Abstract
The binding of Cbl-interacting protein of 85 kDa (CIN85) to c-Cbl is important to endocytosis and degradation of epidermal growth factor receptor (EGFR). The proline-arginine motif PXXXPR in c-Cbl and SH3 domains of CIN85 are essential to this interaction. Here we demonstrated that SH3KBP1 binding protein 1 (SHKBP1), which also contains two PXXXPR motifs, constitutively bound to SH3 domains of CIN85. Importantly, the binding of SHKBP1 prevented the interaction of CIN85 with c-Cbl and inhibited the translocation of CIN85 to EGFR containing vesicles, thus reducing EGFR degradation and enhancing EGF induced SRE transcription activity. Therefore, our results indicated that SHKBP1 could promote EGFR signaling pathway by interrupting c-Cbl-CIN85 complex and inhibiting EGFR degradation.
Keywords: SH3KBP1 binding protein 1, CIN85, c-Cbl, Epidermal growth factor receptor, Down-regulation
Introduction
Epidermal growth factor receptor (EGFR) plays a critical role in the regulation of many biological responses such as cell proliferation, differentiation and cellular adhesion1–4. Thus, the regulation of EGFR signaling is very important to maintain tissue homeostasis5. Aberrations in the EGFR activity could lead to the development of various cancers such as lung, breast and brain cancers3,6. To regulate the lifetime of this signaling, the activated EGFR also triggers negative feedback loop that inhibit its activation by promoting its endocytosis and lysosomal degradation. Casitas B-lineage Lymphoma (Cbl) family member c-Cbl plays a crucial role in this process, acting as an ubiquitin ligase as well as an endocytic adaptor molecule. Upon growth factor stimulation, c-Cbl binds to activated EGFR and acts as a multifunctional adaptor protein to recruit additional endocytic regulatory proteins such as Cbl-interacting protein of 85 kDa (CIN85), which plays important roles in receptor endocytosis and degradation7–9.
Through the tyrosine kinase binding module and the ring-finger domain in its amino terminus, c-Cbl could promote the ubiquitination of various activated receptor tyrosine kinases (RTKs) such as EGFR and c-Met10. The carboxyl terminus of c-Cbl contains a long proline-rich domain, which serves as an adaptor for binding to multiple signaling proteins and transducing intracellular signaling cascades11. CIN85 is a ubiquitously expressed adaptor protein with three SH3 domains at the amino terminus, as well as a proline-rich region and a coiled-coil domain at the carboxyl terminus12. The c-Cbl-CIN85 complex is formed through the interaction between proline-arginine motif PXXXPR (X denotes any amino acid) of c-Cbl and SH3 domains of CIN85. The arginine-to-alanine mutation in PXXXPR motif abolished the formation of the c-Cbl-CIN85 complex and attenuated the negative feedback regulation of EGFR signaling13–14.
Several recent findings indicated that EGFR signaling can be finely-tuned by regulating the functions of c-Cbl-CIN85 complex. For example, Sprouty2 has been shown to sequester c-Cbl away from CIN85 and activated EGFR to inhibit the down-regulation of EGFR signaling15–16. Another protein named cloned-out of library 1 (Cool 1), a nucleotide exchange factor for Cdc42, is able to form a complex with c-Cbl, thus preventing its interaction with activated EGFR and reducing EGFR degradation17. Other proteins such as infected cell protein 0 (ICP0), Arf GTPase-activating proteins ASAP1 and adaptor proteins Huntingtin-interacting protein 1-related protein (Hip1R) and Signal-transducing adaptor protein 1 (STAP1) are also implicated in the regulation of EGFR signaling14,18.
In the present study, we found SH3KBP1 binding protein 1 (SHKBP1) bound to SH3 domains of CIN85 via two PXXXPR motifs in its carboxyl terminus. SHKBP1 could compete with c-Cbl to interact with CIN85 and attenuate the down-regulation of EGFR signaling following receptor activation. Overexpression of SHKBP1 delayed the degradation of EGFR and enhanced EGFR signaling by disturbing the translocation of CIN85 to EGFR containing vesicles. Taken together, these data provide a novel mechanism of the regulation of c-Cbl-CIN85 association and fine-tuning of EGFR activation.
Materials and Methods
Antibodies and Plasmids
EGF (PHG0311) was purchased from Invitrogen (Carlsbad, CA, USA). Rabbit anti-EGFR polyclonal antibody (06–129) and Mouse anti-c-Cbl monoclonal antibody (05–440) were purchased from Millipore (Billerica, MA, USA). Mouse anti-Flag monoclonal antibody (M2) (A2220) and mouse anti-α-Tubulin Clone B-5-1-2 (T5168) were purchased form Sigma (St. Louis, MO, USA). Mouse anti-Myc monoclonal antibody was prepared from a mouse hybridoma clone 9E10 obtained from ATCC (Manassas, VA, USA). Alexa fluor 594 goat anti-mouse IgG (H+L) (A-11005) and Alexa fluor 594 goat anti-rabbit IgG (H+L) (A-11012) were purchased from Invitrogen (Carlsbad, CA, U S A). Full length cDNA of SHKBP1 (MMM1013-9201499) was purchased from Open Biosystems (Huntsville, AL, USA). Full length cDNA of c-Cbl and CIN85 were kindly provided by Dr. Biao Wang (Department of Medicine, University of Minnesota Medical School). EGFR/pRK5 and SRE-Luc reporter plasmids were kindly provided by Prof. Zhengjun Chen (Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences) and Dr. Bernard Roizman (Department of Molecular Genetics and Cell Biology, University of Chicago), respectively. Myc-SHKBP1 and Myc-c-Cbl inserts were prepared by PCR using the corresponding full length cDNA plasmids as template and cloned into the pCMV-3Tag2 vector (Stratagene, Santa Clara, CA, USA). Similarly, Flag-SHKBP1, Flag-CIN85, Flag-CIN85-3SH3 and Flag-CIN85-PCC PCR products were cloned into the pCMV-3Tag1 vector. GFP-CIN85 was prepared by PCR using full length CIN85 cDNA plasmid as template and cloned in to the pEGFPC1 vector. SHKBP1-R623A and SHKBP1-R684A were obtained by site directed mutagenesis of wild type SHKBP1 using the TaKaRa MutanBEST Kit (TaKaRa, Dalian, Liaoning, China). And SHKBP1-R623A/R684A was obtained by site directed mutagenesis of SHKBP1-R623A using the TaKaRa MutanBEST Kit. Sequences of all constructs were verified by sequencing. And the primers used for construction were listed in supplemental table 1.
Cells and Transfection
HEK293T and HeLa cells were purchased from ATCC (Manassas, VA, USA) and cultured in DMEM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Gibco, Grand Island, NY, USA) and 1% antibiotics (Sigma, St. Louis, MO, USA) in a humidified incubator at 37 °C supplied with 5% CO2. For transfection, 0.5×106 cells were seeded onto individual well of 6-well plates and cultured for 24 hours before transfection with DNA using LipofectAMINE reagent (Invitrogen, Carlsbad, CA, USA) following manufacturer’s instructions. Thirty-six hours after transfection, cells were incubated with serum-free DMEM medium for additional twelve hours and subsequently treated with 100 ng/ml EGF for the time indicated. After EGF treatment, cells were washed with cold PBS and lysed in cold lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% glycerol and 1% Triton X-100) supplemented with a cocktail of protease and phosphatase inhibitors as previous reported19.
Yeast Two-Hybrid Experiment
The ProQuest yeast two-hybrid system was used to study protein-protein interaction according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). DNA encoding SHKBP1 and its mutants were cloned into the pDBLeu vector, while DNA of CIN85 was cloned into the pPC86 vector. Yeast cells (MAV203 strain) were co-transformed with the CIN85 plasmid and either a wild type or mutant SHKBP1 plasmid to study protein-protein interaction. SHKBP1 or its mutants co-transformed with empty pPC86 and CIN85 co-transformed with empty pDBLeu were used as control. Meanwhile, co-transformed pPC86 and pDBLeu vectors were used as negative control in this experiment. After transformation yeasts were spread on SC-Leu-Trp plates (synthetic complete culture medium with 1.5% agar that lacks leucine and tryptophan prepared following manufacturer’s instruction) and placed in a 37 °C incubator for 5–7 days. Visible yeast colonies were picked up and analyzed by a LacZ beta-galactosidase assay with X-gal substrate to determine the interaction of CIN85 with wild type or mutant SHKBP1. The interaction was scored and compared to the positive and negative control yeast strains supplied by the manufacturer.
Immuno-precipitation and Western-Blot
Cell lysates were quantitated by Bio-Rad protein assay kit II (Bio-Rad Laboratories, Hercules, CA, USA). Same amount of protein from each lysate was mixed with either mouse anti-Myc or anti-FLAG (M2) monoclonal antibody and Protein A beads (GE Healthcare, Piscataway, NJ, USA) in a microcentrifuge tube, and placed on a rotator overnight at 4°C. Normal mouse IgG was used as control antibody for the immuno-precipitation. The immuno-complexes precipitated with Protein A beads were washing in ice-cold lysis buffer a few times and finally resuspended and boiled in Uli Laemmli sample buffer. The immuno-precipitates were resolved by SDS-PAGE, transferred to PVDF membranes and probed with the indicated primary antibodies as previously reported20.
Immuno-fluorescence
To study the co-localization of SHKBP1 or its mutants with CIN85, HEK293T cells seeded on collagen-coated coverslips were transfected with indicated expression plasmids for 36 hours and starved for additional 12 hours. Transfected cells were then stimulated with or without EGF (100 ng/ml) for 10 minutes and then fixed in 4% formaldehyde for 20 minutes. After permeabilization and blocking, cells were incubated with primary antibodies and then with Alexa Fluor-conjugated secondary antibodies (Invitrogen, Carlsbad, CA, USA). The preparations were mounted and the images were taken with a confocal microscope (Carl Zeiss, Jena, Germany).
Luciferase Reporter Assay
HEK293T or HeLa cells were seeded and cultured in collagen-coated 12-well plates overnight. Cells were transfected with the indicated plasmids, together with SRE-Luc and beta-galactosidase reporter plasmids. Thirty-six hours post-transfection, cells were incubated with serum-free DMEM for an additional 12 hours and then stimulated with 100 ng/ml EGF for the indicated time. Stimulated cells were lysed in 1× lysis buffer and the lysates were mixed with luciferase assay reagent according to the manufacturer’s instructions (Promega, Madison, MI, USA). After incubation in the dark for the indicated time, the luciferase activity was measured by a Victor Wallace luminometer (Turku, Finland) and the luciferase activity was normalized against beta-galactosidase activity measured in parallel21. Samples were prepared in triplicates, measured and the normalized luciferase activity was expressed as the mean ± s.d.
Results
PXXXPR motifs of SHKBP1 constitutively bound to CIN85
CIN85 was found to bind to proline-arginine motif PXXXPR contained in numerous adaptor proteins including c-Cbl, Cbl-b, Sprouty2, Cool1, ASAP1 and STAP12,14,16–17. Through bioinformatic analysis, we found SHKBP1 had two PXXXPR motifs in the carboxyl terminus: PSPSPR (aa: 618–623) and PTPAPR (aa: 679–684), indicating potential interaction with CIN85. Indeed, yeast two-hybrid experiments confirmed the interaction of SHKBP1 with CIN85 (Table 1). The arginine residue within the proline-arginine motif has been shown to be essential for the interaction with SH3 domain, as the arginine-to-alanine mutation abolished this association16. To explore the role of two PXXXPR motifs in the interaction of SHKBP1 with CIN85, arginine resiudes in these two motifs were mutated to alanine residues. Three SHKBP1 mutants - SHKBP1-R623A (denoted as R623A), SHKBP1-R684A (denoted as R684A) and SHKPB1-R623A/R684A (denoted as R623A/R684A) were created and tested in the yeast two-hybrid interaction assay. Yeast colonies with R623A plasmid showed a strong positive signal while colonies containing R684A plasmids only showed a weak positive signal in the LacZ assay (Table 1). In addition, colonies with the double mutant R623A/R684A plasmid showed no signal at all (Table 1). Taken together, these findings indicated that two PXXXPR motifs are responsible for the interaction of SHKBP1 with CIN85, although the contribution of the two motifs to the interaction seemed to be different.
Table 1. Yeast two-hybrid experiment examining the interaction of SHKBP1 or its mutants with CIN85.
MAV203 yeast strain was co-transformed with SHKBP1/pDBLeu or its mutants plasmids and CIN85/pPC86 plasmids. SHKBP1 or its mutants co-transformed with empty pPC86 and CIN85 co-transformed with empty pDBLeu were used as control. And pPC86 and pDBLeu vectors were used as negative control. Transformed yeasts were selected in SC-Leu-Trp plates, and a LacZ assay was performed with colonies grown on the SC-Leu-Trp plates and control yeast strains: a strong positive control A, a weak positive control C and a negative control supplied by manufacturer. (‘+++’ indicates a strong positive signal, ‘+’ indicates a weak positive signal and ‘−’ indicates a negative signal).
| Plasmids | SHKBP1 | SHKBP1- R623A |
SHKBP1- R684A |
SHKBP1- R623A/684A |
pDBLeu |
| CIN85 | CIN85 | CIN85 | CIN85 | pPC86 | |
| LacZ activity | +++ | +++ | + | − | − |
| Plasmids | SHKBP1 | SHKBP1- R623A |
SHKBP1- R684A |
SHKBP1- R623A/684A |
pDBLeu |
| pPC86 | pPC86 | pPC86 | pPC86 | CIN85 | |
| LacZ activity | − | − | − | − | − |
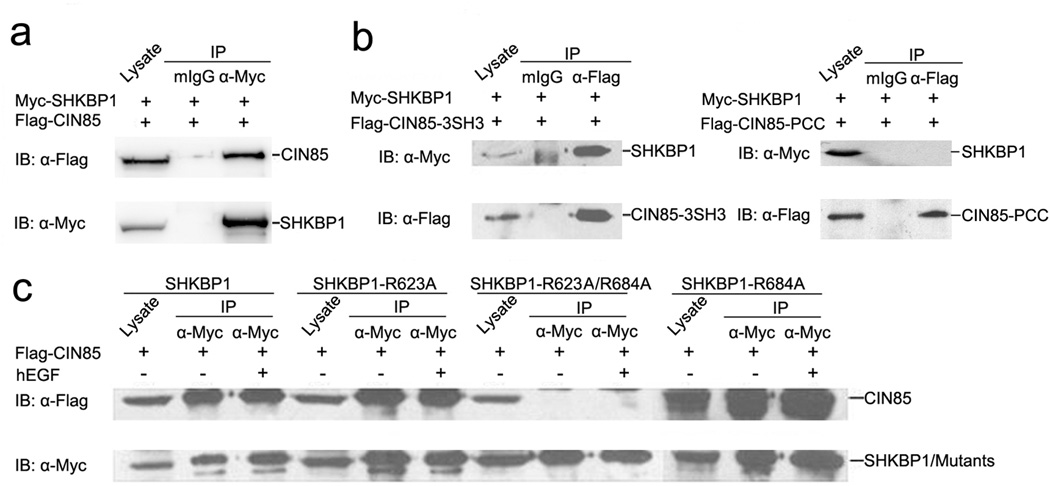
To confirm this interaction also occurs in mammalian cells, SHKBP1 expressing plasmid was co-transfected with CIN85-coding vector into HEK293T cells. Co-immunoprecipitation study demonstrated the interaction between SHKBP1 and CIN85 in the transfected cells (Figure 1a). Furthermore, SH3 domains (CIN85-3SH3) but not the proline-rich region and coiled-coil region (CIN85-PCC) contributed to the binding of CIN85 to SHKBP1 (Figure 1b). While single mutants R623A and R684A could bind to CIN85, the double mutant R623A/R684A failed to interact with CIN85 (Figure 1c). Interestingly, we found that the interaction of SHKBP1 with CIN85 was not affected by EGF stimulation (Figure 1c). The above data suggested a direct and constitutive interaction between SHKBP1 and CIN85, which was mediated by PXXXPR motifs of SHKBP1.
Figure 1. PXXXPR motifs of SHKBP1 constitutively bound to CIN85.
a HEK293T cells co-transfected with Myc-SHKBP1 and Flag-CIN85 plasmids were lysed 48 hours after transfection. Immuno-precipitations were performed with anti-Myc antibody and control mouse IgG (mIgG). Immuno-precipitated proteins were processed for western blot with anti-Flag antibody. The membrane was stripped and re-blotted with anti-Myc antibody.
b HEK293T cells co-transfected with Flag-CIN85-3SH3 or Flag-CIN85-PCC and Myc-SHKBP1 plasmids were lysed 48 hours after transfection and cell lysates were immuno-precipitated with anti-Flag antibody and control mIgG. Immuno-precipitated proteins were processed for western-blot with anti-Myc antibody. The membrane was stripped and re-blotted with anti-Flag antibody.
c HEK293T cells transfected with Myc-SHKBP1 or its mutants and Flag-CIN85 plasmids were starved in serum-free DMEM for twelve hours and then treated with 100 ng/ml hEGF for 10 minutes. The cells were lysed and immuno-precipitated with anti-Myc antibody. Immunoprecipitated proteins were processed for western-blot with anti-Flag antibody and the membrane was then stripped and re-blotted with anti-Myc antibody.
SHKBP1 competed with c-Cbl to bind to CIN85 through its two PXXXPR motifs
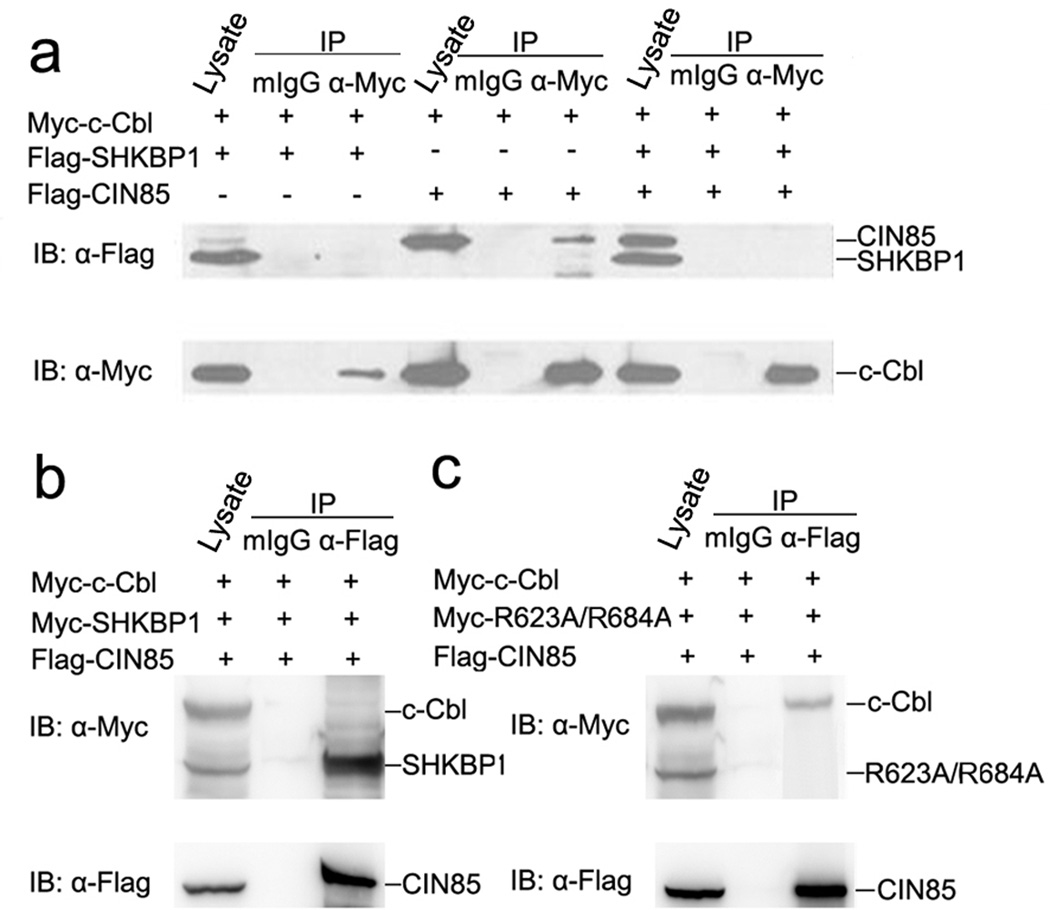
It was known that c-Cbl associated with CIN85 through the PXXXPR motif of c-Cbl and the SH3 domain(s) of CIN85, and EGF stimulation could further promote the assembly of this complex4,9. The dependence of the SHKBP1-CIN85 interaction on PXXXPR motifs of SHKBP1 led us to assume whether SHKBP1 could interfere with the association between c-Cbl and CIN85. As previously reported, c-Cbl could bind to CIN85 (Figure 2a). However, such an association was completely abolished in the presence of ectopically expressed SHKBP1 (Figure 2a). c-Cbl did not bind to SHKBP1 and failed to affect the association of SHKBP1 and CIN85 in co-transfected cells (Figure 2a and 2b). Importantly, the double mutant R623A/R684A failed to affect the interaction between c-Cbl and CIN85 (Figure 2c). These data indicated that SHKBP1 interfered with the formation of the c-Cbl-CIN85 complex through sequestration of CIN85 via its two PXXXPR motifs.
Figure 2. SHKBP1 competed with c-Cbl to bind to CIN85 through its two PXXXPR motifs.
a HEK293T cells co-transfected with expression plasmids encoding Myc-c-Cbl, Flag-SHKBP1 or Flag-CIN85 were lysed 48 hours after transfection and cell lysates were immuno-precipitated with anti-Myc antibody and control mIgG. Immuno-precipitated proteins were processed for western-blot with anti-Flag antibody and the membrane was stripped and re-blotted with anti-Myc antibody.
b HEK293T cells co-transfected with plasmids encoding Myc-c-Cbl, Myc-SHKBP1 and Flag-CIN85 were lysed 48 hours after transfection and cell lysates were immuno-precipitated with anti-Flag antibody and control mIgG. Immuno-precipitated proteins were processed for western-blot with anti-Myc antibody and the membrane was stripped and re-blotted with anti-Flag antibody.
c HEK293T cells co-transfected with plasmids encoding Myc-c-Cbl, Myc-R623A/684A and Flag-CIN85 were lysed 48 hours after transfection and cell lysates were immuno-precipitated with anti-Flag antibody and control mIgG. Immuno-precipitated proteins were processed for western-blot with anti-Myc antibody and the membrane was stripped and re-blotted with anti-Flag antibody.
SHKBP1 inhibited EGFR degradation after EGF stimulation
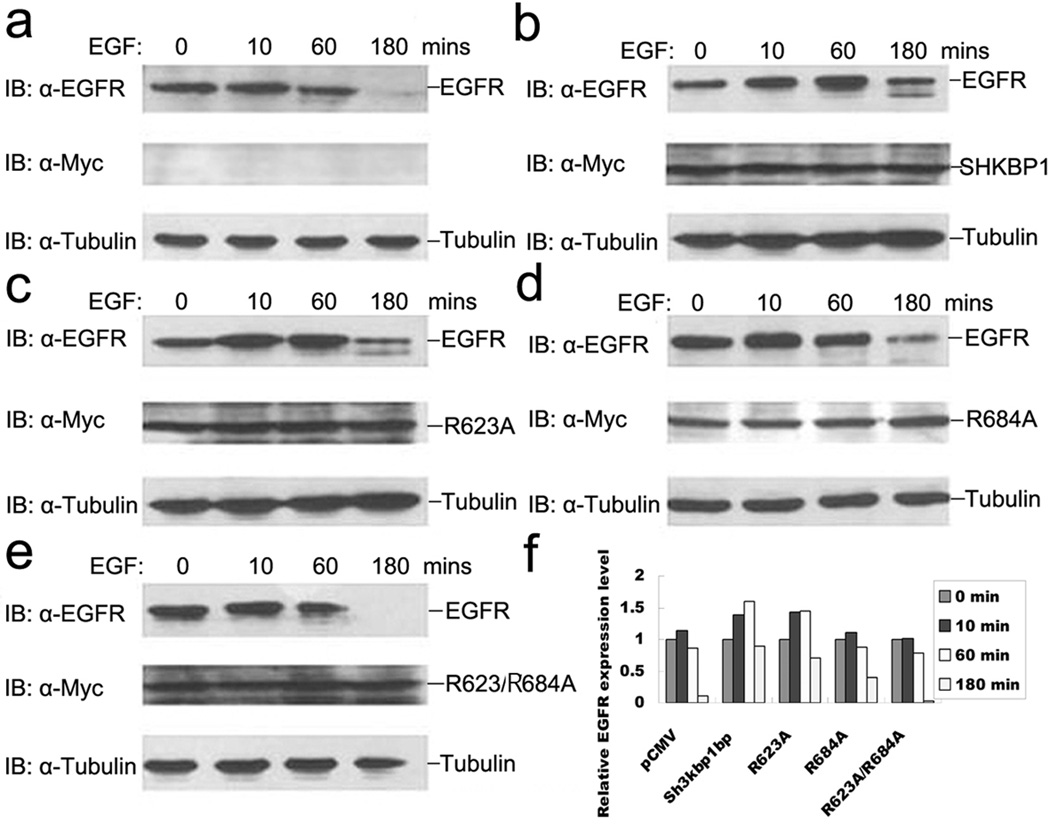
Next, we hypothesized that the direct binding of SHKBP1 to CIN85 would attenuate c-Cbl-CIN85 complex promoted EGFR degradation4. To test it, EGFR level in the transfected cells was measured following EGF stimulation of cells transected with wild type or various mutants of SHKBP1. As shown in Figure 3a, EGFR level was up-regulated immediately upon EGF stimulation and then reduced gradually. The EGFR protein was not detectable about 180 minutes after EGF stimulation in mock vector transfected cells (Figure 3a). Overexpression of wild type SHKBP1 and R623A mutant strongly delayed the degradation of EGFR upon EGF activation (Figure 3b and 3c). In contrast, the receptor degradation was only slightly delayed by the overexpression of R684A mutant (Figure 3d) while the R623A/R684A double mutant showed no effect at all (Figure 3e).
Figure 3. SHKBP1 inhibited EGFR degradation after EGF stimulation.
HeLa cells were transfected with indicated expression plasmids: pCMV (a), Myc-SHKBP1 (b), Myc-R623A (c), Myc-R684A (d) and Myc-R623A/684A (e), respectively. Thirty-six hours post-transfection, cells were starved in serum free DMEM for 12 hours. The cells were then stimulated with 100 ng/ml EGF for the indicated times. Lysates were prepared and analyzed by western-blot with anti-EGFR, anti-Myc or anti-α-tubulin antibodies. The relative level of EGFR (a–e) was quantified by densitometry scan and graphed in (f).
The expression level of SHKBP1 and its various mutants in transfected cells remained the same during the time course of EGF stimulation (Figure 3a–e). In contrast, quantitative analysis of relative EGFR levels in these cells clearly demonstrated changes in EGFR expression (Figure 3f). These data suggested that SHKBP1 interfered with the formation of c-Cbl-CIN85 complex by its binding to CIN85, thereby retarding c-Cbl-CIN85-dependent EGFR degradation after EGF stimulation.
SHKBP1 disturbed the translocation of CIN85 to EGFR containing vesicles
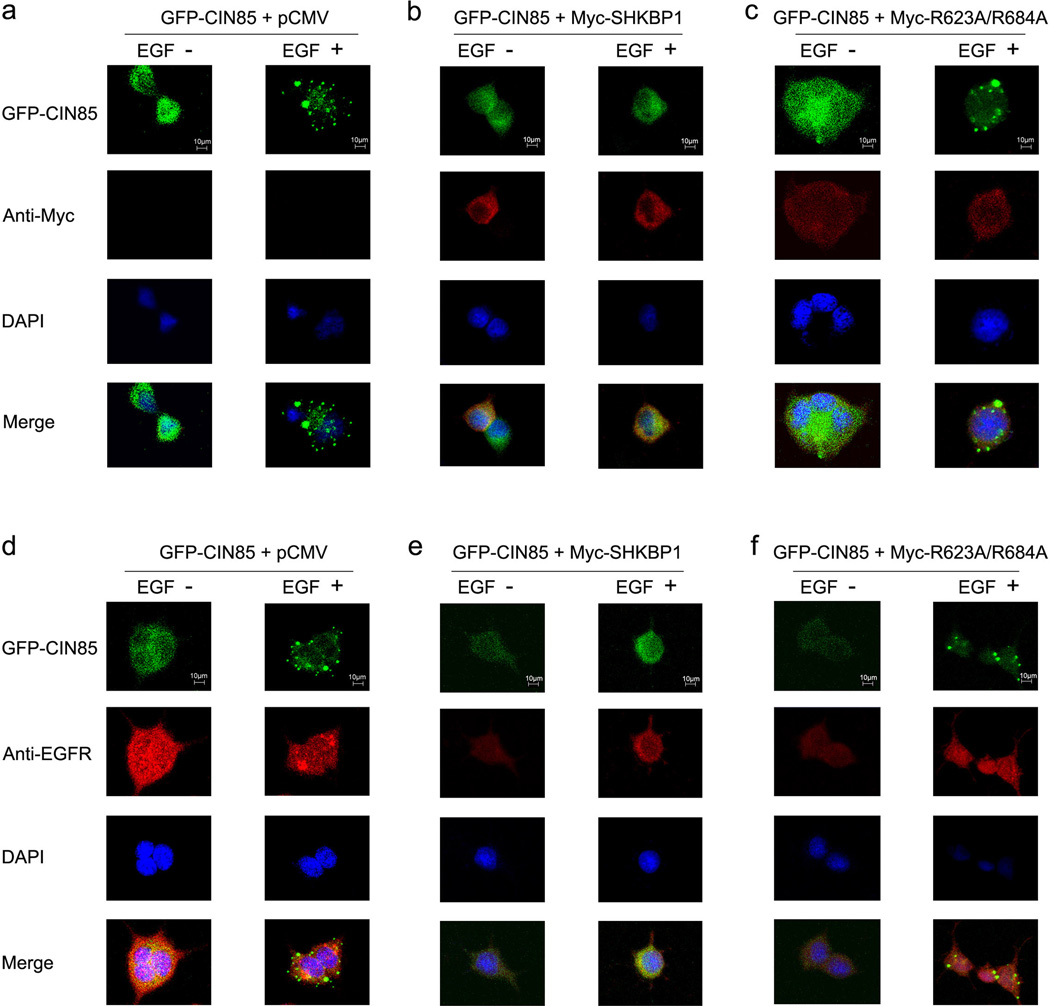
CIN85 was found to co-localize with EGFR and c-Cbl complex upon EGF stimulation which is important to receptor endocytosis and the formation of EGFR containing vesicles during EGFR degradation8,22. We therefore explored the localization of SHKBP1 and its role on CIN85 translocation after EGF stimulation by immuno-fluorescence. CIN85 translocated to EGFR containing vesicles upon EGF stimulation (Figure 4a and 4d). Over-expression of wild type SHKBP1 and single mutant R623A and R684A inhibited the formation of EGFR containing vesicles and the translocation of CIN85 (Figure 4b, 4e and Supplemental Figure 1). In contrast, expression of double mutant R623A/R684A did not disturb the translocation of CIN85 to EGFR containing vesicles (Figure 4c and 4f). These data suggested that SHKBP1 could disturb the translocation of CIN85 to EGFR containing vesicles upon EGF stimulation via the interaction of its PXXXPR motifs with CIN85.
Figure 4. SHKBP1 disturbed the translocation of CIN85 to EGFR degradation vesicles.
HEK293T cells were co-transfected with GFP-CIN85 plasmid and control pCMV mock vector (a, d), Myc-SHKBP1 (b, e) or Myc-R623A/R684A (c, f). Thirty-six hours post-transfection cells were starved in serum free DMEM for 12 hours. The cells were left unstimulated or stimulated with hEGF for 10 minutes. Immunofluorscence studies on cells were performed with antibodies against Myc tag (a, b, c) or EGFR (d, e, f), and the nucleus were stained with DAPI. The merged pictures were shown in the bottom panel, and the staffs (10µm) were shown in the upper panel.
SHKBP1 enhanced EGF induced SRE transcription activity
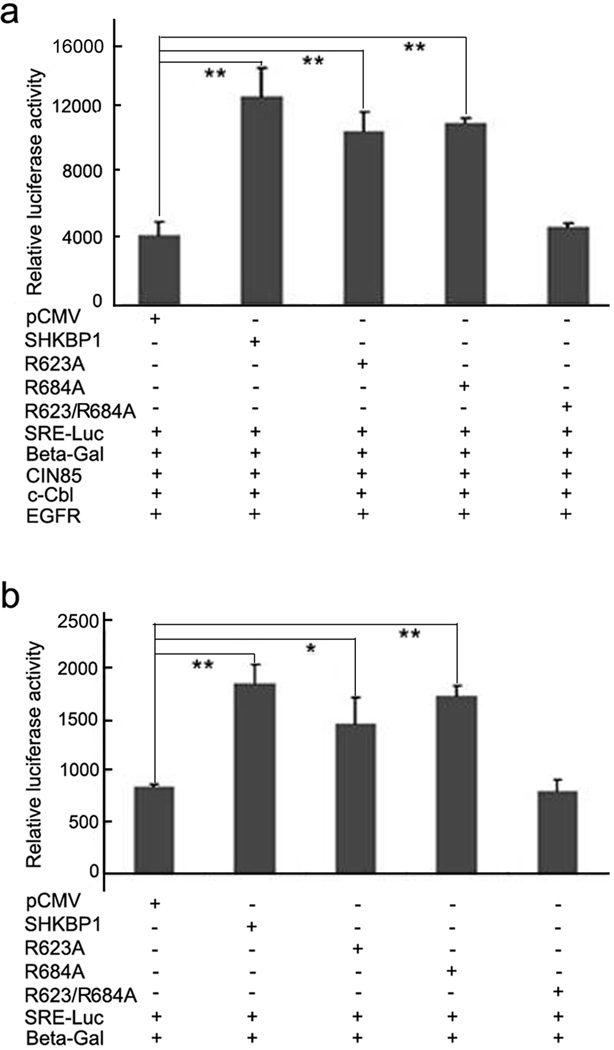
Finally, we examined whether the reduction in EGFR degradation due to over-expression of SHKBP1 would lead to the alteration in the downstream signaling. It was known that activated EGFR can induce transcription activity of promoters with the serum response element (SRE)18. Therefore, the SRE luciferase reporter assay was used to examine the effect of SHKBP1 on EGFR signaling. Different amount of SHKBP1 was co-transfected with the SRE-Luc gene reporter, c-Cbl, CIN85 and EGFR into HEK293T cells and the luciferase activities in transfected cells were measured. The luciferase activities showed a dose-dependent effect of SHKBP1 on SRE transcription (Supplemental Figure 2). Moreover, overexpression of SHKBP1, R623A or R684A mutant could significantly increase SRE transcription activity by 3.0 fold (P=0.001), 2.5 fold (P=0.003) and 2.6 fold (P=0.002), when compared to mock vector group, respectively (Figure 5a). Similarly, overexpression of SHKBP1, R623A or R684A mutants in HeLa cells could increase SRE transcription activity by 2.2 fold (P=0.003), 1.7 fold (P=0.036) and 2.0 fold (P=0.001), respectively (Figure 5b). However, R623A/R684A double mutant had little effect on SRE-Luc reporter transcription in both HEK293T and HeLa cells, indicating that SHKBP1 could enhance EGFR signaling through its interaction with CIN85.
Figure 5. SHKBP1 enhanced EGF induced SRE transcription activity.
a HEK293T cells were co-transfected with EGFR (100ng), c-Cbl (100ng), CIN85 (100ng), SRE-Luc (100ng), β-gal (50ng), and SHKBP1 or its mutants plasmids (300ng). Thirty-six hours post-transfection cells were starved in serum free DMEM for 12 hours. The cells were then stimulated with EGF for another 12 hours and processed for luciferase assay.
b HeLa cells were co-transfected with SRE-Luc (200ng), β-gal (100ng), and SHKBP1 or its mutants plasmids (300ng). Thirty-six hours post-transfection cells were starved in serum free DMEM for 12 hours. The cells were then stimulated with EGF for additional 12 hours and processed for luciferase assay. The luciferase activity was measured in triplicated samples and expressed as the mean ± s.d. Student’s t-test was performed to compare the statistical difference between each group to pCMV control group; ‘**’ indicates p<0.01, ‘*’ indicates p<0.05.
Discussion
In the current study, we found that SHKBP1 could bind to SH3 domains of CIN85 through its PXXXPR motifs in the carboxyl terminus (Figure 1 and Table 1). We further showed that the interaction of SHKBP1 with CIN85 could abolish c-Cbl-CIN85 complex formation (Figure 2), thus retarding EGFR degradation by disturbing the translocation of CIN85 to EGFR containing vesicles following EGF stimulation (Figure 3, Figure 4 and Supplemental Figure 1). Consistent with these observations, SHKBP1 enhanced EGFR signaling induced SRE transcriptional reporter activity which required at least one intact PXXXPR motif (Figure 5 and Supplemental Figure 2).
The binding of EGF to EGFR induces receptor activation and downstream signaling cascades important to cell proliferation, differentiation and migration1–2,4. Aberrant activation or overexpression of EGFR is commonly found in various cancers. It is clear that proper regulation of EGFR signaling is essential for normal physiology1,6. The c-Cbl-CIN85 complex has been shown to play a critical role in the down-regulation of activated EGFR. c-Cbl is associated with the cytoplasmic domain of EGFR upon receptor activation and catalyzes the ubiqutination of EGFR7. c-Cbl acts as a multifunctional adaptor protein to recruit CIN85 to the EGFR-c-Cbl complex, which facilitate the endocytosis of ubiquinitated EGFR and subsequent degradation. Therefore, the perturbation of EGFR-c-Cbl-CIN85 complex could have severe consequences in EGFR-regulated cell growth control. This regulatory mechanism has been reported for many c-Cbl associated proteins. For example, Sprouty2 not only acts at the interface of the c-Cbl-CIN85 complex, but also directly associates with c-Cbl to sequester c-Cbl from activated EGFR, thus inhibiting the down-regulation of EGFR signaling15–16. Additionally, Cool-1, a nucleotide exchange factor for Cdc42, is able to form a complex with c-Cbl, thereby sequestering c-Cbl from activated EGFR and preventing the down-regulation of EGFR signaling17.
We found SHKBP1 as a new regulator in EGFR activation. Unlike Sprouty2 and Cool-1 that promote EGFR signaling through their interaction with c-Cbl, SHKBP1 could retard EGFR degradation and up-regulate EGFR signaling by binding to CIN85 and sequestering CIN85 from c-Cbl-CIN85 complex. PXXXPR motifs were essential for this function of SHKBP1. Therefore, our results may reveal a new mechanism of enhanced EGFR signaling in the development of various diseases such as cancers. It would be interesting to investigate whether the interaction of SHKBP1 with CIN85 is also involved in the regulation of other receptor tyrosine kinases (RTKs) during cancer development.
In summary, our study demonstrated that SHKBP1 inhibited c-Cbl-CIN85 complex mediated down-regulation of EGFR signaling by sequestration of CIN85. This novel mechanism provides an alternative route for the development of drugs targeting the EGFR signaling pathway.
Supplementary Material
Acknowledgments
We thank Dr. Biao Wang, Prof. Zhengjun Chen and Dr. Bernard Roizman for providing cDNAs and constructs, and Dr. Qiu Chen for advising.
Abbreviations
- SHKBP1
SH3KBP1 binding protein 1
- CIN85
Cbl-interacting protein of 85 kDa
- Cbl
Casitas B-lineage Lymphoma
- EGFR
Epidermal growth factor receptor
- SRE
Serum response element
- Leu
Leucine
- Trp
tryptophan
- Arg
Arginine
Contributor Information
Lifeng Feng, Email: zjucrazy@hotmail.com.
Jin-Tao Wang, Email: jwang@umich.edu.
Hongchuan Jin, Email: jinhc@srrsh.com.
Reference
- 1.Dikic I, Giordano S. Negative receptor signalling. Curr Opin Cell Biol. 2003;15:128–135. doi: 10.1016/s0955-0674(03)00004-8. [DOI] [PubMed] [Google Scholar]
- 2.Haglund K, Shimokawa N, Szymkiewicz I, Dikic I. Cbl-directed monoubiquitination of CIN85 is involved in regulation of ligand-induced degradation of EGF receptors. Proc Natl Acad Sci U S A. 2002;99:12191–12196. doi: 10.1073/pnas.192462299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider MR, Sibilia M, Erben RG. The EGFR network in bone biology and pathology. Trends Endocrinol Metab. 2009;20:517–524. doi: 10.1016/j.tem.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 5.Waterman H, Yarden Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 2001;490:142–152. doi: 10.1016/s0014-5793(01)02117-2. [DOI] [PubMed] [Google Scholar]; Shepard HM, Brdlik CM, Schreiber H. Signal integration: a framework for understanding the efficacy of therapeutics targeting the human EGFR family. J Clin Invest. 2008;118:3574–3581. doi: 10.1172/JCI36049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saif MW. Colorectal cancer in review: the role of the EGFR pathway. Expert Opin Investig Drugs. 2010;19:357–369. doi: 10.1517/13543781003593962. [DOI] [PubMed] [Google Scholar]
- 7.Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 8.Szymkiewicz I, Kowanetz K, Soubeyran P, Dinarina A, Lipkowitz S, Dikic I. CIN85 participates in Cbl-b-mediated down-regulation of receptor tyrosine kinases. J Biol Chem. 2002;277:39666–39672. doi: 10.1074/jbc.M205535200. [DOI] [PubMed] [Google Scholar]
- 9.Pennock S, Wang Z. A tale of two Cbls: interplay of c-Cbl and Cbl-b in epidermal growth factor receptor downregulation. Mol Cell Biol. 2008;28:3020–3037. doi: 10.1128/MCB.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan YH, Krishnaswamy S, Nandi S, Kanteti R, Vora S, Onel K, Hasina R, Lo FY, El-Hashani E, Cervantes G, Robinson M, Kales SC, Lipkowitz S, Karrison T, Sattler M, Vokes EE, Wang YC, Salgia R. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS One. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jozic D, Cardenes N, Deribe YL, Moncalian G, Hoeller D, Groemping Y, Dikic I, Rittinger K, Bravo J. Cbl promotes clustering of endocytic adaptor proteins. Nat Struct Mol Biol. 2005;12:972–979. doi: 10.1038/nsmb1000. [DOI] [PubMed] [Google Scholar]
- 12.Take H, Watanabe S, Takeda K, Yu ZX, Iwata N, Kajigaya S. Cloning and characterization of a novel adaptor protein, CIN85, that interacts with c-Cbl. Biochem Biophys Res Commun. 2000;268:321–328. doi: 10.1006/bbrc.2000.2147. [DOI] [PubMed] [Google Scholar]; Watanabe S, Take H, Takeda K, Yu ZX, Iwata N, Kajigaya S. Characterization of the CIN85 adaptor protein and identification of components involved in CIN85 complexes. Biochem Biophys Res Commun. 2000;278:167–174. doi: 10.1006/bbrc.2000.3760. [DOI] [PubMed] [Google Scholar]
- 13.Dikic I. CIN85/CMS family of adaptor molecules. FEBS Lett. 2002;529:110–115. doi: 10.1016/s0014-5793(02)03188-5. [DOI] [PubMed] [Google Scholar]
- 14.Kowanetz K, Husnjak K, Holler D, Kowanetz M, Soubeyran P, Hirsch D, Schmidt MH, Pavelic K, De Camilli P, Randazzo PA, Dikic I. CIN85 associates with multiple effectors controlling intracellular trafficking of epidermal growth factor receptors. Mol Biol Cell. 2004;15:3155–3166. doi: 10.1091/mbc.E03-09-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Bar-Sagi D. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr Biol. 2003;13:308–314. doi: 10.1016/s0960-9822(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 16.Haglund K, Schmidt MH, Wong ES, Guy GR, Dikic I. Sprouty2 acts at the Cbl/CIN85 interface to inhibit epidermal growth factor receptor downregulation. EMBO Rep. 2005;6:635–641. doi: 10.1038/sj.embor.7400453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Q, Baird D, Peng X, Wang J, Ly T, Guan JL, Cerione RA. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat Cell Biol. 2006;8:945–956. doi: 10.1038/ncb1453. [DOI] [PubMed] [Google Scholar]
- 18.Liang Y, Kurakin A, Roizman B. Herpes simplex virus 1 infected cell protein 0 forms a complex with CIN85 and Cbl and mediates the degradation of EGF receptor from cell surfaces. Proc Natl Acad Sci U S A. 2005;102:5838–5843. doi: 10.1073/pnas.0501253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HB, Wang JT, Zhang L, Geng ZH, Xu WL, Xu T, Huo Y, Zhu X, Plow EF, Chen M, Geng JG. P-selectin primes leukocyte integrin activation during inflammation. Nat Immunol. 2007;8:882–892. doi: 10.1038/ni1491. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Xiao Y, Ding BB, Zhang N, Yuan X, Gui L, Qian KX, Duan S, Chen Z, Rao Y, Geng JG. Induction of tumor angiogenesis by Slit-Robo signaling and inhibition of cancer growth by blocking Robo activity. Cancer Cell. 2003;4:19–29. doi: 10.1016/s1535-6108(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 21.Criqui-Filipe P, Ducret C, Maira SM, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 1999;18:3392–3403. doi: 10.1093/emboj/18.12.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Zheng X, Yang X, Liao K. CIN85 associates with endosomal membrane and binds phosphatidic acid. Cell Res. 2009;19:733–746. doi: 10.1038/cr.2009.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.