Abstract
Solid cancers are spatially and temporally heterogeneous. This limits the use of invasive biopsy based molecular assays but gives huge potential for medical imaging, which has the ability to capture intra-tumoural heterogeneity in a non-invasive way. During the past decades, medical imaging innovations with new hardware, new imaging agents and standardised protocols, allows the field to move towards quantitative imaging. Therefore, also the development of automated and reproducible analysis methodologies to extract more information from image-based features is a requirement. Radiomics – the high-throughput extraction of large amounts of image features from radiographic images – addresses this problem and is one of the approaches that hold great promises but need further validation in multi-centric settings and in the laboratory.
Keywords: Imaging, Radiomics, Tumour, Intra tumour, heterogeneity
1. Introduction
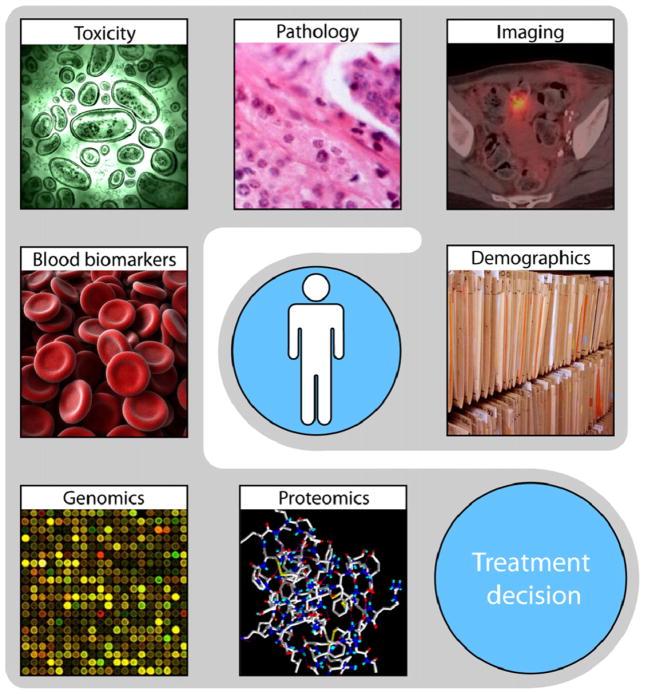
The use and role of medical imaging technologies in clinical oncology has greatly expanded from primarily a diagnostic tool to include a more central role in the context of individualised medicine over the past decade (Fig. 1). It is expected that imaging contains complementary and interchangeable information compared to other sources, e.g. demographics, pathology, blood biomarkers, genomics and that combining these sources of information will improve individualised treatment selection and monitoring.1
Fig. 1.
Different sources of information, e.g. demographics, imaging, pathology, toxicity, biomarkers, genomics and proteomics, can be used for selecting the optimal treatment.
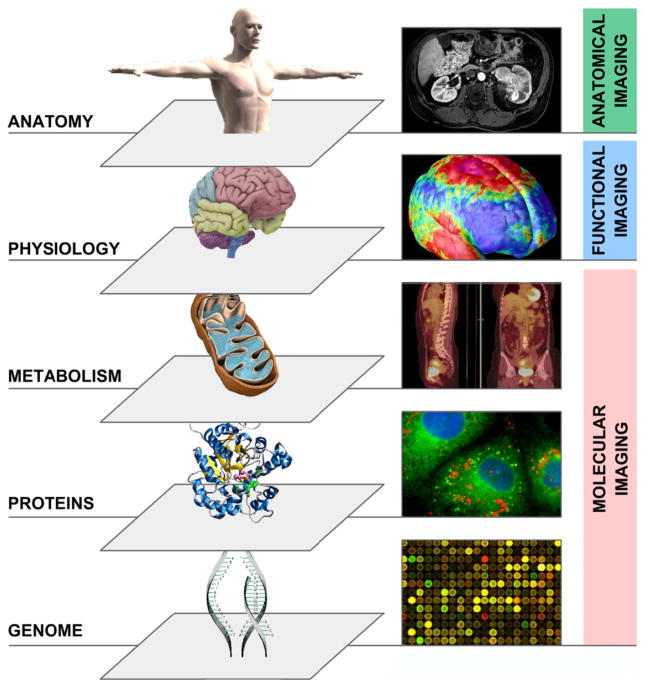
Cancer can be probed in many ways depending on the non-invasive imaging device used or the mode by which it operates (Fig. 2). Classically, anatomical computed tomography (CT) imaging is a often used modality, acquiring images of the ‘anatome’ in high resolution (e.g. 1 mm3). CT imaging is now routinely used and is playing an essential role in all phases of cancer management, including prediction, screening, biopsy guidance for detection, treatment planning, treatment guidance and treatment response evaluation.2,3 CT is used in the assessment of structural features of cancer but it is not perceived to portray functional or molecular details of solid tumours. Functional imaging concerns physiological processes and functions such as diffusion, perfusion and glucose uptake. Here, commonly used methodologies are dynamic contract enhanced-magnetic resonance imaging (DCE-MRI), assessing tumour perfusion and fluoro-2-deoxy-D-glucose (FDG) positron emission tomography (PET) imaging, assessing tumour metabolism, which both often are found to have prognostic value.4–6 Finally, another modality is molecular imaging, visualising at the level of specific pathways or macro-molecule in vivo. For example, there are molecular markers assessing tumour hypoxia or labelled antibodies, assessing receptor expression levels of a tumour.1,7
Fig. 2.
Multilevel imaging: anatomical, functional, and molecular imaging.
Over the past decades, medical imaging has progressed in four distinct ways:
Innovations in medical devices (hardware): This concerns improvements in imaging hardware and the development of combined modality machines. For example, in the last decade we moved from single slice CT to multiple slices CT and CT/PET. More recent developments are dual-source and dual-energy CT. These techniques significantly increase the temporal resolution for 4-D CT reconstructions allowing visualisation of fine structures in tissues, also in several stages in the cardiac or respiration phase. Moreover, dual-energy CT can be used to improve identification of tissue composition and density.
Innovations in imaging agents: Innovations in imaging agents (or imaging biomarker, imaging probe, radiotracer), i.e. molecular substances injected in the body and used as an indicator of a specific biological process occurring in the body. This is achieved by contrast agents, i.e. an imaging agent using positive emission tomography (radiotracer). A common use is to find indications of pathological processes, e.g. hypoxia markers using PET imaging.
Standardised protocol allowing quantitative imaging: Historically radiology has been a qualitative science, perhaps with the exception of the quantitative use of CT based electron densities in radiotherapy treatment planning. The use of standardised protocols like common MRI spin-echo sequences helps to allow multicentric use of imaging as well as transforming radiology to a more quantitative, highly reproducible science.
Innovations in imaging analysis: The analysis of medical images has a large impact on the conclusions of the derived images. More and more software is becoming available, allowing for more quantification and standardisation. This has been illustrated by the development of the computer-assisted detection (CAD systems) that improves the performance of detecting cancer in mammography or in lung diseases.8
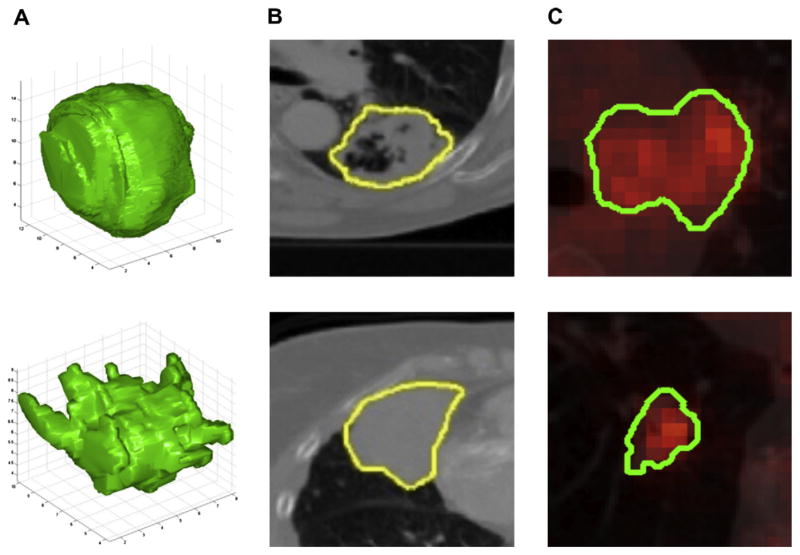
Radiomics focuses on improvements of image analysis, using an automated high-throughput extraction of large amounts (200+) of quantitative features of medical images and belongs to the last category of innovations in medical imaging analysis. The hypothesis is that quantitative analysis of medical image data through automatic or semi-automatic software of a given imaging modality can provide more and better information than that of a physician. This is supported by the fact that patients exhibit differences in tumour shape and texture measurable by different imaging modalities (Fig. 3).
Fig. 3.
(A) Two representative 3-D representations of a round tumour (top) and spiky tumour (bottom) measured by computed tomography (CT) imaging. (B) Texture differences between non-small cell lung cancer (NSCLC) tumours measured using CT imaging, more heterogeneous (top) and more homogeneous (bottom). (C) Differences of FDG-PET uptake, showing heterogeneous uptake.
2. The workflow of Radiomics: a (semi) high-throughput approach
Fig. 4 depicts the processes involved in the Radiomics workflow. The first step involves the acquisition of high quality and standardised imaging, for diagnostic or planning purposes. From this image, the macroscopic tumour is defined, either with an automated segmentation method or alternatively by an experienced radiologist or radiation oncologist. Quantitative imaging features are subsequently extracted from the previously defined tumour region. These features involve descriptors of intensity distribution, spatial relationships between the various intensity levels, texture heterogeneity patterns, descriptors of shape and of the relations of the tumour with the surrounding tissues (i.e. attachment to the pleural wall in lung, differentiation). The extracted image traits are then subjected to a feature selection procedure. The most informative features are identified based on their independence from other traits, reproducibility and prominence on the data. The selected features are then analysed for their relationship with treatment outcomes or gene expression. The ultimate goal is to provide accurate risk stratification by incorporating the imaging traits into predictive models for treatment outcome and to evaluate their added value to commonly used predictors.
Fig. 4.
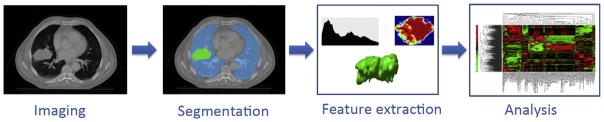
The Radiomics workflow. On the medical images, segmentation is performed to define the tumour region. From this region the features are extracted, e.g. features based on tumour intensity, texture and shape. Finally, these features are used for analysis, e.g. the features are assessed for their prognostic power, or linked with stage, or gene expression.
3. The Radiomics hypothesis: inferring proteo-genomic and phenotypic information from radiological images
The underlying hypothesis of Radiomics is that advanced image analysis on conventional and novel medical imaging could capture additional information not currently used, and more specifically, that genomic and proteomics patterns can be expressed in terms of macroscopic image-based features. If proven, we can infer phenotypes or gene–protein signatures, possibly containing prognostic information, from the quantitative analysis of medical image data.
This hypothesis is supported by image-guided biopsies, which demonstrated that tumours show spatial differences in protein expressions.9 More specifically, it has been demonstrated that major differences in protein expression patterns within a tumour can be correlated to radiographic findings (or radiophenotypes) such as contrast-enhanced and non-enhanced regions based on CT data.10 The authors suggest that image-guided proteomics holds promise for characterising tissues prior to treatment decisions and without imaging there is indeed a risk that the optimum treatment decision could be neglected (i.e. the use or not of a targeted agent). Also, Kuo et al. reported the association of CT-derived imaging traits with histo-pathologic markers, and several pre-defined gene expression modules on liver cancer. 11,12 In ovarian carcinoma, an imaging feature describing the enhancement fraction as proportion of enhancing tumour tissue on a pre-treatment CT scan, was found predictive for outcome after first line chemotherapy. 13 In lung cancer, CT derived information has been limited to pre-treatment assessment of tumour volume and as response evaluation defined as tumour size reduction.14 For PET imaging, the maximum and median FDG uptake has often been investigated, indicating strong prognostic power.6,19 However, more complex descriptions of FDG uptake are only investigated on a limited scale. There was a study of El Naqa et al.15, investigating the predictive power of intensity–volume histogram (IVH) metrics, shape and texture features to assess response to treatment of a limited set of patients with head and neck and cervix cancers. Tixier et al. also explored the potential of SUV based, shape and texture features extracted from baseline FDG-PET, images, to assess response to therapy and prognosis in order to predict response to combined chemo-radiation treatment in oesophageal cancer.16 Also, textural features in FDG PET images exhibited small variations due to different acquisition modes and reconstruction parameters.17 These examples open the question of whether quantitative extraction of additional imaging features on conventional imaging improves the ability of currently used parameters to predict or monitor response to treatment.
Furthermore, Radiomics can be linked with the concept of radio-genomics, which assumes that imaging features are related to gene signatures. An interesting finding in recent literature is that tumours with more genomic heterogeneity are more likely to develop a resistance to treatment and to metastasise.18 This links to the concept that more heterogeneous tumours have a worse prognosis. According to the Radiomics hypothesis, the genomic heterogeneity could translate to an expression in an intra-tumoural heterogeneity that could be assessed through imaging and that would ultimately exhibit worse prognosis. This hypothesis has been sustained by Jackson et al.19 and as well as by Diehn et al.20 who quite convincingly showed that proliferation and hypoxia gene expression patterns can be predicted by mass effect and tumour contrast enhancement, respectively. They also showed that a specific imaging pattern could predict overexpression of epidermal growth factor receptor (EGFR), a known therapeutic target. Moreover, in their analysis the presence of certain image features was highly predictive of outcome. The authors concluded that imaging in this case MR provided an ‘in vivo portrait’ of genome-wide gene expression in glioblastoma multiform. Similar findings have been found in hepatocellular carcinomas by Segal et al.21, showing that the combination of only 28 imaging traits was sufficient to reconstruct the variation of 116 gene expression modules.
These types of studies will need to be extended, by including more patients with external validation datasets, more tumour types that exhibit phenotypes such as invasiveness. This will be the focus of the QuIC-ConCePT consortium, to confirm experimentally the Radiomics hypothesis, namely to establish a causal relationship between gene expression patterns and image features.
4. Conclusions
Solid cancers have extraordinarily spatial and temporal heterogeneity at different levels: genes, proteins, cells, microenvironment, tissues and organs. This limits the use of biopsy based molecular assays but in contrast gives a huge potential for non-invasive imaging, which has the ability to capture intra-tumoural heterogeneity in a non-invasive way. Medical imaging innovations with new hardware, new imaging agents and standardised protocol now allow for quantitative imaging but require the development of ‘smart’ automated software to extract more information from image-based features. Radiomics – the high-throughput extraction of image features from radiographic images – is one approach that holds great promises but needs further validation in a multi-centric setting and in the laboratory.
Acknowledgments
The authors are members of the QuIC-ConCePT project partly funded by EFPIA companies and the Innovative Medicine Initiative Joint Undertaking (IMI JU) under Grant Agreement No. 115151. We also acknowledge financial support from the CTMM framework (AIRFORCE project), EU 6th and 7th framework program (Euroxy and Metoxia program), Interreg (www.eurocat.info), and the Dutch Cancer Society (KWF UM 2011-5020, KWF UM 2009-4454).
Appendix A
QuIC-ConCePT Consortium participants include: AstraZeneca, European Organisation for Research and Treatment of Cancer (EORTC), Cancer Research UK, University of Manchester, Westfälische Wilhelms-Universität Münster, Radboud University Nijmegen Medical Center, Institut National de la Santé et de la Recherche Médical, Stichting Maastricht Radiation Oncology ‘Maastro Clinic’, VUmc Amsterdam, King’s College London, Universitair Ziekenhuis Antwerpen, Institute of Cancer Research – Royal Cancer Hospital, Erasmus Universitair Medisch Centrum Rotterdam, Imperial College of Science Technology and Medicine, Keosys S.A.S., Eidgenössische Technische Hochschule Zürich, Amgen NV, Eli Lilly and Company Ltd., GlaxoSmithKline Research & Development Limited, Merck KGa, Pfizer Limited, F. Hoffmann – La Roche Ltd., Sanofi–Aventis Research and Development.
Footnotes
Conflict of interest statement
None declared.
References
- 1.Lambin P, et al. The ESTRO Breur Lecture 2009. From population to voxel-based radiotherapy: exploiting intra-tumour and intra-organ heterogeneity for advanced treatment of non-small cell lung cancer. Radiother Oncol. 2010;96:145–52. doi: 10.1016/j.radonc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Fass L. Imaging and cancer: a review. Mol Oncol. 2008;2:115–52. doi: 10.1016/j.molonc.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1. 1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 4.van Baardwijk A, et al. The maximum uptake of (18)F-deoxyglucose on positron emission tomography scan correlates with survival, hypoxia inducible factor-1alpha and GLUT-1 in non-small cell lung cancer. Eur J Cancer. 2007;43:1392–8. doi: 10.1016/j.ejca.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Berghmans T, et al. Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC Lung Cancer Staging Project. J Thorac Oncol. 2008;3:6–12. doi: 10.1097/JTO.0b013e31815e6d6b. [DOI] [PubMed] [Google Scholar]
- 6.Paesmans M, et al. Primary tumor standardized uptake value measured on fluorodeoxyglucose positron emission tomography is of prognostic value for survival in non-small cell lung cancer: update of a systematic review and meta-analysis by the European Lung Cancer Working Party for the International Association for the Study of Lung Cancer Staging Project. J Thorac Oncol. 2010;5:612–9. doi: 10.1097/JTO.0b013e3181d0a4f5. [DOI] [PubMed] [Google Scholar]
- 7.Aerts HJWL, et al. Disparity between in vivo EGFR expression and 89Zr-labeled cetuximab uptake assessed with PET. J Nucl Med. 2009;50:123–31. doi: 10.2967/jnumed.108.054312. [DOI] [PubMed] [Google Scholar]
- 8.Li H, et al. Evaluation of computer-aided diagnosis on a large clinical full-field digital mammographic dataset. Acad Radiol. 2008;15:1437–45. doi: 10.1016/j.acra.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Meter T, et al. Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn Mol Pathol. 2006;15:195–205. doi: 10.1097/01.pdm.0000213464.06387.36. [DOI] [PubMed] [Google Scholar]
- 10.Hobbs SK, et al. Magnetic resonance image-guided proteomics of human glioblastoma multiforme. J Magn Reson Imaging. 2003;18:530–6. doi: 10.1002/jmri.10395. [DOI] [PubMed] [Google Scholar]
- 11.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol. 2007;18:821–31. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–41. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 13.O’Connor JPB, et al. Enhancing fraction predicts clinical outcome following first-line chemotherapy in patients with epithelial ovarian carcinoma. Clin Cancer Res. 2007;13:6130–5. doi: 10.1158/1078-0432.CCR-07-0331. [DOI] [PubMed] [Google Scholar]
- 14.Dehing-Oberije C, et al. Development and validation of a prognostic model using blood biomarker information for prediction of survival of non-small-cell lung cancer patients treated with combined chemotherapy and radiation or radiotherapy alone (NCT00181519, NCT00573040, and NCT00572325) Int J Radiat Oncol Biol Phys. 2010 doi: 10.1016/j.ijrobp.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 15.El Naqa I, et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recognit. 2009;42:1162–71. doi: 10.1016/j.patcog.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tixier F, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med. 2011;52:369–78. doi: 10.2967/jnumed.110.082404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010;49:1012–6. doi: 10.3109/0284186X.2010.498437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson A, O’Connor JPB, Parker GJM, Jayson GC. Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging. Clin Cancer Res. 2007;13:3449–59. doi: 10.1158/1078-0432.CCR-07-0238. [DOI] [PubMed] [Google Scholar]
- 20.Diehn M, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105:5213–8. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segal E, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–80. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]