s0162-0134(02)00438-5. [DOI] [PubMed] [Google Scholar]
- 29.Sun J, Loehr TM, Wilks A, Ortiz de Montellano PR. Identification of histidine 25 as the heme ligand in human liver heme oxygenase. Biochemistry. 1994;33:13734–13740. doi: 10.1021/bi00250a026. [DOI] [PubMed] [Google Scholar]
- 30.Caillet-Saguy C, Piccioli M, Turano P, Lukat-Rodgers G, Wolff N, Rodgers KR, Izadi-Pruneyre N, Delepierre M, Lecroisey A. Role of the iron axial ligands of heme carrier HasA in heme uptake and release. J. Biol. Chem. 2012;287:26932–26943. doi: 10.1074/jbc.M112.366385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egeberg KD, Springer BA, Martinis SA, Sligar SG, Morikis D, Champion PM. Alteration of sperm whale myoglobin heme axial ligation by site-directed mutagenesis. Biochemistry. 1990;29:9783–9791. doi: 10.1021/bi00494a004. [DOI] [PubMed] [Google Scholar]
- 32.Mokry DZ, Nadia-Albete A, Johnson MK, Lukat-Rodgers GS, Rodgers KR, Lanzilotta WN. Spectroscopic evidence for a 5-coordinate oxygenic ligated high spin ferric heme moiety in the Neisseria meningitidis hemoglobin binding receptor. Biochim. Biophys. Acta. 2014;1840:3058–3066. doi: 10.1016/j.bbagen.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagai K, Kagimoto T, Hayashi A, Taketa F, Kitagawa T. Resonance Raman studies of hemoglobins M: evidence for iron-tyrosine charge-transfer interactions in the abnormal subunits of Hb M Boston and Hb M Iwate. Biochemistry. 1983;22:1305–1311. doi: 10.1021/bi00274a048. [DOI] [PubMed] [Google Scholar]
- 34.Nagai M, Yoneyama Y, Kitagawa T. Characteristics in tyrosine coordinations of four hemoglobins M probed by resonance Raman spectroscopy. Biochemistry. 1989;28:2418–2422. doi: 10.1021/bi00432a012. [DOI] [PubMed] [Google Scholar]
- 35.Li XY, Czernuszewicz RS, Kincaid JR, Su YO, Spiro TG. Consistent porphyrin force field. 1. Normal-mode analysis for nickel porphine and nickel tetraphenylporphine from resonance Raman and infrared spectra and isotope shifts. J. Phys. Chem. 1990;94:31–47. [Google Scholar]
- 36.Li XY, Czernuszewicz RS, Kincaid JR, Spiro TG. Consistent porphyrin force field. 3. Out-of-plane modes in the resonance Raman spectra of planar and ruffled nickel octaethylporphyrin. J. Am. Chem. Soc. 1989;111:7012–7023. [Google Scholar]
- 37.Li XY, Czernuszewicz RS, Kincaid JR, Stein P, Spiro TG. Consistent porphyrin force field. 2. Nickel octaethylporphyrin skeletal and substituent mode assignments from nitrogen-15, meso-d 4 and methylene-d 16 Raman and infrared isotope shifts. J. Phys. Chem. 1990;94:47–61. [Google Scholar]
- 38.Hu S, Smith KM, Spiro TG. Assignment of protoheme resonance Raman spectrum by heme labeling in myoglobin. J. Am. Chem. Soc. 1996;118:12638–12646. [Google Scholar]
- 39.Pond AE, Roach MP, Sono M, Rux AH, Franzen S, Hu R, Thomas MR, Wilks A, Dou Y, Ikeda-Saito M, Ortiz de Montellano PR, Woodruff WH, Boxer SG, Dawson JH. Assignment of the heme axial ligand(s) for the ferric myoglobin (H93G) and heme oxygenase (H25A) cavity mutants as oxygen donors using magnetic circular dichroism. Biochemistry. 1999;38:7601–7608. doi: 10.1021/bi9825448. [DOI] [PubMed] [Google Scholar]
- 40.Nakamoto K. Infrared Spectra of Inorganic and Coordination Compounds. New York: John Wiley & Sons Inc; 1970. [Google Scholar]
- 41.Yu NT, Kerr EA, Spiro TG. Biological Applications of Raman Spectroscopy. Vol. 3. New York: Wiley; 1988. pp. 39–95. [Google Scholar]
- 42.Vogel KM, Kozlowski PM, Zgierski MZ, Spiro TG. Role of the axial ligand in heme-CO backbonding; DFT analysis of vibrational data. Inorg. Chim. Acta. 2000;297:11–17. [Google Scholar]
- 43.Feis A, Rodriguez-Lopez JN, Thorneley RN, Smulevich G. The distal cavity structure of carbonyl horseradish peroxidase as probed by the resonance Raman spectra of His 42 Leu and Arg 38 Leu mutants. Biochemistry. 1998;37:13575–13581. doi: 10.1021/bi981399v. [DOI] [PubMed] [Google Scholar]
- 44.Lukat-Rodgers GS, Rodgers KR, Caillet-Saguy C, Izadi-Pruneyre N, Lecroisey A. Novel heme ligand displacement by CO in the soluble hemophore HasA and its proximal ligand mutants: implications for heme uptake and release. Biochemistry. 2008;47:2087–2098. doi: 10.1021/bi7019518. [DOI] [PubMed] [Google Scholar]
- 45.Linder DP, Silvernail NJ, Barabanschikov A, Zhao J, Alp EE, Sturhahn W, Sage JT, Scheidt WR, Rodgers KR. The diagnostic vibrational signature of pentacoordination in heme carbonyls. J. Am. Chem. Soc. 2014;136:9818–9821. doi: 10.1021/ja503191z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho WW, Li H, Eakanunkul S, Tong Y, Wilks A, Guo M, Poulos TL. Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J. Biol. Chem. 2007;282:35796–35802. doi: 10.1074/jbc.M706761200. [DOI] [PubMed] [Google Scholar]
- 47.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. Haem recognition by a Staphylococcus aureus NEAT domain. Mol. Microbiol. 2007;63:139–149. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 48.Sharp KH, Schneider S, Cockayne A, Paoli M. Crystal structure of the heme–IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus . J. Biol. Chem. 2007;282:10625–10631. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 49.Mattle D, Zeltina A, Woo J-S, Goetz BA, Locher KP. Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis . J. Mol. Biol. 2010;404:220–231. doi: 10.1016/j.jmb.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 50.Villareal VA, Pilpa RM, Robson SA, Fadeev EA, Clubb RT. The IsdC protein from Staphylococcus aureus uses a flexible binding pocket to capture heme. J. Biol. Chem. 2008;283:31591–31600. doi: 10.1074/jbc.M801126200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnoux P, Haser R, Izadi N, Lecroisey A, Delepierre M, Wandersman C, Czjzek M. The crystal structure of HasA, a hemophore secreted by Serratia marcescens . Nat. Struct. Biol. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 52.Wolff N, Izadi-Pruneyre N, Couprie J, Habeck M, Linge J, Rieping W, Wandersman C, Nilges M, Delepierre M, Lecroisey A. Comparative analysis of structural and dynamic properties of the loaded and unloaded hemophore HasA: functional implications. J. Mol. Biol. 2008;376:517–525. doi: 10.1016/j.jmb.2007.11.072. [DOI] [PubMed] [Google Scholar]
- 53.Kumar R, Lovell S, Matsumura H, Battaile KP, Moënne-Loccoz P, Rivera M. The hemophore HasA from Yersinia pestis (HasAyp) coordinates hemin with a single residue, Tyr75, and with minimal conformational change. Biochemistry. 2013;52:2705–2707. doi: 10.1021/bi400280z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Decatur SM, Franzen S, DePillis GD, Dyer RB, Woodruff WH, Boxer SG. Trans effects in nitric oxide binding to myoglobin cavity mutant H93G. Biochemistry. 1996;35:4939–4944. doi: 10.1021/bi951661p. [DOI] [PubMed] [Google Scholar]
- 55.Meyer C, Schmid R, Scriba PC, Wehling M. Purification and partial sequencing of high-affinity progesterone-binding site(s) from porcine liver membranes. Eur. J. Biochem. 1996;239:726–731. doi: 10.1111/j.1432-1033.1996.0726u.x. [DOI] [PubMed] [Google Scholar]
- 56.Peluso JJ. Progesterone receptor membrane component 1 and its role in ovarian follicle growth. Front. Neurosci. 2013;7:99. doi: 10.3389/fnins.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohe HJ, Ahmed IS, Twist KE, Craven RJ. PGRMC1 (progesterone receptor membrane component 1): a targetable protein with multiple functions in steroid signaling, P450 activation and drug binding. Pharmacol. Ther. 2009;121:14–19. doi: 10.1016/j.pharmthera.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Létoffé S, Deniau C, Wolff N, Dassa E, Delepelaire P, Lecroisey A, Wandersman C. Haemophore-mediated bacterial haem transport: evidence for a common or overlapping site for haem-free and haem-loaded haemophore on its specific outer membrane receptor. Mol. Microbiol. 2001;41:439–450. doi: 10.1046/j.1365-2958.2001.02530.x. [DOI] [PubMed] [Google Scholar]
- 59.Caillet-Saguy C, Turano P, Piccioli M, Lukat-Rodgers GS, Czjzek M, Guigliarelli B, Izadi-Pruneyre N, Rodgers KR, Delepierre M, Lecroisey A. Deciphering the structural role of histidine 83 for heme binding in hemophore HasA. J. Biol. Chem. 2007;283:5960–5970. doi: 10.1074/jbc.M703795200. [DOI] [PubMed] [Google Scholar]
- 60.Tong Y, Guo M. Cloning and characterization of a novel periplasmic heme-transport protein from the human pathogen Pseudomonas aeruginosa . J. Biol. Inorg. Chem. 2007;12:735–750. doi: 10.1007/s00775-007-0226-x. [DOI] [PubMed] [Google Scholar]
- 61.Mallory JC, Crudden G, Johnson BL, Mo C, Pierson CA, Bard M, Craven RJ. Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae . Mol. Cell. Biol. 2005;25:1669–1679. doi: 10.1128/MCB.25.5.1669-1679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ray GB, Li XY, Ibers JA, Sessler JL, Spiro TG. How far can proteins bend the FeCO unit? Distal polar and steric effects in heme proteins and models. J. Am. Chem. Soc. 1994;116:162–176. [Google Scholar]
- 63.Spiro TG, Wasbotten IH. CO as a vibrational probe of heme protein active sites. J. Inorg. Biochem. 2005;99:34–44. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.