Abstract
Background
The Caldicellulosiruptor bescii genome encodes a potent set of carbohydrate-active enzymes (CAZymes), found primarily as multi-domain enzymes that exhibit high cellulolytic and hemicellulolytic activity on and allow utilization of a broad range of substrates, including plant biomass without conventional pretreatment. CelA, the most abundant cellulase in the C. bescii secretome, uniquely combines a GH9 endoglucanase and a GH48 exoglucanase in one protein. The most effective commercial enzyme cocktails used in vitro to pretreat biomass are derived from fungal cellulases (cellobiohydrolases, endoglucanases and a β-d-glucosidases) that act synergistically to release sugars for microbial conversion. The C. bescii genome contains six GH5 domains in five different open reading frames. Four exist in multi-domain proteins and two as single catalytic domains. E1 is a GH5 endoglucanase reported to have high specific activity and simple architecture and is active at the growth temperature of C. bescii. E1 is an endo-1,4-β-glucanase linked to a family 2 carbohydrate-binding module shown to bind primarily to cellulosic substrates. We tested if the addition of this protein to the C. bescii secretome would improve its cellulolytic activity.
Results
In vitro analysis of E1 and CelA shows synergistic interaction. The E1 gene from Acidothermus cellulolyticus was cloned and expressed in C. bescii under the transcriptional control of the C. bescii S-layer promoter, and secretion was directed by the addition of the C. bescii CelA signal peptide sequence. The vector was integrated into the C. bescii chromosome at a site previously showing no detectable detrimental consequence. Increased activity of the secretome of the strain containing E1 was observed on both carboxymethylcellulose (CMC) and Avicel. Activity against CMC increased on average 10.8 % at 65 °C and 12.6 % at 75 °C. Activity against Avicel increased on average 17.5 % at 65 °C and 16.4 % at 75 °C.
Conclusions
Expression and secretion of E1 in C. bescii enhanced the cellulolytic ability of its secretome. These data agree with in vitro evidence that E1 acts synergistically with CelA to digest cellulose and offer the possibility of engineering additional enzymes for improved biomass deconstruction with the knowledge that C. bescii can express a gene from Acidothermus, and perhaps other heterologous genes, effectively.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-015-0296-x) contains supplementary material, which is available to authorized users.
Background
The ability to deconstruct plant biomass without conventional pretreatment is an important attribute for any organism being considered for the consolidated bioprocessing of plant biomass to fuels and chemicals. Existing methods for biomass pretreatment typically rely on physical, chemical and enzymatic hydrolysis, and the cost of enzyme cocktails is a major economic barrier to the use of plant biomass as a substrate [1–4]. The most effective commercial cocktails of enzymes used in vitro to pretreat biomass contain cellobiohydrolase I (CBH I), cellobiohydrolase II (CBH II), β-d-glucosidase and endoglucanase I (EG I) that act synergistically to release sugars for microbial conversion to products [1, 4]. In nature, microbes produce enzymes in structures like the Clostridium thermocellum cellulosome [5–8] or free enzymes that act independently, such as those produced by most fungi and cellulolytic bacteria [9]. Caldicellulosiruptor species contain a variety of enzymes predicted to be involved in plant biomass deconstruction, and the synergistic activities of these enzymes are likely responsible for their ability to utilize plant biomass without conventional pretreatment [10, 11]. The C. bescii genome contains 52 annotated glycoside hydrolases; of these CelA is the most abundant protein in the secretome [12] and the only enzyme to combine both a GH9 and GH48 catalytic domain [13]. Deletion of CelA in the C. bescii genome, in fact, resulted in a significant reduction of its ability to deconstruct plant biomass likely because of the reduced exoglucanase activity (GH48 activity) in the mutant [14]. Cellulolytic microorganisms in nature possess high endoglucanase activity conferred by one or two highly active GH9 or GH5 endoglucanases [15, 16]. The C. bescii genome contains six GH5 endoglucanases. Three similar GH5 domains are in the same cluster annotated as CbMan5A, ManB, and CbMan5C [17]. Four of the six GH5 domains are present as a part of multi-domain enzymes (one each in Cbes1859 and Cbes1865, and two domains in Cbes1866; Additional file 1: Table S1). Two are present as singular domains and found outside of the cluster (Cbes0234 and Cbes0594; Additional file 1: Table S1). Several of these genes are upregulated on switchgrass [18]: Cbes0234 (GH5 only), 17-fold; Cbes1865 (GH9–CBM–CBM–CBM–GH5), 23-fold; Cbes1866 (GH5–CBM–CBM–CBM–GH5), 7-fold. Thus, most of the GH5 endoglucanases in C. bescii are multi-domain proteins and that, given the accepted mode of action of endoglucanases, could potentially limit their contribution when compared to highly active and smaller endoglucanases [19]. For example, biochemical and mutational analyses of Cbes1865 (GH9–CBM–CBM–CBM–GH5) showed that after deletion of the GH9 module, the truncated protein had an increased apparent Kcat value 2- to 3-fold higher on several mannan substrates, including locust bean gum, guar gum, and konjac glucomannan [20]. The potent GH5 family endo-1,4-β-glucanase from A. cellulolyticus 11B, is different from those found in C. bescii. E1 is linked to a family 2 CBM instead of a family 3 CBM and contains a single catalytic domain [21, 22]. This structure might allow a versatility prevented by the large, multi-modular enzymes found in abundance in C. bescii. For instance, E1 may have more accessibility to the substrate within the cavities formed by enzymes such as CelA [23]. We also note that the C. bescii genome does not contain a CBM2. In addition, the linker region between the GH5 domain and the CBM2 of E1 is different from those in C. bescii in terms of sequence homology and amino acid composition. In C. bescii, only one GH5 domain, the C-terminus portion of Cbes1866 (CelB), exhibits high amino acid sequence homology (98 % of sequence query cover and 35 % sequence identity) with the catalytic domain of E1. The low sequence homology of the other GH5 domains in C. bescii (Additional file 1: Table S1) suggests they probably contain catalytic activities differing from E1 making it an ideal candidate to supplement the GH5 containing enzymes in the C. bescii secretome.
There are also several potential advantages to expressing E1 in C. bescii to improve its performance in biomass deconstruction. Cultures can be grown at higher temperatures matching the optimum cellulolytic activities of thermostable E1 (Topt about 81 °C) [21, 22]. E1 is active over a broad range of pH and still quite active at a low pH (~pH 5.5) [21, 22], which is similar to the optimum pH range for CelA and covers the spectrum of pH changes of media acidification during C. bescii growth (pH drop from pH 7.2 to ~5.0) [24]. The resistance of engineered E1 to feedback inhibition by cellobiose has been reported [25] which may be important for C. bescii cellulolytic activity. Moreover, no extracellular β-glucosidase has been identified in the C. bescii genome. While C. bescii does contain a β-glucosidase (Cbes0458), it does not contain an identifiable signal sequence.
Here we report synergistic activity between E1 and CelA in vitro and in vivo. The E1 gene from A. cellulolyticus was cloned and expressed in C. bescii and the secretome of the resulting strain showed increased activity on both CMC and Avicel. These data suggest that while extremely effective, the ability of the C. bescii secretome to deconstruct plant biomass may be improved with the addition of key enzymes.
Results and discussion
Synergy between E1 and CelA, the primary exoglucanase in the C. bescii secretome, in vitro
For action on crystalline cellulose, cellulase systems are typically weak in exoglucanase activity, not endoglucanase activity [26]. Endoglucanases are needed catalytically and usually account for no more than ~20 % of most cellulase secretomes. EI is more than a typical endoglucanase, because it has some processive qualities. To investigate the possibility that the C. bescii CelA and the A. cellulolyticus E1 enzymes might act synergistically in vitro, we tested both CelA and E1 separately on Avicel at an enzyme loading of 15 and 4 mg/g, respectively. In the absence of an exoglucanase, E1, converts only 8 % of the Avicel in 7 days. CelA alone converts 51 % (Fig. 1). The combination of CelA with E1 at the same total enzyme loading as CelA alone, allows the conversion of more than 69 % of the Avicel over the course of 7 days (Fig. 1). This represents an improvement of more than 10 % that can be directly attributed to synergy between the two enzymes in vitro.
Fig. 1.
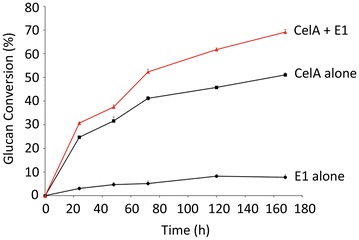
Synergistic effect of purified C. bescii CelA and A. cellulolyticus E1 enzymes in vitro. Avicel hydrolysis by C. bescii CelA (black squares) at an enzyme loading of 15 mg/g, A. cellulolyticus E1 (black dots) at an enzyme loading of 4 mg/g, and a mixture of C. bescii CelA and A. cellulolyticus E1 (red triangles) at an enzyme loading of 11 and 4 mg/g, respectively.
Heterologous expression and secretion of the E1 protein from A. cellulolyticus in C. bescii
To express full length E1 protein (Acel 0614) from A. cellulolyticus (Fig. 2a) in C. bescii, the gene was amplified by PCR from A. cellulolyticus chromosomal DNA and cloned into an integrational expression vector, pDCW175 (Fig. 2b, Additional file 1: Figure S1) under the transcriptional control of the C. bescii S-layer promoter [27]. The C. bescii CelA signal peptide sequence was added upstream of the GH5 domain of the E1 protein, and a C-terminal histidine-tag was added to facilitate future protein purification. The signal peptide derived from the C. bescii celA gene was used for secretion of E1. CelA is the most abundant extracellular protein produced by C. bescii [11, 19], suggesting that its signal sequence works well. We recently demonstrated the use of this signal peptide for the expression of full length CelA in C. bescii [28]. The native E1 signal sequence shows no sequence homology with any signal peptides in C. bescii. The CelA signal sequence was fused to the 5′ end of the coding sequence of the E1 catalytic domain, replacing the E1 signal peptide, to create the E1 expression/secretion cassette in pDCW175.
Fig. 2.
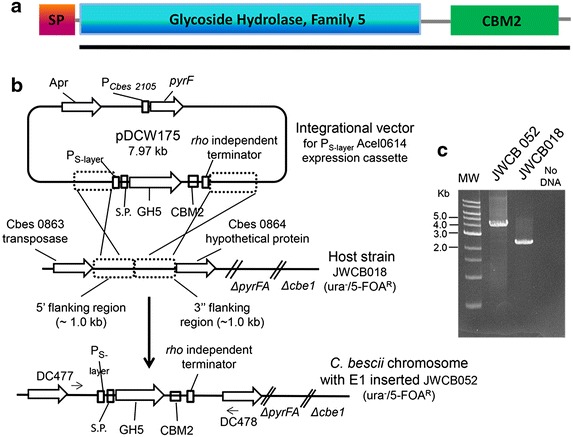
Chromosomal integration of the E1 gene into the C. bescii genome. a A diagram of native E1 protein: SP, signal peptide; a family 5 endoglucanase; and CBM2, a family 2 carbohydrate-binding module/domain. The black bar beneath the diagram represents the portion of the pDCW175 construct derived from Acel_0614. b A depiction of the chromosomal location and integration event of the E1 expression cassette/secretion cassette. c Agarose gel showing PCR products amplified using primers DC477 and DC478 annealing to regions outside the site of integration in the newly constructed strain JWCB052 ΔpyrFA ldh::ISCbe4 Δcbe1::PS-layer acel0614 E1 containing the E1 expression cassette, 4.079 kb (lane 2) and the parent strain, JWCB018 ΔpyrFA ldh::ISCbe4 Δcbe1, 2.440 kb (lane 3); DNA MW standards (lane 1); no template PCR control (lane 4).
This plasmid, that contained a wild-type copy of the C. bescii pyrF gene, was transformed into JWCB018, a strain containing a deletion of pyrF resulting in uracil auxotrophy, and transformants were selected for uracil prototrophy (Fig. 2b; Table 1) [29, 30]. JWCB018 also contains a deletion of CbeI, a restriction endonuclease, to facilitate transformation of DNA from E. coli [30, 31]. The plasmid was targeted to integrate into an inter-cistronic region on the C. bescii genome previously determined to be available without affecting growth or resulting in a detectable phenotype [27]. Transformants containing an integrated vector were then treated with 5-FOA to select for excision of the plasmid sequences leaving the EI expression cassette in the chromosome. Integration of the plasmid into the targeted site in the C. bescii chromosome as well as the presence of the PS-layeracel0614 (E1) cassette after strain construction was confirmed by PCR amplification of the relevant chromosome regions (Fig. 2c) as well as sequencing of the PCR products. The resulting strain JWCB052 (ΔpyrFA ldh::ISCbe4 Δcbe1 PS-layerE1) was used for further analysis.
Table 1.
Strains and plasmids used in this work
| Strains/plasmids | Strain and genotype/phenotype | References |
|---|---|---|
| C. bescii | ||
| JWCB001 | Wild type (ura +/5-FOAS) | DSMZa |
| JWCB018 | ΔpyrFA ldh::ISCbe4 Δcbe1 (ura −/5-FOAR) | [29] |
| JWCB052 | ΔpyrFA ldh::ISCbe4 Δcbe1::PS-layer acel0614 (E1) b (ura -/5-FOAR) | This study |
| Escherichia coli | ||
| JW314 | DH5α containing pDCW144 (ApramycinR) | [27] |
| JW336 | DH5α containing pDCW174 (ApramycinR) | This study |
| JW337 | DH5α containing pDCW175 (ApramycinR) | This study |
| Plasmids | ||
| pDCW144 | Integrational vector for Caldicellulosiruptor bescii (ApramycinR) | [27] |
| pDCW174 | Integrational vector containing signal peptide of CelA for Caldicellulosiruptor bescii (ApramycinR) | This study |
| pDCW175 | Integrational vector containing the PS-layer acel0614 (E1) b expression cassette (ApramycinR) | This study |
a German Collection of Microorganisms and Cell Cultures.
b acel0614 (E1) [glycoside hydrolase (family 5) with cellulose-binding (family 2) derived from Acidothermus cellulolyticus 11B].
Expression and secretion of E1 by C. bescii was confirmed by Western blot analysis using a mouse anti-E1 monoclonal antibody to detect the EI protein from the concentrated supernatant of a verified transformant (JWCB052). Concentrated culture supernatants from the E1 transformant (JWCB052) grown at varying temperatures (65, 70, and 75 °C) were electrophoresed on a 4–15 % gradient Mini-Protean TGX gel (BIO-RAD) and transferred to a PVDF membrane and developed with the E1 antibody. The positive control for detection of E1 protein was a truncated fragment of the E1 catalytic domain (40 kDa) produced in Streptomyces lividans (Fig. 3). A ~60 kDa band was visible in the culture supernatants corresponding to the predicted size for full length E1 protein (Fig. 3). These data confirm expression and extracellular localization of E1 in C. bescii.
Fig. 3.
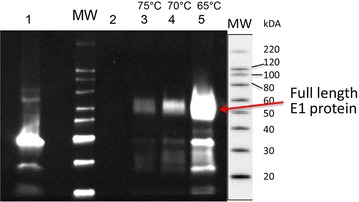
Confirmation of E1 expression and secretion in C. bescii using Western blot analysis. Concentrated extracellular proteins (10 µg) were electrophoresed in a 15 % gradient Mini-Protean TGX gel (BIO-RAD) and electro-transferred to a PVDF membrane (ImmobilonTM-P; Millipore). The membrane was then probed with an E1 monoclonal antibody. Truncated version of E1 produced in Streptomyces lividans (lane 1); parent strain JWCB018 ΔpyrFA ldh::ISCbe4 Δcbe1 grown at 75 °C (lane 2); E1 expression strain JWCB052 ΔpyrFA ldh::ISCbe4 Δcbe1::P S-layer acel0614 (E1) grown at 75 °C (lane 3), 70 °C (lane 4), and 65 °C (lane 5); MW, MagicMark™ molecular weight marker (Invitrogen).
As shown in Fig. 3, the most abundant amount of extracellular E1 protein was detected in cells grown at 65 °C, with lesser amounts from cells grown at either 70 or 75 °C. E1 originates from A. cellulolyticus, which has an optimum growth temperature of 55 °C, whereas C. bescii has an optimum growth temperature of 78 °C. There are a number of issues with the heterologous expression and secretion of proteins from mesophilic sources in thermophiles, like C. bescii. In addition to messenger RNA instability and post-translational proteolysis by host proteases, protein stability at high temperature is often a problem. In addition, there is severe codon usage bias due to differences in the GC content of the E1 gene (61 %) and the C. bescii genome (35 %) [32]. This bias can affect E1 expression due to limitations in supply of certain tRNA species. Insufficient tRNA pools can lead to translational stalling and premature termination. We observed a ~30 % increase in doubling time at 65 °C (2.3 h) compared to 75 °C (1.6 h) (Fig. 4). Slower growth might help overcome the codon usage bias by slowing down transcription and translation rates to allow the incorporation of rare tRNAs which corresponds to our observed increase in protein at the lower temperature (Fig. 3).
Fig. 4.
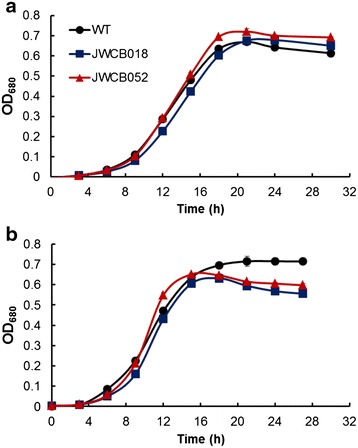
Growth of the wild type, parent strain (JWCB018), and E1 expression strain (JWCB052) on cellobiose. Growth of the wild type (black circles); JWCB018, ΔpyrFA ldh::ISCbe4 Δcbe1 (blue squares); and JWCB052, ΔpyrFA ldh::ISCbe4 Δcbe1::P S-layer acel0614 (E1) (red triangles) strains on cellobiose. a Growth at 65 °C and b growth at 75 °C.
The E1 endoglucanase has limited activity on the disaccharide, cellobiose [33], so to test whether or not expression of E1 in C. bescii resulted in a growth defect, growth of the wild type, the parent strain, and the E1 producing strain on cellobiose were compared. To examine the overall effect of E1 production and secretion on the growth of C. bescii, growth on cellobiose was monitored by OD680 at 65 and 75 °C for the E1 producing strain (JWCB052), the parent strain (JWCB018), and the wild-type strain (Fig. 4). Expression of E1 in C. bescii had no detectable effect on the growth of cells at either 65 or 75 °C (Fig. 4).
Expression of E1 in C. bescii results in enhanced activity of extracellular fractions on both Avicel and carboxymethylcellulose
To test if expression of E1 in C. bescii affected the cellulolytic activity of the extracellular enzyme fraction, enzyme assays were performed from concentrated culture supernatants of C. bescii cells producing E1 protein (JWCB052) compared to the C. bescii parent strain (JWCB018). Cells were grown at 65 °C as E1 was produced at the highest levels at that temperature (Fig. 3). Even though A. cellulolyticus grows optimally at the temperature of the hot spring from which it was collected, 55 °C [34], the E1 protein has been shown to be active at considerably higher temperatures, with an optimum of 81 °C (in vitro) [21, 22, 35]. Supernatants were then concentrated and exchanged with buffer before assaying on CMC for 1 h or on Avicel for 24 h at 65 and 75 °C. CMC is traditionally used as an assay for endoglucanase activity while Avicel is used to assay exoglucanase activity [36]. Increased activity of concentrated culture supernatants from the E1 expression strain (JWCB052) was observed on both CMC and Avicel, and at both assay temperatures (65 and 75 °C) compared to the parent strain (JWCB018). These data are based on two independent biological replicates (Fig. 5, Additional file 1: Figure S2). Activity on Avicel increased on average 17.5 % at 65 °C and 16.4 % at 75 °C (Fig. 5b, Additional file 1: Figure S2B), while activity on CMC increased on average 10.8 % at 65 °C and 12.6 % at 75 °C (Fig. 5a, Additional file 1: Figure S2A). The fact that there is a larger increase in exoglucanase activity (on Avicel) in the E1 containing strain than endoglucanase activity (on CMC) suggests that the addition of the E1 endoglucanase results in the generation of increased chain ends to improve cellulolytic activity. We conclude from these data that the endoglucanase activity of the C. bescii secretome, and likely CelA, is limiting in its activity on cellulose.
Fig. 5.
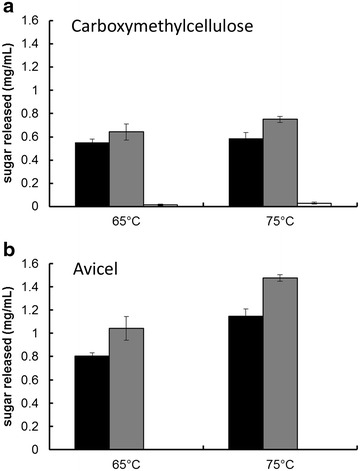
Relative quantification of enzymatic activity of the extracellular fraction of C. bescii expressing E1 (Acel0614) on Avicel and carboxymethylcellulose. Carboxymethylcellulose (CMC) or Avicel was used as substrate at either 65 or 75 °C. a Activity of the extracellular fraction (25 µg/mL concentrated protein) on CMC from the parent strain JWCB018 ΔpyrFA ldh::ISCbe4 Δcbe1 (black), the E1 expression strain, JWCB052 ΔpyrFA ldh::ISCbe4 Δcbe1::P S-layer acel0614 (E1) (grey), and no enzyme control (white). b Activity of the extracellular fraction (25 µg/mL of concentrated protein) on Avicel from the parent strain JWCB018 (black), the E1 expression strain, JWCB052 (grey), and no enzyme control (white).
Conclusions
The E1 and CelA enzymes act synergistically in vitro to digest Avicel despite the fact that CelA contains an endoglucanase activity, albeit a tethered one. This suggests that the ability to generate new cellulose chain ends may be rate limiting in cellulose hydrolysis and that the addition of the smaller E1 augments this activity resulting in increased activity. The increase in activity observed when E1 is added to the secretome of C. bescii is very promising. Despite an abundance of cellulolytic enzymes, including several endoglucanases in C. bescii, the addition of E1 showed a measurable difference in cellulolytic activity. Further modifications could lead to improvements for C. bescii as both a microorganism for consolidated bioprocessing and as an expression host for secretion of heterologous proteins especially those originating from thermophiles. The increase in protein yields at lower growth temperatures suggests that codon usage bias is likely an issue for expression of E1 in C. bescii. Codon optimization might well improve expression at higher temperatures and increase cellulolytic activity on biomass substrates. Opportunities also remain for improving the E1 protein itself. Linker regions within multi-domain enzymes like CelA have been implicated in protein stability, glycosylation, and flexibility for enzymatic activity. The linker region of E1 shows no homology to those found in the multi-domain proteins of C. bescii and swapping the linker region of E1 with ones found in CelA, for example, might increase its activity. Likewise, since there is no homology between the CBM2 of E1 with those in C. bescii, swapping the CBM domains of C. bescii cellulases with the CBM2 of E1 might improve the substrate binding versatility of these enzymes. This work also offers the possibility of constructing hybrid proteins by gene fusions between various GH domains and CBMs, such as those found in CelA to produce new multi-domain enzymes with novel catalytic activity.
Methods
Strains, media and culture conditions
Caldicellulosiruptor bescii strains, E. coli strains, and plasmids used in this study are listed in Table 1. C. bescii strains were grown anaerobically in liquid or on solid surface in low osmolarity defined (LOD) medium [24] with maltose (0.5 % wt/v; catalog no. M5895, Sigma) as the carbon source, final pH 6.8, at 75 °C as previously described [29, 30]. Liquid cultures were grown from a 0.5 % inoculum or a single colony and incubated at 75 °C in anaerobic culture bottles degassed with five cycles of vacuum and argon. For uracil auxotrophs, JWCB018 and JWCB052, the LOD medium was supplemented with 40 μM uracil. E. coli strain DH5α was used for plasmid DNA constructions and preparations. Techniques for E. coli were performed as described [37]. E. coli cells were grown in LB broth supplemented with apramycin (50 μg/mL) and plasmid DNA was isolated using a Qiagen Mini-prep Kit. Chromosomal DNA from C. bescii strains was extracted using the Quick-gDNA™ MiniPrep (Zymo) or using the DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s instructions. E. coli strain DH5α cells were transformed by electroporation in a 2-mm-gap cuvette at 2.5 V and transformants were selected for apramycin resistance.
Vector construction for the knock-in of Acel_0614 into C. bescii
The plasmids described below were generated using Q5 High-Fidelity DNA polymerase (New England BioLabs) for PCR reactions, restriction enzymes (New England BioLabs), and the Fast-link DNA Ligase kit (Epicentre Biotechnologies) according to the manufacturer’s instructions. Plasmid pDCW174 (Table 1) was constructed by inserting the sequence of the CelA (Cbes1867) signal peptide into pDCW144 [27], which also contains the regulatory region of Cbes2303 and encodes a C-terminal 6X histidine-tag and a rho-independent transcription terminator. The 6.3 kb DNA fragment was amplified with primers DC464 (adding a BamHI site) and DC466 (adding an SphI site) using pDCW144 as template. A 1.752 kb DNA fragment containing the coding sequence of the N-terminus portion (including the 72 bp signal peptide sequence) of Cbes1867 was amplified with DC368 (with SphI site) and DC560 (with BamHI site) using C. bescii gDNA as a template. These two linear DNA fragments were digested with BamHI and SphI, and then ligated to construct pDCW174 (8.05 kb) (Table 1). The 6.41 kb DNA fragment, containing the Cbes 2303 regulatory region, the 72 bp signal peptide sequence, a C-terminal 6X histidine-tag and the rho-independent transcription terminator, was amplified from pDCW174 using primers DC579 (adding an ApaI site) and DC580 (adding a XmaI site) and was used as a back-bone fragment to construct pDCW175 (Fig. 2b, Additional file 1: Figure S1; Table 1). A 1.6 kb DNA fragment containing the coding sequence of Acel_0614 was amplified with DC581 (adding an ApaI site) and DC582 (adding an XmaI site) using A. cellulolyticus 11B gDNA as template. These two linear DNA fragments were digested with ApaI and XmaI, and then ligated to construct pDCW175 (7.97 kb) (Additional file 1: Figure S1). The DNA sequences of the primers are shown in Additional file 1: Table S2. The sequences of pDCW174 and pDCW175 were verified by Automatic sequencing (Macrogen USA, Maryland). Plasmids are available upon request.
Transformation, screening, purification, and sequence verification of engineered C. bescii
To construct strain JWCB052, one microgram of pDCW175 DNA was used for electroporation into JWCB018 (ΔpyrFA ldh::ISCbe4 ΔcbeI) as previously described [29, 30]. Cells were then plated onto solid LOD medium and uracil prototrophic transformants were inoculated into liquid medium for genomic DNA extraction and subsequent PCR screening of the targeted region of the chromosome. Confirmed transformants were inoculated into nonselective liquid defined medium, with 40 μM uracil, and incubated overnight at 75 °C to allow loop-out of the plasmid. The cultures were then plated onto 5-FOA (8 mM) containing solid medium. Transformants containing the knock-in were further purified by two additional passages under selection on solid medium and screened a second time by PCR. The insertion of the Acel_0614 expression cassette at the targeted chromosome region was verified by PCR amplification and sequence analysis using primers DC462 and DC463. A PCR product was generated from genomic DNA using primers (DC477 and DC478) outside the homologous regions used to construct the knock-in. Primers and sequences used in this study are listed in Additional file 1: Table S2.
Preparation of extracellular fractions and Western blotting
Extracellular protein (ECP) was collected from 2 L of culture grown at a range of temperatures (65, 70, or 75 °C) in closed bottles shaking at 150 rpm to an OD680 of 0.25–0.3. LOD media was supplemented with 40 μM uracil and 40 mM MOPS. Cultures were centrifuged (6,000×g at 4 °C for 15 min), filtered (glass fiber, 0.7 µm) to separate out cells, and concentrated with a 3 kDa molecular weight cut-off column (Hollow Fiber Cartridge, GE Healthcare). Protein concentrations were determined using the Bio-Rad protein assay kit with bovine serum albumin (BSA) as the standard. ECP samples (10 µg) were electrophoresed in 4–15 % gradient Mini-Protean TGX gels (BIO-RAD) for either Coomassie blue-staining or for electrotransfer to PVDF membranes (ImmobilonTM-P; Millipore) using a Bio-Rad Mini-Protean 3 electrophoretic apparatus. The membrane was probed with E1 monoclonal antibody (1:1,500 dilution, provided by Bill Adney and Steve Decker, NREL) using the ECL Western Blotting substrate Kit (Thermo Scientific) as specified by the manufacturer.
Enzyme activity assays
Enzymatic assays using pure enzyme preparations of CelA and E1, their isolation and purification were conducted as previously described [23], were performed in 20 mM acetate buffer pH 5.5 with 100 mM NaCl and 10 mM CaCl2. The enzyme loadings were 15 mg/g for CelA, 4 mg/g for E1, and 11 and 4 mg/g for CelA and E1, respectively, in the mixture. Avicel digestion experiments were conducted for 7 days with constant mixing at 75 °C with sampling at various time points. Enzymes were inactivated by boiling for 15 min after which samples were filtered through 0.45 µm Acrodisc syringe filters. The released sugars were analyzed by HPLC. Samples were injected at 20 mL and run on an Agilent 1100 HPLC system equipped with a BioRad Aminex HPX-87H 300 mm × 7.8 mm column heated to 55 °C. A constant flow of 0.6 mL/min was used with 0.1 M H2SO4 in water as the mobile phase to give optimal sugar separation. Glucose and cellobiose were quantified against independent standard curves and converted to anhydrous glucan equivalent and the results are reported as anhydrous glucan converted. All experiments were performed in triplicate and the resulting extents of conversion are shown as percent glucan converted.
To assess the activity of the secretome of the C. bescii strain containing E1, concentrated extracellular protein (ECP) was buffer exchanged with 20 mM MES/2 mM β-mercaptoethanol (pH 5.5). Protein concentrations were determined using the Bio-Rad protein assay kit with bovine serum albumin (BSA) as the standard. Cellulolytic activity was determined using 10 g/L of either CMC or Avicel in MES reaction buffer (pH 5.5) as previously described [38]. Twenty five microgram per milliliter of extracellular protein was added to each reaction and incubated at 65 or 75 °C (1 h for CMC and 24 h for Avicel). Controls were incubated for the same length of time without added enzyme. Reducing sugars in the supernatant were measured using dinitrosalicylic acid (DNS). Samples and standards (glucose) were mixed 1:1 with DNS and boiled for 2 min and measured at OD575. Activity was reported as mg/mL of sugar released.
Authors’ contributions
DC, JY, MC designed and carried out experiments, analyzed results and participated in the writing of the manuscript. RB did the experiments on in vitro analysis of purified proteins. YB, MH and JW participated in design and coordination of the work and contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Elise Snyder for outstanding technical assistance, Sidney Kushner for expert technical advice, Joe Groom for critical review of the manuscript, Charles Wyman and Rajeev Kumar for information about commercial enzyme cocktails. The BioEnergy Science Center is a US Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Abbreviations
- C. bescii
Caldicellulosiruptor bescii
- A. cellulolyticus
Acidothermus cellulolyticus
- CBM
carbohydrate-binding module
- GH
glycoside hydrolase
- CAZy
carbohydrate-active enzymes
- CMC
carboxymethylcellulose
- YE
yeast extract
- 5-FOA
5-fluoroorotic acid
- LOD
low osmolarity defined
- LB broth
Luria Bertani broth
- ECP
extracellular protein
- BSA
bovine serum albumin
- DNS
dinitrosalicylic acid
Additional file
Figure S1. Plasmid map of chromosomal knock-in vector in C. bescii for extracellular expression of E1 (Acel0614). Figure S2. Relative quantification of enzymatic activity of the extracellular fraction of C. bescii expressing E1 (Acel0614) on Avicel and carboxymethylcellulose. Table S1. The list of Glycoside Hydrolase Family 5 (GH5) catalytic domains in Caldicellulosiruptor bescii and their sequence homology with the GH5 domain in E1 from A. cellulolyticus. Table S2. Primers used in this study.
Contributor Information
Daehwan Chung, Email: chung301@gmail.com.
Jenna Young, Email: jmy715@uga.edu.
Minseok Cha, Email: mcha@uga.edu.
Roman Brunecky, Email: roman.brunecky@nrel.gov.
Yannick J Bomble, Email: yannick.bomble@nrel.gov.
Michael E Himmel, Email: mike.himmel@nrel.gov.
Janet Westpheling, Email: janwest@uga.edu.
References
- 1.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315(5813):804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 2.McCann MC, Carpita NC. Designing the deconstruction of plant cell walls. Curr Opin Plant Biol. 2008;11(3):314–320. doi: 10.1016/j.pbi.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DB. Three microbial strategies for plant cell wall degradation. Ann NY Acad Sci. 2008;1125:289–297. doi: 10.1196/annals.1419.026. [DOI] [PubMed] [Google Scholar]
- 4.Klein-Marcuschamer D, Oleskowicz-Popiel P, Simmons BA, Blanch HW. The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol Bioeng. 2012;109(4):1083–1087. doi: 10.1002/bit.24370. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert HJ. Cellulosomes: microbial nanomachines that display plasticity in quaternary structure. Mol Microbiol. 2007;63(6):1568–1576. doi: 10.1111/j.1365-2958.2007.05640.x. [DOI] [PubMed] [Google Scholar]
- 6.Bayer EA, Morag E, Lamed R. The cellulosome—a treasure trove for biotechnology. Trends Biotechnol. 1994;12(9):379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 7.Bayer EA, Setter E, Lamed R. Organization and distribution of the cellulosome in Clostridium thermocellum. J Bacteriol. 1985;163(2):552–559. doi: 10.1128/jb.163.2.552-559.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamed R, Kenig R, Setter E, Bayer EA. Major characteristics of the cellulolytic system of Clostridium thermocellum coincide with those of the purified cellulosome. Enzyme Microb Technol. 1985;7(1):37–41. doi: 10.1016/0141-0229(85)90008-0. [DOI] [Google Scholar]
- 9.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66(3):506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MWW, Kelly RM. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr Opin Biotechnol. 2008;19(3):210–217. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, et al. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol. 2009;75(14):4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zverlov V, Mahr S, Riedel K, Bronnenmeier K. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile ‘Anaerocellum thermophilum’ with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology. 1998;144(Pt 2):457–465. doi: 10.1099/00221287-144-2-457. [DOI] [PubMed] [Google Scholar]
- 13.Blumer-Schuette SE, Giannone RJ, Zurawski JV, Ozdemir I, Ma Q, Yin YB, et al. Caldicellulosiruptor core and pangenomes reveal determinants for noncellulosomal thermophilic deconstruction of plant biomass. J Bacteriol. 2012;194(15):4015–4028. doi: 10.1128/JB.00266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young J, Chung D, Bomble YJ, Himmel M, Westpheling J. Deletion of Caldicellulosiruptor bescii CelA reveals its crucial role in the deconstruction of lignocellulosic biomass. Biotechnol Biofuels. 2014;7:142. doi: 10.1186/s13068-014-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson DB. Processive and nonprocessive cellulases for biofuel production—lessons from bacterial genomes and structural analysis. Appl Microbiol Biotechnol. 2012;93(2):497–502. doi: 10.1007/s00253-011-3701-9. [DOI] [PubMed] [Google Scholar]
- 16.Kostylev M, Wilson D. Synergistic interactions in cellulose hydrolysis. Biofuels. 2012;3(1):61–70. doi: 10.4155/bfs.11.150. [DOI] [Google Scholar]
- 17.Dam P, Kataeva I, Yang SJ, Zhou FF, Yin YB, Chou WC, et al. Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res. 2011;39(8):3240–3254. doi: 10.1093/nar/gkq1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kataeva I, Foston MB, Yang SJ, Pattathil S, Biswal AK, Poole FL, et al. Carbohydrate and lignin are simultaneously solubilized from unpretreated switchgrass by microbial action at high temperature. Energy Environ Sci. 2013;6(7):2186–2195. doi: 10.1039/c3ee40932e. [DOI] [Google Scholar]
- 19.Lochner A, Giannone RJ, Rodriguez M, Jr, Shah MB, Mielenz JR, Keller M, et al. Use of label-free quantitative proteomics to distinguish the secreted cellulolytic systems of Caldicellulosiruptor bescii and Caldicellulosiruptor obsidiansis. Appl Environ Microbiol. 2011;77(12):4042–4054. doi: 10.1128/AEM.02811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su X, Mackie RI, Cann IK. Biochemical and mutational analyses of a multidomain cellulase/mannanase from Caldicellulosiruptor bescii. Appl Environ Microbiol. 2012;78(7):2230–2240. doi: 10.1128/AEM.06814-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker JO, Adney WS, Nieves RA, Thomas SR, Wilson DB, Himmel ME. A new thermostable endoglucanase, Acidothermus cellulolyticus E1—synergism with Trichoderma reesei Cbh-I and comparison to Thermomonospora fusca E(5) Appl Biochem Biotechnol. 1994;45–6:245–256. doi: 10.1007/BF02941803. [DOI] [Google Scholar]
- 22.Sakon J, Adney WS, Himmel ME, Thomas SR, Karplus PA. Crystal structure of thermostable family 5 endocellulase E1 from Acidothermus cellulolyticus in complex with cellotetraose. Biochemistry. 1996;35(33):10648–10660. doi: 10.1021/bi9604439. [DOI] [PubMed] [Google Scholar]
- 23.Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, et al. Revealing nature’s cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science. 2013;342(6165):1513–1516. doi: 10.1126/science.1244273. [DOI] [PubMed] [Google Scholar]
- 24.Farkas J, Chung DW, Cha M, Copeland J, Grayeski P, Westpheling J. Improved growth media and culture techniques for genetic analysis and assessment of biomass utilization by Caldicellulosiruptor bescii. J Ind Microbiol Biotechnol. 2013;40(1):41–49. doi: 10.1007/s10295-012-1202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Himmel ME, Adney WS, Baker JO, Vinzant TB, Thomas SR, Sakon J,et al (2000) E1 endoglucanase variants y245 g, y82r and w42r. Google Patents
- 26.Baker JO, Ehrman CI, Adney WS, Thomas SR, Himmel ME. Hydrolysis of cellulose using ternary mixtures of purified celluloses. Appl Biochem Biotechnol. 1998;70–2:395–403. doi: 10.1007/BF02920154. [DOI] [PubMed] [Google Scholar]
- 27.Chung D, Cha M, Guss AM, Westpheling J. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc Natl Acad Sci USA. 2014;111(24):8931–8936. doi: 10.1073/pnas.1402210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung D, Young J, Bomble YJ, Vander Wall TA, Groom J, Himmel ME, et al. Homologous expression of the Caldicellulosiruptor bescii CelA reveals that the extracellular protein is glycosylated. PloS One. 2015;10(3):e0119508. doi: 10.1371/journal.pone.0119508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung D, Cha M, Farkas J, Westpheling J. Construction of a stable replicating shuttle vector for Caldicellulosiruptor species: use for extending genetic methodologies to other members of this genus. PloS One. 2013;8(5):e62881. doi: 10.1371/journal.pone.0062881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung D, Farkas J, Huddleston JR, Olivar E, Westpheling J. Methylation by a unique alpha-class N4-cytosine methyltransferase is required for DNA transformation of Caldicellulosiruptor bescii DSM6725. PloS One. 2012;7(8):e43844. doi: 10.1371/journal.pone.0043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung DH, Huddleston JR, Farkas J, Westpheling J. Identification and characterization of CbeI, a novel thermostable restriction enzyme from Caldicellulosiruptor bescii DSM 6725 and a member of a new subfamily of HaeIII-like enzymes. J Ind Microbiol Biotechnol. 2011;38(11):1867–1877. doi: 10.1007/s10295-011-0976-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kataeva IA, Yang SJ, Dam P, Poole FL, Yin Y, Zhou FF, et al. Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium “Anaerocellum thermophilum” DSM 6725. J Bacteriol. 2009;191(11):3760–3761. doi: 10.1128/JB.00256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rignall TR, Baker JO, McCarter SL, Adney WS, Vinzant TB, Decker SR, et al. Effect of single active-site cleft mutation on product specificity in a thermostable bacterial cellulase. Appl Biochem Biotechnol. 2002;98:383–394. doi: 10.1385/ABAB:98-100:1-9:383. [DOI] [PubMed] [Google Scholar]
- 34.Mohagheghi A, Grohmann K, Himmel M, Leighton L, Updegraff DM. Isolation and characterization of Acidothermus cellulolyticus gen. nov., sp. nov., a new genus of thermophilic, acidophilic, cellulolytic bacteria. Int J Syst Bacteriol. 1986;36(3):435–443. doi: 10.1099/00207713-36-3-435. [DOI] [Google Scholar]
- 35.Tucker MP, Mohagheghi A, Grohmann K, Himmel ME. Ultra-thermostable cellulases from Acidothermus cellulolyticus—comparison of temperature optima with previously reported cellulases. Bio Technol. 1989;7(8):817–820. doi: 10.1038/nbt0889-817. [DOI] [Google Scholar]
- 36.Himmel ME, Adney WS, Rivard CJ, Baker JO. Cellulase Assays: A Review. In: Klass D, editor. Energy from biomass and wastes. Chicago: Institute of Gas Technology; 1993. p. 1268. [Google Scholar]
- 37.Sambrook JaR DW. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 38.Kanafusa-Shinkai S, Wakayama J, Tsukamoto K, Hayashi N, Miyazaki Y, Ohmori H, et al. Degradation of microcrystalline cellulose and non-pretreated plant biomass by a cell-free extracellular cellulase/hemicellulase system from the extreme thermophilic bacterium Caldicellulosiruptor bescii. J Biosci Bioeng. 2013;115(1):64–70. doi: 10.1016/j.jbiosc.2012.07.019. [DOI] [PubMed] [Google Scholar]


