Abstract
Histone gene transcription is actively downregulated after completion of DNA synthesis to avoid overproduction. However, the precise mechanistic details of the cessation of histone mRNA synthesis are not clear. We found that histone H2B phosphorylation at Tyr37 occurs upstream of histone cluster 1, Hist1, during the late S phase. We identified WEE1 as the kinase that phosphorylates H2B at Tyr37. Loss of expression or inhibition of WEE1 kinase abrogated H2B Tyr37 phosphorylation with a concomitant increase in histone transcription in yeast and mammalian cells. H2B Tyr37 phosphorylation excluded binding of the transcriptional coactivator NPAT and RNA polymerase II and recruited the histone chaperone HIRA upstream of the Hist1 cluster. Taken together, our data show a previously unknown and evolutionarily conserved function for WEE1 kinase as an epigenetic modulator that marks chromatin with H2B Tyr37 phosphorylation, thereby inhibiting the transcription of multiple histone genes to lower the burden on the histone mRNA turnover machinery.
Core histone mRNA levels are exquisitely regulated within cells1,2. Histone transcription is initiated at the G1/S phase and is actively downregulated during the late S and G2 phases to avoid overproduction soon after the completion of DNA synthesis in the S phase3–8. Eukaryotic cells have evolved multiple mechanisms to maintain appropriate histone abundance during the cell cycle. One of the most extensively studied mechanisms that operates in all higher eukaryotes is the increased turnover of histone mRNAs9. Recently, another mechanism was reported in Schizosaccharomyces pombe in which the activator for histone transcription is actively degraded, resulting in histone transcriptional downregulation10. It is probable that in higher eukaryotes, histone transcript turnover may not be sufficient to accomplish a rapid decrease in histone transcript abundance. Cells would also need to shut off histone transcription at the end of DNA synthesis to avoid burdening the histone turnover machinery to downregulate histone abundance. The importance of histone transcriptional shutdown was noted as early as the late 1980s11; however, little progress has since been made in terms of how this crucial process is precisely regulated in a cell cycle–dependent manner.
Tyrosine kinases regulate key cellular processes, including cell growth, proliferation and differentiation. Although the cytosolic effectors of most tyrosine kinases have been well studied, direct epigenetic modulation by tyrosine phosphorylation is a relatively new subject of study12,13. Multiple well-conserved tyrosine residues are present in histones. However, the phosphorylation status and physiological functions of the majority of these tyrosine residues are unknown. We therefore sought to address this relatively understudied epigenetic modification. Using a mass spectrometry–based approach, we discovered that in mammalian cells, histone H2B is phosphorylated at Tyr37 during the late S phase upstream of the Hist1 cluster, where about 80% of the histone genes are located. In higher eukaryotic cells, histone transcription is regulated at the transcriptional and post-transcriptional levels3,9. For example, in synchronized HeLa cells, the rate of histone mRNA synthesis increases approximately three-fold during the initial 2.5 h after release from the thymidine and aphidicolin block at the G/S boundary2. The abundance of nuclear histone mRNA reaches its maximum by the second hour after entry into S phase and remains at that level until the end of the S phase; mRNA levels then drop steeply at the end of the S phase2. Soon after transcription, histone mRNAs are subjected to post-transcriptional processing and increase in stability9. A cumulative effect of these two processes is an increase of approximately 15- to 20-fold in steady-state histone mRNA levels in the S phase. At the end of the S phase, cells switch off histone transcription. How cells terminate histone transcription is unknown. Here we report that phosphorylation of H2B at Tyr37 by WEE1 leads to coordinated transcriptional suppression of replication-dependent core histone genes in the late S/G2 phase.
Wee1, discovered for its small size as a cdc (cell division cycle) mutant in the fission yeast S. pombe, is evolutionarily conserved. WEE1 is a nuclear tyrosine kinase that negatively regulates the activity of the cyclin-dependent kinase Cdc2 by tyrosine phosphorylation to prevent mitotic entry before the completion of DNA synthesis14–16. The expression of WEE1 protein is tightly regulated in cells. Its expression peaks during the S phase and is quickly degraded in the G2 phase. We show here for the first time that WEE1 kinase, previously established as a cell-cycle regulator, is a new epigenetic modifier of histone H2B.
RESULTS
Identification of Tyr37-phosphorylated H2B in late S phase
To delineate the functional consequences of histone tyrosine phosphorylation, we subjected purified histones to mass spectrometry–based identification of post-translational modifications. Using this method, we identified a previously unreported phosphorylation at Tyr37 in histone H2B (Supplementary Fig. 1). As the functional role of Tyr37-phosphorylated H2B (pTyr37 H2B) is unknown, we raised phospho-specific antibodies to pTyr37 H2B, which we extensively validated (Supplementary Figs. 2–4). We compared the recognition of different H2B peptides by the antibodies to pTyr37 H2B and found that these antibodies recognized only the H2B phosphopeptide that was phosphorylated at Tyr37, but not the nonphosphorylated H2B peptide or the H2B peptide harboring a Y37F substitution (Supplementary Fig. 2a). Further, competition of pTyr37 H2B antibodies with phosphopeptide resulted in almost complete loss of recognition of Tyr37 H2B phosphopeptide (Supplementary Fig. 2b,c). We also screened the pTyr37 H2B antibodies for cross-reactivity with 59 distinct acetylation, methylation, phosphorylation and citrullination modifications on core histones using histone peptide arrays. The pTyr37 H2B antibody did not cross-react with any of these post-translational modifications; however, when we hybridized the same blots with histone H3 trimethyl Lys9 (H3K9me3) antibodies, the expected pattern of hybridization was produced (Supplementary Fig. 2d). In addition, immunoblotting with whole-cell extracts revealed a band of ~15 kDa (Supplementary Fig. 2e). Collectively, these data indicate that the antibodies are selective for Tyr37-phosphorylated H2B.
A mass spectrometry–based analysis revealed that in asynchronously growing cultures, the amount of pTyr37 H2B is 0.21% that of the total H2B protein (Supplementary Table 1), which suggested that H2B could be tyrosine phosphorylated transiently during the cell cycle and then rapidly dephosphorylated. To assess the cell cycle–specific expression of pTyr37 H2B, we synchronized mouse embryo fibroblasts (MEFs) by double thymidine block, which arrests cells at the G1/S boundary, released them into fresh medium and collected the aliquots after various time intervals. We confirmed the synchronization of the cells by immunoblotting with cyclin A2, cyclin E and pTyr15 Cdc2 antibodies, as well as by flow cytometry (Supplementary Fig. 3a–c). These experiments collectively indicate that the majority of the cells were arrested at the G1/S boundary by the thymidine block; the cells progressed into the cell cycle synchronously to reach mid S phase by about 4 h, late S phase by 6.5–7 h and the G2/M phase by about 8–9 h after thymidine release into fresh medium.
We detected pTyr37 H2B at 6.25–6.75 h after thymidine release, and the amount of pTyr37 H2B peaked at 6.5 h after release (Fig. 1a and Supplementary Fig. 3a). Synchronized H1975, a human lung cancer cell line, also contained pTyr37 H2B at 6.5 h after thymidine release (data not shown). An 5-ethynyl-2-deoxyuridine (EdU) incorporation assay of MEFs at 6.5 h after thymidine release revealed that a substantial proportion were in the late S phase (Supplementary Fig. 3c and Supplementary Note).
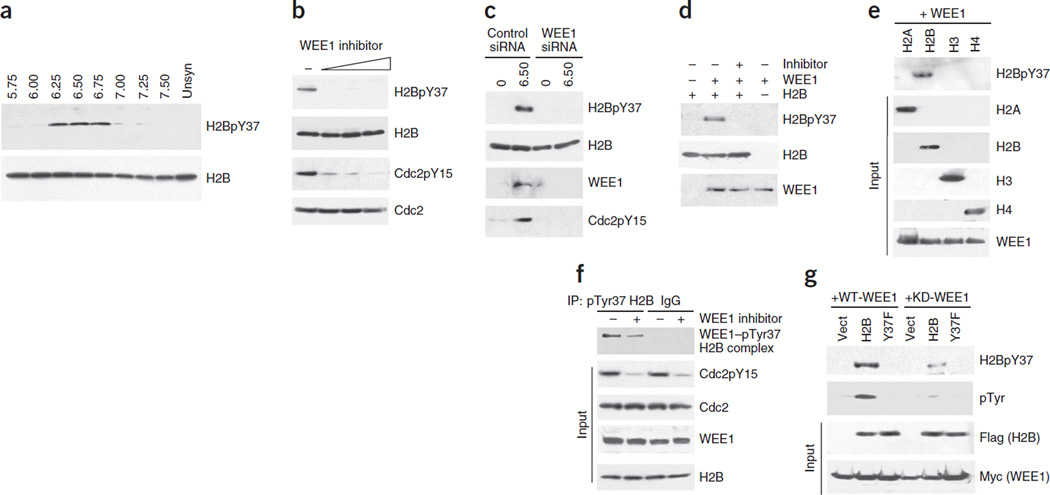
Figure 1.
WEE1 phosphorylates histone H2B at Tyr37. (a) Synchronized MEF lysates immunoprecipitated with pTyr37 H2B antibodies and immunoblotted with H2B antibody (top). Total levels of histone H2B are shown in the bottom blot. Unsyn, unsynchronized. (b) H1975 cells treated with the WEE1 inhibitor MK-1775 (0.31, 0.62 and 1.25 µM) had loss of H2B Tyr37 phosphorylation. (c) MEFs transfected with WEE1 siRNA had loss of H2B Tyr37 phosphorylation compared to cells transfected with control siRNAs. (d) In vitro kinase assay with purified proteins showing direct phosphorylation of H2B by WEE1 kinase. (e) In vitro kinase assay with purified proteins indicating that WEE1 kinase phosphorylates H2B but not other core histones. (f) Coimmunoprecipitation revealing endogenous WEE1–pTyr37 H2B complexes in MEFs. (g) MEFs coexpressing Myc-tagged WEE1 (WT-WEE1) or kinase-dead mutant WEE1 (KD-WEE1) and empty vector, Flag-tagged H2B or Y37F mutant H2B were immunoprecipitated with pTyr37 H2B antibodies and then immunoblotted with Flag antibody, revealing that WEE1 specifically phosphorylated H2B at Tyr37.
WEE1 kinase phosphorylates H2B at Tyr37
To identify a kinase that can target H2B for Tyr37 phosphorylation, we assessed multiple kinases, including receptor tyrosine kinases such as EGFR, HER2, PDGFR, FGFR and IR. We also assessed nonreceptor tyrosine kinases, including Src, Abl and Ack1 (also known as TNK2), for their ability to phosphorylate H2B. However, none of these kinases could phosphorylate H2B in vivo (data not shown). During the S phase, temporal regulation of the expression of WEE1 kinase and the tyrosine phosphorylation of its substrate, Cdc2, is well established14,16,17. To examine whether H2B is a WEE1 kinase substrate, we treated H1975 cells with increasing concentrations of a WEE1 inhibitor, MK-1775 (ref. 18). Even treatment with a 0.3 µM concentration of this WEE1 inhibitor resulted in the complete loss of the H2B Tyr37 phosphorylation mark (H2BpY37) (Fig. 1b). Similarly, transfection of the cells with WEE1 short interfering RNA (siRNA) also resulted in the complete loss of H2BpY37 (Fig. 1c). To test whether WEE1 directly phosphorylates H2B, we performed an in vitro kinase assay19. When we incubated purified WEE1 and H2B together, H2B was phosphorylated at Tyr37, and this phosphorylation was abrogated by treatment with the WEE1 inhibitor (Fig. 1d). Further, WEE1 specifically phosphorylated wild-type H2B peptide but not the Y37F mutant peptide (Supplementary Fig. 4a). Moreover, WEE1 specifically phosphorylated H2B but not three other core histones, H2A, H3 and H4 in vitro (Fig. 1e). To determine the direct binding of WEE1 to H2B in vivo, we coimmunoprecipitated WEE1 and H2B, which revealed formation of endogenous WEE1–pTyr37 H2B complexes (Fig. 1f). We also confirmed the binding of WEE1 to endogenous H2B as well as the phosphorylation of H2B by transfecting Myc-tagged WEE1 (Supplementary Fig. 4b).
To further validate H2B as a substrate for WEE1 kinase in vivo, we generated Flag-tagged constructs expressing either wild-type or Y37F mutant H2B. Coexpression of the two forms of H2B with wild-type (WT) WEE1 or kinase-dead WEE1 revealed that WT WEE1 specifically phosphorylated H2B but did not phosphorylate the Y37F H2B mutant (Fig. 1g). To determine whether WEE1 kinase can phosphorylate other core histones in vivo, we transfected cells with the WEE1 kinase and immunoprecipitated the core histones. Immunoblotting with antibodies to pTyr37 H2B revealed that the WEE1 kinase phosphorylated H2B but not other core histones under these conditions (Supplementary Fig. 4c). Collectively, these data indicate that WEE1 kinase specifically phosphorylates H2B at Tyr37.
H2BY37 phosphorylation occurs upstream of Hist1
To identify regions in chromatin where H2B Tyr37 phosphorylation occurs, we performed a native chromatin immunoprecipitation (ChIP)-coupled DNA microarray analysis (ChIP-on-chip)20 and native ChIP sequencing (Supplementary Note). We immunoprecipitated the sheared chromatin from synchronized MEFs (0 h and 6.5 h after thymidine release) using the pTyr37 H2B antibody or IgG and then sequenced the resulting DNA. This led to the identification of 3,524 potential sites and regions where pTyr37 H2B is present in the mouse genome; l,240 of these sites were within genes, whereas 351 sites were located upstream and 326 sites were located downstream of genes (Supplementary Tables 2 and 3). The MEME motif search tool found 20 unique motifs in our ChIP data (Supplementary Table 4). An analysis of these 20 motifs using FIMO is shown in Supplementary Table 5. Based on the GC-rich content and highest E value, we used the motif termed ‘motif 3’ to search all the ChIP data using the MAST program. This screen revealed 511 genes that could potentially be regulated by H2B Tyr37 phosphorylation (Supplementary Table 6). In addition, using MetaCore, we ran several enrichment analyses to cluster genes adjacent to the ChIP sequencing peaks. The gene clusters are shown in Supplementary Figure 5b based on their functional role.
A native ChIP-on-chip analysis revealed two sites containing pTyr37 H2B, named here site 1 and 2, upstream of histone-encoding genes of Hist1 (Supplementary Table 3). Notably, motif 3 was also present in both site 1 and site 2 (Supplementary Fig. 5a). Collectively, these data opened the possibility that histone expression is regulated by a modified histone. We chose to investigate this finding further because of the novelty of the observation. The genes for the five histones H1, H2A, H2B, H3 and H4 are clustered together in the genomes of all metazoans. There are 10–20 functional copies of these genes for each of the histone proteins, and each individual gene encodes a fraction of the total histone protein. In mammals, the genes for the core histones are clustered together in two loci, Hist1 and Hist2. The largest cluster, HIST1, is located on chromosome 6 in humans and chromosome 13 in mice21–23. The mouse and human histone gene clusters have strikingly similar gene numbers and organizations. The mouse Hist1 cluster contains the majority of the histone-coding genes: 45 core histone genes and 6 histone H1 genes1,23. The histone genes in the Hist1 cluster are arranged in three subclusters. About a third of the histone genes located in subcluster 1 are closest to sites 1 and 2 (Fig. 2a); we analyzed some of these genes in this study.
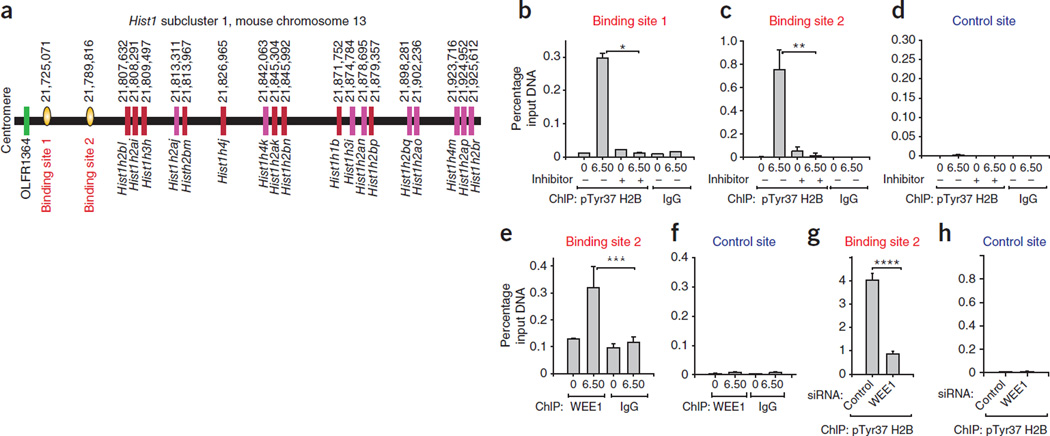
Figure 2.
H2BpY37 occurs upstream of the histone gene cluster Hist1. (a) Shown are the positions of histone genes in the mouse Hist1 subcluster 1 and two pTyr37 H2B binding sites. The starting nucleotide position is shown above. Subcluster 1 is conserved between human and mouse. (b–d) Native ChIP with pTyr37 H2B or IgG antibodies followed by qPCR using primers corresponding to site 1 (b), site 2 (c) or the control site (d) validated the presence of H2BpY37 at sites 1 and 2, upstream of the Hist1 cluster. Data shown in the bar graphs are the mean ± s.d. from three independent experiments. *P = 0.001, **P = 0.006. (e,f) ChIP using WEE1 or IgG antibodies followed by qPCR for site 2 (e) or the control site (f) revealing recruitment of WEE1 to site 2. Data shown in the bar graphs are the mean ± s.d. from three independent experiments. ***P = 0.02. (g,h) ChIP performed in cells transfected with WEE1 siRNAs showed loss of H2B Tyr37 phosphorylation at site 2. (g) but not at the control site (h). Data shown in the bar graphs are the mean ± s.d. from three independent experiments. ****P = 0.001. Student’s t test was used to calculate statistical significance throughout the paper.
To validate the presence of pTyr37 H2B upstream of the Hist1 cluster, we immunoprecipitated sheared chromatin from synchronized MEFs (0 h and 6.5 h after thymidine release) using the pTyr37 H2B antibody, followed by quantitative PCR (ChIP-qPCR)24,25. The primers used for ChIP DNA amplification are shown in Supplementary Table 7. This method revealed that pTyr37 H2B is present at both site 1 and site 2; however, phosphorylation at these sites was significantly reduced by treatment with the WEE1 inhibitor (Fig. 2b,c). As a control, we used primers corresponding to the ‘gene desert’ on chromosome 6 where we did not detect any pTyr37 H2B (control site, Fig. 2d). To assess whether WEE1 regulates H2B Tyr37 phosphorylation upstream of the Hist1 cluster, we performed ChIP-qPCR using WEE1 antibodies. The amount of WEE1 at site 2, but not at the control site, was significantly increased in MEFs at 6.5 h after thymidine release (Fig. 2e,f). To further validate the role of WEE1, we transfected cells with WEE1 or control siRNAs and performed ChIP-qPCR using pTyr37 H2B antibodies. There was a significant decrease in site 2–specific chromatin immunoprecipitated DNA in samples transfected with WEE1 siRNA compared to the sample transfected with control siRNA (Fig. 2g,h), indicating a functional, WEE1-kinase–dependent association of pTyr37 H2B with the Hist1 gene cluster.
H2BTyr37 phosphorylation regulates transcription of histones
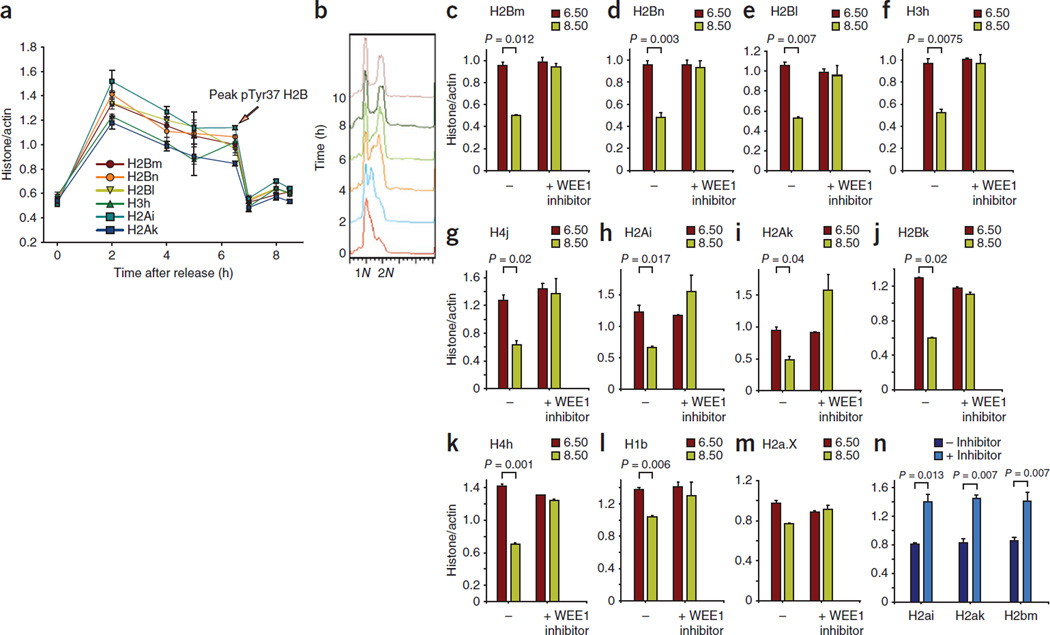
Whereas the modification of core histones has been shown to regulate transcription26–29, the transcription of the histones themselves is not known to be regulated by histone tyrosine phosphorylation. We assessed the expression of histone mRNA in synchronized MEFs and observed that core histone mRNA levels increased rapidly within 2 h after thymidine release, which is consistent with data from synchronized HeLa cells2. However, at 6.5 h after thymidine release, when cells are in the late S phase and H2B is phosphorylated at Tyr37, we found a sharp decline in histone mRNA levels (Fig. 3a,b). To investigate whether pTyr37 H2B has a direct role in regulating the transcription of histone genes, we treated synchronized MEFs with the WEE1-specific inhibitor MK-1775 (or left the MEFs untreated as a control group) and then performed qRT-PCR for the individual histone genes. In untreated cells, we found a rapid decrease in the number of transcripts of multiple core histone genes after 6.5 h after thymidine release (Fig. 3c–k). Notably, histone mRNA levels did not decrease in the cells treated with the WEE1 inhibitor (Fig. 3c–g,j,k). Indeed, for H2Ai and H2Ak, we found a significant increase in histone mRNA levels (Fig. 3h,i). Histones H2Bk and H4h, which are located in the histone subclusters 2 and 3, respectively, also had significant increases in the number of transcripts after treatment with the WEE1 inhibitor (Fig. 3j,k). The histone H1b had a modest increase in its transcript levels after treatment with the WEE1 inhibitor (Fig. 3l). As a control, we assessed the transcript abundance of a variant histone, H2AX, whose transcription is regulated in a cell cycle–independent manner. Histone H2AX transcript levels did not change significantly as a result of treatment with the WEE1 inhibitor (Fig. 3m). We also confirmed that S-phase progression was not overtly affected by WEE1 inhibition by performing a flow cytometric analysis of these cells (Supplementary Fig. 3b). To directly show a role of H2B Tyr37 phosphorylation in suppressing histone transcription, we performed a nuclear run-on assay (Supplementary Note). We labeled the nascent transcripts with biotin-UTP, followed by purification using streptavidin beads. qRT-PCR revealed a significant increase in histone mRNA synthesis in cells treated with the WEE1 inhibitor (Fig. 3n). Collectively, these data indicate that H2B Tyr37 phosphorylation may have a repressive effect on the transcription of the core histone genes located in the Hist1 cluster.
Figure 3.
H2BpY37 suppresses the transcription of histone genes located in the Hist1 cluster. (a) qRT-PCR of RNA prepared from synchronized MEFs indicating a rapid decrease in histone mRNA levels after 6.5 h after thymidine release. The y axis label “histone/actin” indicates the amount of histone transcript compared to that of actin. Error bars indicate the standard error. (b) Synchronized MEFs were harvested, stained with propidium iodide and then subjected to flow cytometry, which indicated that cells were in the late S/G2 phase at 6–8 h after thymidine release. (c–m) qRT-PCR of RNA prepared from synchronized MEFs indicating an increase in histone mRNA levels at 8.5 h in samples treated with WEE1 inhibitor compared to untreated samples. The histograms include data from two independent biological experiments, each done in triplicates. (n) The nuclei were prepared from synchronized MEFs treated with the WEE1 inhibitor MK-1775 (0.625 µM, 14 h) or left untreated. Run-on assay was performed, revealing that WEE1 regulates histone transcription. Data in all the bar graphs are the mean ± s.d. Data are representative of two independent biological experiments.
H2B Tyr40 phosphorylation in Saccharomyces cerevisiae
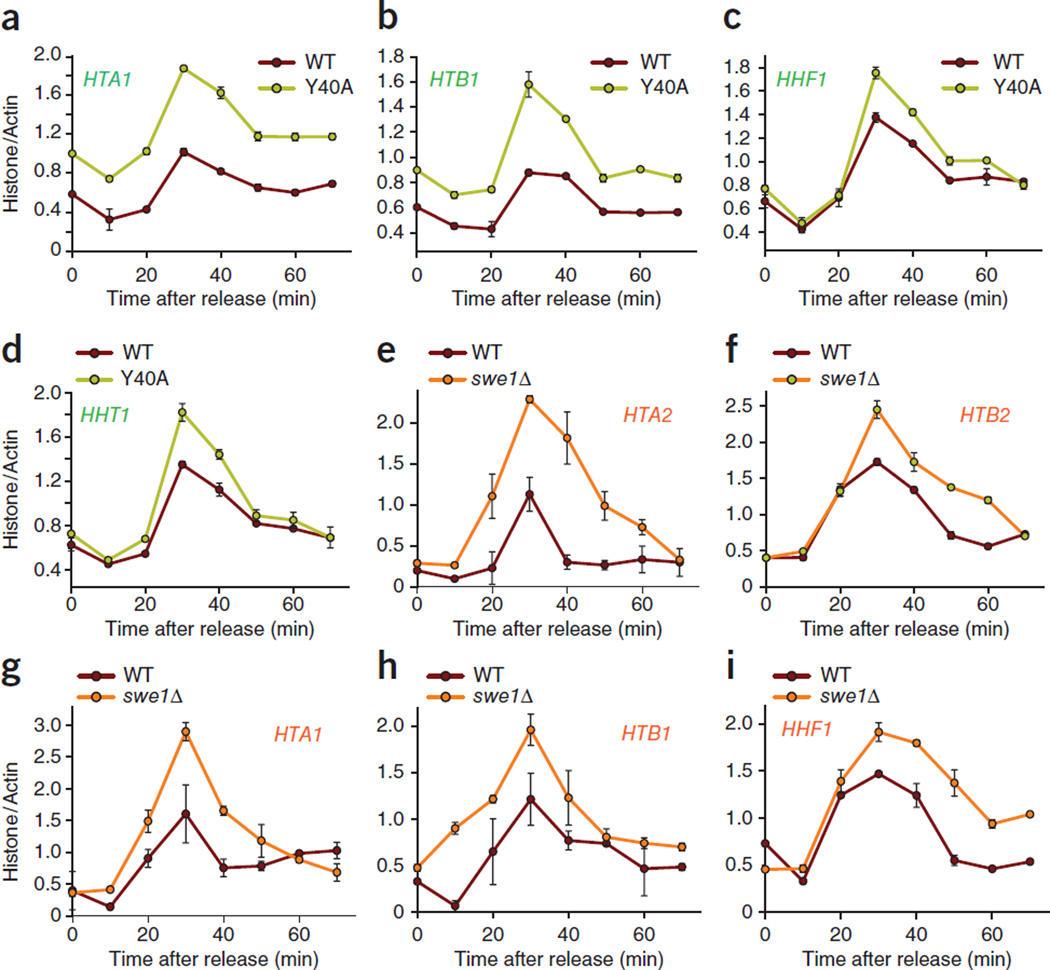
Because multiple functional genes are available for each of the core histones in higher eukaryotes, performing mutational studies is challenging. The budding yeast S. cerevisiae has been used extensively as a model organism to study the roles of a variety of post-translational modifications in histones30–32 and of histone transcriptional activation3,5– 8,11. In S. cerevisiae, histones are transcribed from four sets of gene pairs, HTA1-HTB1 and HTA2-HTB2 for H2A and H2B and HHF1-HHT1 and HHT2-HHF2 for H3 and H4 (refs. 3,5–7). Similarly to what occurs in metazoans, transcription is activated at the G1/S transition and is repressed in the G1 and G2/M phases of the cell cycle in yeast. We observed that the Tyr37 site is evolutionarily conserved, including in unicellular eukaryotes—for example, Tyr40 in S. cerevisiae (Supplementary Fig. 6a). As Tyr37 is invariant from human to yeast and a point mutant of H2B lacking the Tyr40 site is available in S. cerevisiae33, we assessed histone transcription in yeast cells expressing the Y40A mutant of H2B. We synchronized WT and Y40A H2B mutant S. cerevisiae cells by adding α-factor, after which we resuspended them in fresh medium and harvested them at different time points. We found H2B Tyr40 phosphorylation at approximately 30 min after release from α-factor; in contrast, H2B Tyr40 phosphorylation was abrogated in the Y40A H2B mutant yeast (Supplementary Fig. 6b). To examine histone transcription, we isolated mRNAs from WT and Y40A H2B mutant yeast and then performed qRT-PCR. The WT cells had peak histone mRNA synthesis at 30 min after α-factor release, which was followed by a rapid decrease in mRNA synthesis (Fig. 4a–d). In contrast, H2B Y40A mutant yeast had significantly higher HTA1, HTB1, HHF1 and HHT1 mRNA levels than WT yeast at 30 min after α-factor release, which remained elevated until the end of cell cycle at ~50 min after α-factor release (Fig. 4a–d).
Figure 4.
Histone tyrosine phosphorylation by SWE1 is required for the transcriptional suppression of histones in yeast. (a–i) WT and Y40A mutant yeast cells (a–d) and WT and swe1Δ mutant yeast cells (e–i) were synchronized followed by RNA preparation and qRT-PCR. The mutants Y40A and swe1Δ showed marked increases in histone transcript abundances. The data shown are derived from individual experiments that are representative of at least three independent replicates. All data are the mean ± s.d.
To determine whether H2B Tyr40 phosphorylation occurs in yeast histone promoters, we performed native ChIP followed by qPCR using primers corresponding to the promoter of the HTA1-HTB1 loci (Supplementary Fig. 6c). H2B Tyr40 phosphorylation was enriched in the promoter region containing the NEG site at 30 min after α-factor release in WT cells but not in the Y40A H2B mutant cells (Supplementary Fig. 6d). Collectively, these data indicate that H2B Tyr40 phosphorylation is required for the suppression of yeast histone transcription. We found that WEE1 kinase regulates histone transcriptional repression in mammalian cells. To assess whether yeast cells use a similar strategy, we used a yeast mutant lacking the homolog of WEE1 in S. cerevisiae, SWE1 (refs. 34,35). H2B was phosphorylated in the WT but not in the SWE1 mutant (swe1Δ) yeast cells (Supplementary Fig. 6e). To determine whether loss of SWE1, and thus loss of Tyr40 phosphorylation of H2B, would also lead to the loss of transcriptional suppression of yeast histones, we performed qRT-PCR on RNA isolated from WT and swe1Δ mutant yeast. As seen in mammalian cells, loss of SWE1 in yeast resulted in a marked increase in HTA2, HTB2, HTA1, HHF1 and HTB1 mRNA levels (Fig. 4e–i). Taken together, these data indicate that the WEE1 kinase, its substrate and the post-translational modification are all conserved in yeast.
We performed a fluorescence-activated cell sorting analysis to determine whether the increase in histone gene expression was the result of an altered cell-cycle profile of the Y40A H2B mutant cells (Supplementary Note). We released synchronized WT and Y40A H2B mutant S. cerevisiae cells into yeast extract peptone dextrose (YPD) medium, harvested them at the indicated time points, stained them with SYTOX green and then performed flow cytometry. WT and Y40A H2B mutant cells had similar cell-cycle kinetics (Supplementary Fig. 7). We also performed a growth analysis of Y40A H2B cells in complete growth medium and found their growth to be indistinguishable from that of WT yeast cells (data not shown), which suggests that the higher histone mRNA levels observed in the Y40A H2B mutant are not caused by altered cell growth or an altered cell cycle.
pTyr37 H2B suppresses NPAT and RNA polymerase II binding
Histone transcriptional activation in mammalian cells has been well studied: for example, NPAT has been shown to have an essential role in the transcriptional activation of histone genes by its recruitment, along with that of RNA polymerase II, at both Hist clusters, Hist1 and Hist2 (refs. 36,37). On the basis of its histone mRNA profile, we reasoned that H2B Tyr37 phosphorylation could potentially modulate histone transcription by its interaction with NPAT and RNA polymerase II. To test this hypothesis, we immunoprecipitated chromatin from synchronized MEFs at 6.5 h after thymidine release with NPAT or RNA polymerase II antibodies, which we followed with qPCR. Although we did not find recruitment of NPAT or RNA polymerase II when H2B Tyr37 phosphorylation was optimal, we did detect binding of both NPAT and RNA polymerase II when the phosphorylation was abrogated by treatment with the WEE1 inhibitor (Fig. 5a–c). The recruitment of NPAT and RNA polymerase II was minimal at the control site (Supplementary Fig. 8a,b). To confirm that the presence of RNA polymerase II at site 2 was dependent on NPAT, we transfected MEFs with control and NPAT siRNAs and immunoprecipitated chromatin with RNA polymerase II antibodies, which we followed with qPCR. The binding of RNA polymerase II at site 2 was markedly decreased by the depletion of NPAT by siRNA (Supplementary Fig. 8c,d).
Figure 5.
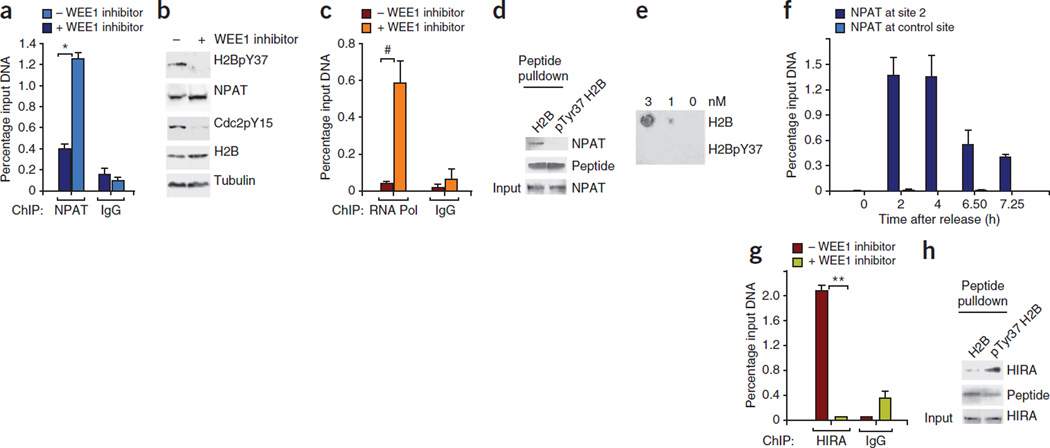
H2BpY37 differentially regulates NPAT and HIRA recruitment. (a) ChIP performed using NPAT and IgG antibodies followed by qPCR revealed loss of NPAT recruitment at site 2 after deposition of H2BpY37 marks. *P = 0.0018. (b) Immunoblotting revealed that NPAT protein amounts were not altered in MEFs treated with WEE1 inhibitor. (c) ChIP performed using RNA polymerase II (Pol) and control IgG antibodies followed by qPCR revealed loss of RNA polymerase II recruitment at site 2 in presence of H2BpY37. #P = 0.01. (d) Immobilized unmodified H2B and Tyr37-phosphorylated H2B25–49 peptides were incubated with HEK 293 cell lysates followed by immunoblotting with NPAT antibodies, indicating that NPAT binds unmodified H2B. (e) A filter-binding assay confirming the selectivity of NPAT binding to unmodified H2B. (f) Synchronized MEFs were harvested and ChIP was performed using NPAT antibodies followed by qPCR, revealing decreased NPAT binding to site 2 after 6.5 h. (g) Recruitment of HIRA at site 2 is dependent on H2B Tyr37 phosphorylation. Data shown in the bar graphs are the mean ± s.d. from three independent experiments. **P = 0.017. (h) Pull-down assay with immobilized unmodified and Tyr37-phosphorylated H2B25–49 peptides revealing selective binding of HIRA to Tyr37-phosphorylated H2B25–49 peptide.
To further understand the mechanism underlying the loss of NPAT binding to site 2 when H2B is phosphorylated at Tyr37, we performed a pull-down assay with biotin-conjugated H2B peptides spanning amino acids 25–49 (referred to here as H2B25–49) and pTyr37 H2B25–49 peptides. We immobilized the H2B25–49 and pTyr37 H2B25–49 peptides on streptavidin-Sepharose beads and then incubated them with whole-cell extracts of HEK 293 cells. We washed the beads and analyzed the bound protein by immunoblotting with NPAT antibodies. Although NPAT specifically bound to the unmodified H2B peptide, this binding was abolished when the peptide was phosphorylated at Tyr37 (Fig. 5d). We further confirmed the specificity of NPAT for unphosphorylated H2B using a filter-binding assay wherein the membrane spotted with peptides was incubated with HEK 293 lysates and probed with NPAT antibodies (Fig. 5e).
To assess the dynamics of NPAT binding, we performed ChIP using NPAT antibodies for the samples collected at 0 h, 2 h, 4 h, 6.5 h or 7.25 h after thymidine release, followed by qPCR for site 2 and the control site. These analyses revealed optimal recruitment of NPAT at site 2 at 2 h after release, and this recruitment continued at 4 h after release; however, at 6.5 h after thymidine release, when H2B is phosphorylated at Tyr37 (as well as after that time point), NPAT binding at site 2 was markedly diminished (Fig. 5f). To determine whether NPAT could interact with WEE1 kinase, we performed a coimmunoprecipitation assay. NPAT bound to WEE1 but did not interact with a WEE1 deletion mutant lacking the N-terminal regulatory domain (Supplementary Fig. 9a,b). This finding, together with the ChIP data, suggests that NPAT recruits WEE1 and that WEE1 phosphorylates H2B and excludes NPAT and RNA polymerase II from binding upstream of Hist1 cluster.
H2B Tyr37 phosphorylation recruits HIRA
Researchers from a previous study identified a negative regulatory site in S. cerevisiae, named the NEG or CCR region, in the promoters of six of the eight yeast histone genes (HTA1-HTB1, HHT1-HHF1 and HHT2-HHF2)38. They identified four evolutionarily conserved trans-acting factors, HIR1, HIR2, HIR3 and HPC2, that bind the negative regulatory site to repress histone transcription8,39. These four proteins form the HIR co-repressor complex, also known as the histone chaperone complex40,41. The concentrations of the HIR proteins are not regulated by the cell cycle42–44, which has led to the identification of regulation wherein histone transcription outside of the S phase is actively inhibited by the interaction of the HIR complex with two other H3 and H4 histone chaperones, Asf1 and Rtt106 (ref. 45). Unlike yeast, humans have a single HIRA protein, histone regulatory homolog A, which is also a component of the histone chaperone complex that spreads across silenced domains to enforce transcriptional silencing46,47. We observed that H2B Tyr40 phosphorylation is enriched in the HTA1-HTB1 promoter region containing the NEG site, where members of the HIR complex localize (Supplementary Fig. 6c,d). Ectopic overexpression of HIRA has been reported to suppress histone transcription48. On the basis of these data, we hypothesized that HIRA could be involved in the transcriptional suppression mediated by pTyr37 H2B in mammalian cells. To assess whether H2B Tyr37 phosphorylation at site 2 resulted in the recruitment of HIRA, we immunoprecipitated chromatin from synchronized MEFs with HIRA antibodies, which we followed with qPCR. HIRA was recruited at site 2 when H2B was phosphorylated at Tyr37; however, the binding of HIRA was abrogated when we added the WEE1 inhibitor, which suppressed the phosphorylation of H2B (Fig. 5g). To further confirm HIRA binding to site 2 when H2B is phosphorylated at Tyr37, we performed a pull-down assay with biotin-conjugated H2B peptides, as described above. HIRA specifically associated with the Tyr37-phosphorylated H2B peptide; however, the binding of HIRA to the phosphorylated peptide was markedly lower than its binding to the unmodified H2B peptide (Fig. 5h).
To perform a comparative assessment of WEE1 and HIRA knockdown on histone transcription, we transfected MEFs with control siRNA, HIRA-specific siRNA or WEE1-specific siRNA. Consistently with earlier data (Fig. 3), we found a marked decrease in histone mRNA levels in the samples transfected with control siRNA at 6.5 h after thymidine release but not in the samples transfected with WEE1 siRNA (Supplementary Fig. 9c–f). Notably, samples transfected with HIRA siRNA had overall higher histone mRNA levels than the samples transfected with control siRNA. Collectively, these data indicate that WEE1-mediated H2B Tyr37 phosphorylation at 6.5 h after thymidine release dislodges NPAT, leading to a drop in transcription in the samples transfected with control siRNA. HIRA binding to Tyr37-phosphorylated H2B might prevent NPAT rebinding, and, thus, when HIRA was knocked down, because Tyr37 phosphorylation was intact, NPAT would be excluded, leading to a decrease in histone mRNA levels after 6.5 h after thymidine release. However, it is possible that NPAT exclusion is not efficiently accomplished in the absence of HIRA, leading to overall increased levels of histone mRNAs.
DISCUSSION
Collectively, our data unveil a previously unknown mechanism wherein marking chromatin specifically upstream of a major histone gene cluster, Hist1, with H2B Tyr37 phosphorylation by WEE1 tyrosine kinase suppressed the transcription of mammalian histone genes by excluding NPAT and RNA polymerase II and promoting HIRA recruitment (Fig. 6). This mechanism seems to be evolutionarily conserved, including in yeast, where phosphorylation of H2B Tyr40 (the equivalent of the mammalian H2B Tyr37 site) suppressed the transcription of the HTA1-HTB1, HTA2-HTB2 and HHF1-HHT1 loci. Notably, there were marginally higher HTT2 mRNA levels in Y40A and swe1Δ mutant yeast cells compared to WT yeast cells (data not shown), suggesting that H2B Tyr40 phosphorylation has a modest role in the transcriptional suppression of the HHT2-HHF2 locus.
Figure 6.
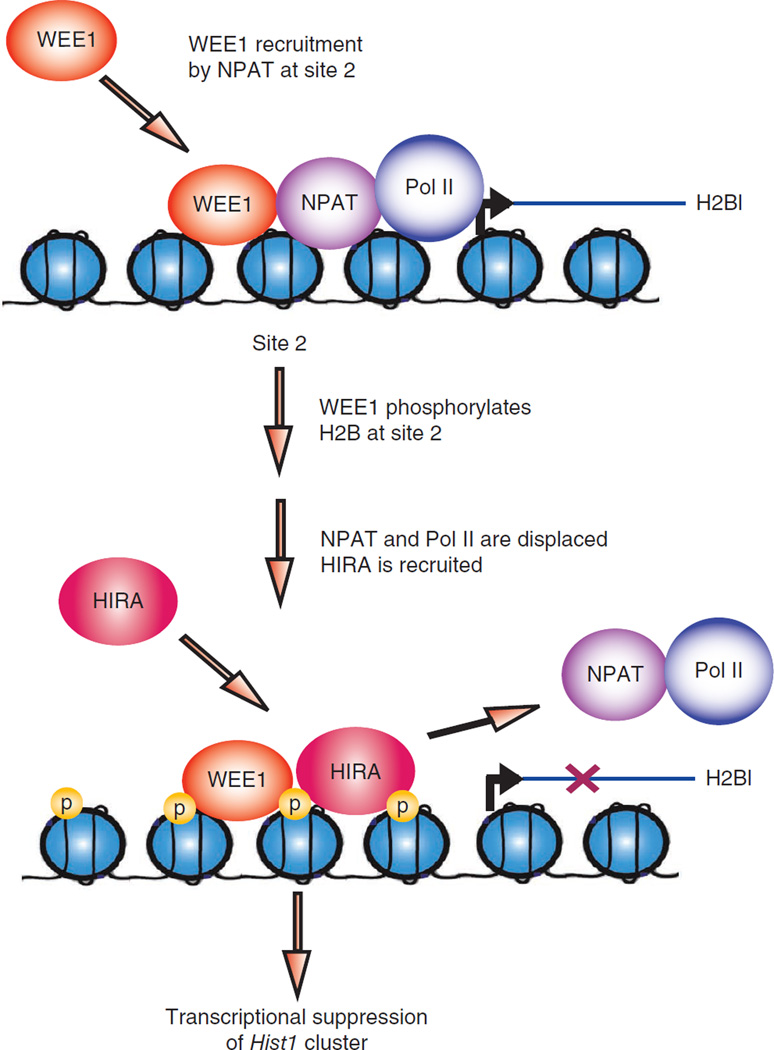
A model of H2B Tyr37 phosphorylation upstream of the major histone gene cluster Hist1 suppresses histone mRNA transcription. WEE1 phosphorylates H2B at Tyr37 upstream of histone cluster 1, Hist1. NPAT is excluded from binding to the Hist1 cluster because of its inability to recognize Tyr37-phosphorylated H2B, which in turn inhibits RNA polymerase II (Pol II) recruitment. Subsequently, HIRA is recruited to Tyr37-phosphorylated H2B, preventing NPAT rebinding and effectively resulting in the suppression of histone mRNA synthesis.
The signaling mechanisms and effectors of most tyrosine kinases are well understood, but the majority of these effectors are transmembrane or cytoplasmic proteins. How tyrosine kinases regulate gene expression by directly modulating chromatin is not fully understood. So far there have been three reports of histone tyrosine phosphorylation, one in the variant histone H2AX and two in the core histone H3 (ref. 12). The N-terminal domain of WSTF (Williams-Beuren syndrome transcription factor) phosphorylates H2AX at Tyr142, which was shown to be crucial for the DNA damage response49, apoptosis and survival50. JAK2, a nonreceptor tyrosine kinase, has been shown to phosphorylate histone H3 at Tyr41, preventing the binding of heterochromatin protein 1α51. Histone H3 was also shown to be phosphorylated at Tyr99 in yeast, and H3 phosphorylation at Tyr99 is crucial for H3 turnover by ubiquitylation and degradation52.
In our quest to identify the kinase that could robustly phosphorylate H2B in vivo, we tested a large number of mammalian kinases, both membrane bound and nonreceptor type, but none of them could robustly phosphorylate H2B in vivo. Our MEF data revealed that H2B Tyr37 phosphorylation is highly regulated by the cell cycle (Figs. 1a and 3a,b). In our studies with the yeast Tyr40 mutant, we observed that it had a significantly higher histone mRNA level than WT yeast. These two observations together gave an important clue: there is one well studied tyrosine kinase in S. cerevisiae, SWE1, that is activated in a cell cycle–dependent manner. Therefore, SWE1 (and, hence, WEE1) could be the candidate kinase for H2B tyrosine phosphorylation. Subsequent experiments confirmed the role of WEE1 and SWE1 in mammalian and yeast histone mRNA transcription, respectively.
When we assessed the HTA2-HTB2 gene pair in the swe1Δ mutant, it also showed a robustly higher mRNA level than WT yeast (Fig. 4). HIR-mediated repression of transcription relies on a specific DNA sequence: the negative regulatory element (NEG), which is found in the promoters of the three HIR-regulated gene pairs but is absent in HTA2-HTB2 (refs. 8,38). There could be two possible scenarios for transcriptional increase of the HTA2-HTB2 gene pair: it has been shown that that constitutive expression of Hir1 and Hir2 can bypass the requirement for the NEG site, leading to suppression of the transcription of HTA2-HTB2 (ref. 44). Based on our data showing HIRA binding to pTyr37 H2B in mammalian cells (Fig. 5g,h), it is not too farfetched to think that H2B Tyr40 phosphorylation could recruit HIR1 and HIR2 protein efficiently at upstream activation elements in the HTA2-HTB2 promoter, resulting in transcriptional suppression. Another potential scenario could be the involvement of Spt10 and Spt21 proteins, deletion of which reduces the levels of all core histone gene transcripts to some degree53,54. Spt10 in complex with Spt21 binds to the HTA2-HTB2 promoter during the S phase. At the end of the S phase, the Spt10–Spt21 complex is displaced from the HTA2-HTB2 promoter to shut down mRNA synthesis. The mechanism of the displacement of the Spt10–Spt21 complex from the HTA2-HTB2 promoter is not fully understood. We anticipate that H2B Tyr40 phosphorylation in the HTA2-HTB2 promoter has a role in this displacement, similar to the way the NPAT and RNA polymerase II complex is displaced because of H2B Tyr37 phosphorylation at site 2 in mammalian cells (Fig. 6).
Overall, whether in mammals or in yeast, H2B Tyr37 phosphorylation seems to be vital to cellular homeostasis; in combination with histone mRNA turnover, repression of histone transcription could efficiently and rapidly lower the amount of transcript, eliminating overload of the core histones after DNA synthesis. To our knowledge, this report provides the first demonstration of tyrosine phosphorylation of H2B and the elucidation of its role in maintaining histone mRNA levels in eukaryotic cells. Further, this work also uncovers a previously unknown function of WEE1, a cell-cycle regulator that has a dual role as an epigenetic modifier that also maintains the amount of histone transcripts.
ONLINE METHODS
Mass spectrometric identification of H2B tyrosine phosphorylation sites
The purified histones were subjected to SDS-PAGE electrophoresis and stained with Coomassie Brilliant Blue-R250 (Bio-Rad), and a band of ~14 kDa was excised. Trypsin in-gel digestion was performed, and the extracted peptides were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). A nanoflow liquid chromatograph coupled to an electrospray ion trap mass spectrometer (LTQ-Orbitrap) was used for MS/MS peptide-sequencing experiments. Five tandem mass spectra were collected in a data-dependent manner after each survey scan. The MS scans were performed in Orbitrap to obtain accurate peptide mass measurement, and the MS/MS scans were performed a in linear ion trap using a 60-s exclusion for previously sampled peptide peaks. Sequest55 and Mascot56 searches were performed against the Swiss-Prot human database.
Generation and affinity purification of the pTyr37 H2B antibody
Two H2B peptides coupled to immunogenic carrier proteins were synthesized as shown below, and pTyr37 H2B antibodies were custom synthesized by 21st Century Biochemicals, MA. Ahx is aminohexanoic acid, which is six-carbon spacer used to distance the carrier protein KLH from immunogen.
Phosphopeptide: acetyl-KRSRKES[pY]SVYVYKVL-Ahx-C-amide
Nonphosphopeptide: acetyl-KRSRKESYSVYVYKVL-Ahx-C-amide
Two rabbits were immunized twice with the phosphopeptide, and the sera from these rabbits was affinity purified. Two antigen-affinity columns were used to purify the phospho-specific antibodies. The first column was the nonphosphopeptide affinity column. Antibodies recognizing the unphosphorylated residues of the peptide bound to the column. The flow-through fraction was collected and then applied to the second column, the phosphopeptide column. Antibodies recognizing the phospho residue bound to the column and were eluted as phospho-specific antibodies. The monoclonal pTyr37 H2B antibody expressing hybridoma was custom generated by ProMab Biotechnologies, CA.
Cell synchronization using a double thymidine block
MEFs or H1975 cells (obtained from American Type Culture Collection) were grown to 60–70% confluency. Thymidine was added at a final concentration of 2 mM, and the cells were incubated for 17 h. Cells were then washed, and fresh serum-containing medium was added. After 10 h, thymidine was added again, and the cells were incubated for 17 h; cells were then washed, replaced with fresh serum-containing medium, and data were collected at the indicated time points.
Native ChIP sequencing and analysis
Synchronized MEFs were harvested at indicated time points after thymidine release (5 × 107 cells). Cells pellets were resuspended in receptor lysis buffer57,58 and sonicated for 25 s to shear the DNA to an average length of 300–500 bp. The soluble chromatin was incubated overnight at 4 °C with pTyr37 H2B antibody and 20 µl of protein-G magnetic beads (Active motif). The complexes were washed with ChIP buffer 1 and 2 (Active Motif) and eluted with elution buffer. Immunoprecipitated DNA was subjected to proteinase K treatment for 2 h. Chromatin immunoprecipitated DNAs were subjected to sequencing. The sequencing yield was very good, with almost 40 million reads in each sample, of which 27.7 and 23.6 million for samples at 6.5 h and 0 h, respectively, mapped uniquely to the mouse mm9 genome.
Sequence analysis
The 36-nt sequence reads (‘tags’) identified by the Sequencing Service (using Illumina’s Genome Analyzer 2) were mapped to the genome using the ELAND algorithm. Alignment information for each tag was stored in the output file *_export.txt. Only tags that mapped uniquely, had no more than two mismatches and that passed quality control filtering were used in the subsequent analysis.
Determination of fragment density
Because the 5′ ends of the sequence tags represent the end of the ChIP or immunoprecipitation fragments, the tags were extended in silico (using Active Motif software) at their 3′ ends to a length of 110–200 bp, depending on the average fragment length in the size-selected library. This information is stored in a BAR (Binary Analysis Results) file that can be viewed in a browser such as Affymetrix’ Integrated Genome Browser (IGB).
Interval analysis (‘peak finding’)
An ‘interval’ is a discrete genomic region defined by the chromosome number and a start and end coordinate. For each BAR file, intervals are calculated and compiled into BED (Browser Extensible Data) files. A typical threshold setting is in the range of 10–20 but may be adjusted depending on the number of tags sequenced or based on the information on positive and negative test sites, independent estimates for the false discovery rate and/or the intent to generate a stringent or relaxed analysis. For an interval to be called, it must contain three consecutive bins with fragment densities greater than the threshold.
Alternative and/or optional analysis steps
(i) Tag normalization: when samples had uneven tag counts, the tag numbers of all the samples were truncated to the number of tags present in the smallest sample. (ii) False peak filtering: input or IgG control samples (which represent false peaks) were used to remove corresponding intervals in ChIP samples or to mark them as probable false positives.
Active-region analysis
To compare peak metrics between two or more samples, overlapping intervals are grouped into ‘active regions’, which are defined by the start coordinate of the most upstream interval and the end coordinate of the most downstream interval (equal to the union of overlapping intervals).
In vitro kinase assay
For the in vitro kinase assay, 340 ng of purified glutathione S-transferase (GST)-WEE1 (Invitrogen) and 1 µg of H2A, H2B, H3 or H4 (NEB) were incubated in the presence or absence of the WEE1 inhibitor MK-1775 (0.625 µM) in WEE1 kinase assay buffer containing 50 mM HEPES (pH 7.5), 15 mM MgCl2, 1 mM EGTA, 10% glycerol, 10 mM DTT and 0.1 mM ATP at 30 °C. After 60 min, the reaction was separated by SDS-PAGE (18%), followed by immunoblottings with pTyr37 H2B, total H2B, H2A, H3, H4 and WEE1 antibodies.
Pull-down assay
Two human histone H2B peptides spanning amino acids 25–49 were synthesized with Tyr37 at middle of the peptide. The sequences were as follows:
H2B25–49: DGKKRKRSRKESYSVYVYKVLKQVH
pTyr37 H2B25–49: DGKKRKRSRKESpYSVYVYKVLKQVH
Both the peptides were biotinylated at the C terminus and immobilized on streptavidin-Sepharose beads. The beads were incubated with HEK 293 cell lysates made in Tris glysine NaCl (TGN) buffer containing 50 mmol/l Tris (pH 7.5), 50 mmol/l glycine, 150 mmol/l NaCl, 1% Triton X-100, 10% glycerol, phosphatase inhibitors (10 mmol/l NaF and 1 mmol/l Na2VO4) and protease inhibitor mix (Roche). The beads were extensively washed with TGN buffer, and bound NPAT or HIRA was resolved by SDS-PAGE, followed by immunoblotting with the respective antibodies. Equal loading of peptide was determined by Coomassie blue staining.
Filter-binding assay
NPAT binding to unphosphorylated H2B was confirmed by filter-binding assay. Two concentrations of H2B25–49 or pTyr37 H2B25–49 peptides were spotted on nitrocellulose membrane, which was incubated with HEK 293 cell lysates prepared in TGN buffer. The blot was washed extensively, followed by immunoblotting with NPAT antibodies.
Yeast strains, culture conditions and cell synchronization
The yeast strains used in this study were pp30-Swe1HA (SWE1HA6-HIS3MX6) and pp30-swe1Δ (SWE1HIS3MX6), which were obtained from M. Mollapour35. WT (MATa (hta1-htb1)Δ::LEU2, (hta2-htb2)Δ::TRP1, his3Δ200 leu2Δ1 ura3-52 trp1Δ63 lys2-128Δ<pZS145-HTA1-Flag-HTB1-HIS3>) and H2B Tyr40A mutant of S. cerevisiae were obtained from A. Shilatifard. Yeast cells were grown in YPD medium at 30 °C for 24 h. The culture was diluted 1:20 and grown for 1 h, after which α-factor was added for 3 h. Cells were harvested by centrifugation (5,000 r.p.m. for 5 min), washed with sterile water and resuspended in fresh medium. Cells were harvested at different time points.
Yeast RNA isolation
S. cerevisiae cells were grown and spheroplasts were obtained as described above. The RNAs were isolated using YeaStar RNA kit as per the manufacturer’s protocol (Zymo Research). To obtain ultra-clean RNA that was DNA free, RNAs were passed through Fast-Spin Columns (DNA-Free RNA kit) as per the manufacturer’s protocol (Zymo Research). RNAs were quantified and used for qRT-PCR.
Bioinformatics analysis
Using the MEME program (http://meme.sdsc.edu/meme/intro.html), we found 20 unique motifs in our ChIP data. Seven motifs out of these 20 were considered significant by their E values, which are shown in Supplementary Table 4. The motifs with high GC-rich contents were considered as the potential transcription-factor–binding sites, and the motifs with AT-rich contents are considered as potential histone-binding sites. Thus, motifs 1, 2 and 7 are potential histone-binding sites and are abundant in all the sequences that we identified in ChIP dataset. The motifs 3, 4 and 5 are potential transcription factor–binding sites, and, based on highest E value, we used motif 3 and the MAST program to search the entire ChIP dataset to see where it is present. The presence of motif 3 at sites 1 and 2 is shown in Supplementary Figure 5a.
FIMO was used to map the 20 motifs we found in MEME onto 3,524 sequences of interest. Because for each motif, one sequence can have more than one mapping position, the number of sequences that have at least one mapping and the number of sequences that have only one mapping are listed for each motif. The FIMO analysis of all the 20 motifs is shown in Supplementary Table 5. We then screened for the nearest genes, which are adjacent to ChIP sequencing peaks and contain motif 3. This analysis revealed 511 genes that could potentially be regulated by H2B Tyr37 phosphorylation, which are listed in Supplementary Table 6. Moreover, using MetaCore (GeneGo Inc.), we ran several enrichment analyses to cluster genes that are adjacent to ChIP sequencing peaks. This enrichment analysis clustered the genes based on their functional role (http://www.geneontology.org/GO.doc.shtml). The Gene Ontology molecular function analysis is shown in Supplementary Figure 5b.
Supplementary Material
Acknowledgments
We thank A. Shilatifard (Stowers Institute) for WT and the Y40A mutant of S. cerevisiae, M. Mollapour (US National Institutes of Health) for WT and the swe1Δ mutant of S. cerevisiae and G. Enders (Fox Chase Cancer Center) for myc-tagged WEE1 constructs. We thank X. Qu for bioinformatics analysis; L. Hall and J. Repass for qRT-PCR analysis; K. Shapland and J. Kroeger for flow cytometry; and E. Seto for critical reading of the manuscript. We thank the Moffitt Flow Cytometry, Molecular Biology core facilities, Lung Cancer Spore and the Comprehensive Melanoma Research Center. The project was supported by a Moffitt Support grant to N.P.M.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
K.M. and N.P.M. conceived the idea and designed all the experiments. K.M. and N.P.M. performed all the experiments except the mass spectrometric identification of H2B Tyr37 phosphorylation, which was performed by B.F. and J.M.K. K.M. and N.P.M. analyzed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- 2.Heintz N, Sive HL, Roeder RG. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol. Cell. Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osley MA. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 4.Borun TW, Gabrielli F, Ajiro K, Zweidler A, Baglioni C. Further evidence of transcriptional and translational control of histone messenger RNA during the HeLa S3 cycle. Cell. 1975;4:59–67. doi: 10.1016/0092-8674(75)90134-8. [DOI] [PubMed] [Google Scholar]
- 5.Hereford L, Bromley S, Osley MA. Periodic transcription of yeast histone genes. Cell. 1982;30:305–310. doi: 10.1016/0092-8674(82)90036-8. [DOI] [PubMed] [Google Scholar]
- 6.Hereford LM, Osley MA, Ludwig TR, II, McLaughlin CS. Cell-cycle regulation of yeast histone mRNA. Cell. 1981;24:367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- 7.Osley MA, Hereford L. Identification of a sequence responsible for periodic synthesis of yeast histone 2A mRNA. Proc. Natl. Acad. Sci. USA. 1982;79:7689–7693. doi: 10.1073/pnas.79.24.7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osley MA, Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takayama Y, et al. Hsk1- and SCF(Pof3)-dependent proteolysis of S. pombe Ams2 ensures histone homeostasis and centromere function. Dev. Cell. 2010;18:385–396. doi: 10.1016/j.devcel.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto S, Yanagida M, Nurse P. Histone transcription in cell cycle mutants of fission yeast. EMBO J. 1987;6:1093–1097. doi: 10.1002/j.1460-2075.1987.tb04863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh RK, Gunjan A. Histone tyrosine phosphorylation comes of age. Epigenetics. 2011;6:153–160. doi: 10.4161/epi.6.2.13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger SL. Cell signaling and transcriptional regulation via histone phosphorylation. Cold Spring Harb. Symp. Quant. Biol. 2010;75:23–26. doi: 10.1101/sqb.2010.75.044. [DOI] [PubMed] [Google Scholar]
- 14.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 15.Featherstone C, Russell P. Fission yeast p107wee1 mitotic inhibitor is a tyrosine/serine kinase. Nature. 1991;349:808–811. doi: 10.1038/349808a0. [DOI] [PubMed] [Google Scholar]
- 16.McGowan CH, Russell P. Cell cycle regulation of human WEE1. EMBO J. 1995;14:2166–2175. doi: 10.1002/j.1460-2075.1995.tb07210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundgren K, et al. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- 18.Hirai H, et al. Small-molecule inhibition of Wee1 kinase by MK–1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- 19.Mahajan K, et al. Ack1 mediated AKT/PKB tyrosine 176 phosphorylation regulates its activation. PLoS ONE. 2010;5:e9646. doi: 10.1371/journal.pone.0009646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum. Genet. 1997;101:284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- 22.Albig W, Kioschis P, Poustka A, Meergans K, Doenecke D. Human histone gene organization: nonregular arrangement within a large cluster. Genomics. 1997;40:314–322. doi: 10.1006/geno.1996.4592. [DOI] [PubMed] [Google Scholar]
- 23.Wang ZF, et al. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1 Mb. Genome Res. 1996;6:688–701. doi: 10.1101/gr.6.8.688. [DOI] [PubMed] [Google Scholar]
- 24.Mahajan K, et al. Effect of Ack1 tyrosine kinase inhibitor on ligand-independent androgen receptor activity. Prostate. 2010;70:1274–1285. doi: 10.1002/pros.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahajan K, et al. Ack1-mediated androgen receptor phosphorylation modulates radiation resistance in castration-resistant prostate cancer. J. Biol. Chem. 2012;287:22112–22122. doi: 10.1074/jbc.M112.357384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 28.Laribee RN, Fuchs SM, Strahl BD. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes Dev. 2007;21:737–743. doi: 10.1101/gad.1541507. [DOI] [PubMed] [Google Scholar]
- 29.Bungard D, et al. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 31.Gardner KE, Zhou L, Parra MA, Chen X, Strahl BD. Identification of lysine 37 of histone H2B as a novel site of methylation. PLoS ONE. 2011;6:e16244. doi: 10.1371/journal.pone.0016244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakanishi S, et al. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mollapour M, et al. Swe1Wee1-dependent tyrosine phosphorylation of Hsp90 regulates distinct facets of chaperone function. Mol. Cell. 2010;37:333–343. doi: 10.1016/j.molcel.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y, Jin J, Harper JW. The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain. Mol. Cell. Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 38.Osley MA, Gould J, Kim S, Kane MY, Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986;45:537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- 39.Xu H, Kim UJ, Schuster T, Grunstein M. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prochasson P, Florens L, Swanson SK, Washburn MP, Workman JL. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 2005;19:2534–2539. doi: 10.1101/gad.1341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green EM, et al. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 2005;15:2044–2049. doi: 10.1016/j.cub.2005.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol. Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 43.Sherwood PW, Tsang SV, Osley MA. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spector MS, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fillingham J, et al. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell. 2009;35:340–351. doi: 10.1016/j.molcel.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 46.Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamane K, et al. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol. Cell. 2011;41:56–66. doi: 10.1016/j.molcel.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nelson DM, et al. Coupling of DNA synthesis and histone synthesis in S phase independent of cyclin/cdk2 activity. Mol. Cell. Biol. 2002;22:7459–7472. doi: 10.1128/MCB.22.21.7459-7472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao A, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cook PJ, et al. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dawson MA, et al. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dollard C, Ricupero-Hovasse SL, Natsoulis G, Boeke JD, Winston F. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:5223–5228. doi: 10.1128/mcb.14.8.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hess D, Liu B, Roan NR, Sternglanz R, Winston F. Spt10-dependent transcriptional activation in Saccharomyces cerevisiae requires both the Spt10 acetyltransferase domain and Spt21. Mol. Cell. Biol. 2004;24:135–143. doi: 10.1128/MCB.24.1.135-143.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yates JR, III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 56.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 57.Mahajan NP, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc. Natl. Acad. Sci. USA. 2007;104:8438–8443. doi: 10.1073/pnas.0700420104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mahajan NP, Whang YE, Mohler JL, Earp HS. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res. 2005;65:10514–10523. doi: 10.1158/0008-5472.CAN-05-1127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.