Abstract
Background
Unlike mammals, zebrafish have the ability to regenerate damaged parts of their central nervous system (CNS) and regain functionality of the affected area. A better understanding of the molecular mechanisms involved in zebrafish regeneration may therefore provide insight into how CNS repair might be induced in mammals. Although many studies have described differences in gene expression in zebrafish during CNS regeneration, the regulatory mechanisms underpinning the differential expression of these genes have not been examined.
Results
We used microarrays to analyse and integrate the mRNA and microRNA (miRNA) expression profiles of zebrafish retina after optic nerve crush to identify potential regulatory mechanisms that underpin central nerve regeneration. Bioinformatic analysis identified 3 miRNAs and 657 mRNAs that were differentially expressed after injury. We then combined inverse correlations between our miRNA expression and mRNA expression, and integrated these findings with target predictions from TargetScan Fish to identify putative miRNA-gene target pairs. We focused on two over-expressed miRNAs (miR-29b and miR-223), and functionally validated seven of their predicted gene targets using RT-qPCR and luciferase assays to confirm miRNA-mRNA binding. Gene ontology analysis placed the miRNA-regulated genes (eva1a, layna, nefmb, ina, si:ch211-51a6.2, smoc1, sb:cb252) in key biological processes that included cell survival/apoptosis, ECM-cytoskeleton signaling, and heparan sulfate proteoglycan binding,
Conclusion
Our results suggest a key role for miR-29b and miR-223 in zebrafish regeneration. The identification of miRNA regulation in a zebrafish injury model provides a framework for future studies in which to investigate not only the cellular processes required for CNS regeneration, but also how these mechanisms might be regulated to promote successful repair and return of function in the injured mammalian brain.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1772-1) contains supplementary material, which is available to authorized users.
Keywords: Zebrafish, microRNA, mRNA, Optic nerve, Regeneration, CNS
Background
The regenerative capacity of the CNS in adult mammals is limited, with minimal axonal re-growth, death of damaged neurons and long-term loss of function [1]. By contrast, fish have the remarkable ability to repair most functional components of the CNS [2, 3] making them a valuable model in which to identify key molecular mechanisms involved in neural regeneration [4–6]. Anamniotes (eg fish) and amniotes (eg mammals) have a high degree of conservation at the level of both nucleotide sequence and amino acid functional domains, resulting in similar molecular and cellular processes in neural development and function of both clades. Therefore, it is possible that genes required for successful CNS regeneration are present in both fish and mammals, but that key regulatory differences in the expression of these genes underpin the differing levels of neuronal survival and axonal regeneration in different species.
MicroRNAs (miRNAs) are an extensive subclass of regulatory non-coding RNAs that repress gene expression at a post-transcriptional level by affecting mRNA translation and stability [7]. miRNAs have been implicated in many aspects of development and homeostatic pathways, with their actions often becoming more pronounced under conditions of physiological or pathological stress [8]. miRNAs are highly abundant in the CNS, which is perhaps unsurprising given the cellular and transcriptional complexity of this tissue [9]. The high degree of conservation of miRNAs across species, combined with their ability to target multiple genes, make them a likely regulator in fundamental processes, such as the ability to regenerate neural pathways in the CNS [10, 11].
To gain further insight into the regulatory mechanisms activated following a CNS lesion, the present study sought to identify miRNA(s) that were altered in zebrafish retina after an optic nerve (ON) crush. This model provides easy access to CNS-derived retinal ganglion cells (RGCs) and their axons that connect with the optic tectum, thereby allowing examination of both cell survival and axonal repair. Similar tissue structure across species has allowed comparison between ON injury in a variety of vertebrate species (e.g. mammals, reptiles, amphibians, fish) as a means to investigate the molecular basis of CNS regeneration [12–15]. The most dynamic gene changes occur approximately 3–4 days post injury suggesting that this phase is a critical point that sets the scene for successful regeneration or failure [12, 13]. During this period in zebrafish, previous studies have suggested that the injured RGCs are geared towards maintaining and enhancing survival whilst simultaneously preparing for axonal outgrowth [13, 16, 17].
In this study we performed a bioinformatic integration of mRNA and miRNA expression profiles of the zebrafish retina 3 days after injury, with the experimental outline represented in Fig. 1. We assessed changes in miRNA expression within the same RNA pool used for mRNA profiling as means of increasing the likelihood of finding true biological miRNA-gene relationships. The integration of these lists revealed three miRNAs that were significantly over-expressed after injury. We focused on two of these, miR-29b and miR-223, and validated seven of their target genes that were under-expressed in our dataset. Our results identified apoptotic signalling, cytoskeletal dynamics and extracellular matrix interactions as key processes activated in zebrafish following regeneration. Our data suggest that miRNAs are potential molecular targets that may be used to regulate these multiple processes in an orchestrated fashion to promote CNS regeneration.
Fig. 1.
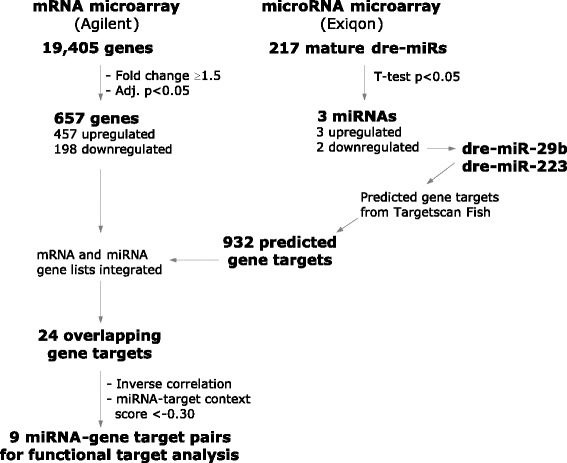
Schematic of experimental outline. Schematic flow diagram of procedures used to identify inversely correlated putative target genes of the differentially expressed miRNAs
Results
Injury-induced changes in gene expression and pathways
To assess the changes in gene expression after an optic nerve crush injury, we examined mRNA from whole zebrafish retinae 3 days after injury. Using an absolute log2-fold change cut-off of 1.5 and an adjusted p-value of ≤0.05, 804 transcripts were identified as differentially expressed between uninjured (control) and injured (crush) tissue and this equated to 657 genes (Additional file 1: Figure S1). Of these, 459 genes were over-expressed due to nerve injury, whilst 198 genes were under-expressed. The microarray data are available from the Gene Expression Omnibus GSE70261, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70261, and full analyses provided in the Additional Files. The top over- and under-expressed genes, ranked by a combination of p-value and absolute log fold change ≥1.5, are listed in Tables 1 and 2 respectively.
Table 1.
Top upregulated genes after optic nerve crush
| Ensembl ID | Gene symbol | Gene name | Fold change | adjusted p-value | GO process or functiona |
|---|---|---|---|---|---|
| ENSDART00000081039 | sb:cb252 | sb:cb252 | 52.9 | 0.000065 | Associated with mitochondria |
| ENSDART00000025036 | gap43 | Growth associated protein 43 | 33.9 | 0.000161 | Tissue regeneration |
| ENSDART00000140944 | cremb | cAMP responsive element modulator b | 20.8 | 0.000065 | DNA-dependent transcription |
| ENSDART00000022060 | atf3 | Activating transcription factor 3 | 13.5 | 0.000279 | DNA-dependent transcription |
| ENSDART00000101970 | CU571382.1 | Uncharacterised protein | 10.7 | 0.000151 | |
| ENSDART00000126441 | lepa | Leptin a | 9.8 | 0.000161 | Nervous system development |
| ENSDART00000110691 | wnt6b | Wingless-type MMTV integration site family, member 6b | 14.6 | 0.000690 | wnt receptor signaling; neuron differentiation |
| ENSDART00000127420 | mdp1 | Magnesium-dependent phosphatase 1 | 6.6 | 0.000065 | Protein tyrosine phosphatase activity |
| ENSDART00000105597 | si:ch211-129c21.1 | si:ch211-129c21.1 | 9.7 | 0.000279 | Multicellular organismal development |
| ENSDART00000062845 | mmp9 | Matrix metalloproteinase 9 | 11.1 | 0.000558 | Proteolysis |
| ENSDART00000060765 | BX323876.3 | Brain natriuretic peptide-like | 9.7 | 0.000396 | Inflammatory response |
| ENSDART00000077197 | tmsb | Thymosin, beta | 11.1 | 0.000801 | Organization of cytoskeleton |
| ENSDART00000034377 | cpa5 | Carboxypeptidase A5 | 10.5 | 0.000690 | Proteolysis |
| ENSDART00000064789 | txn | Thioredoxin | 5.0 | 0.000065 | Cell redox homeostasis |
| ENSDART00000033494 | klf6a | Kruppel-like factor 6a | 4.7 | 0.000065 | Optic nerve formation |
| ENSDART00000064509 | stmn4l | Stathmin-like 4, like | 5.8 | 0.000161 | Regulation of microtubule (de)polymerization |
| ENSDART00000129989 | C14HXorf65 | Chromosome X open reading frame 65 | 5.0 | 0.000100 | |
| ENSDART00000144946 | adcyap1b | Adenylate cyclase activating polypeptide 1b | 7.3 | 0.000409 | Brain development |
| ENSDART00000020673 | f3a | Coagulation factor IIIa | 5.5 | 0.000161 | Blood coagulation; integral to membrane functioning |
| ENSDART00000133512 | fosl1b | FOS-like antigen 1b | 6.6 | 0.000409 | DNA-dependent transcription |
| ENSDART00000045410 | thy1 | Thy-1 cell surface antigen | 4.8 | 0.000161 | Organization of cytoskeleton; focal adhesion |
| ENSDART00000123518 | tuba1 | Tubulin, alpha 1 | 4.7 | 0.000161 | Optic nerve formation; regulation of microtubule processes |
| ENSDART00000101424 | nefma | Neurofilament, medium polypeptide a | 4.5 | 0.000161 | Organization of cytoskeleton |
| ENSDART00000121861 | prph | Peripherin | 10.0 | 0.002781 | Organization of cytoskeleton; tissue regeneration |
| ENSDART00000082153 | itga6a | Integrin, alpha 6a | 5.4 | 0.000409 | Cell adhesion membrane functioning; integrin signalling |
| ENSDART00000129227 | b3gnt2 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 | 4.8 | 0.000279 | Protein glycosylation |
| ENSDART00000141068 | sox11b | SRY-box containing gene 11b | 5.2 | 0.000381 | Neuron differentiation; response to wounding |
Ranked by combined –log10 p-value and ≥1.5 absolute fold change
aGO terms determined by ZFIN and Uniprot databases
Table 2.
Top downregulated genes after optic nerve crush
| Ensembl ID | Gene symbol | Gene name | Fold change | Adjusted p-value | GO process or functiona |
|---|---|---|---|---|---|
| ENSDART00000055936 | isl2b | islet2b | 0.18 | 0.000315 | DNA-dependent transcription |
| ENSDART00000137322 | kcnip3 | Kv channel interacting protein 3, calsenilin | 0.29 | 0.000151 | Potassium channel activity; neuronal apoptotic processes |
| ENSDART00000051693 | irx4a | Iroquois homeobox protein 4a | 0.15 | 0.002945 | DNA-dependent transcription |
| ENSDART00000082745 | emb | Embigin | 0.26 | 0.001049 | Cell adhesion; integral to membrane functioning |
| ENSDART00000090267 | scn4ba | Sodium channel, voltage-gated, type IV, beta a | 0.28 | 0.000848 | Voltage-gated sodium channel activity |
| ENSDART00000052338 | irx4b | Iroquois homeobox protein 4b | 0.24 | 0.001560 | DNA-dependent transcription |
| ENSDART00000052802 | calb2b | Calbindin 2b, (calretinin) | 0.26 | 0.001408 | Calcium ion binding; neuronal excitability |
| ENSDART00000027398 | kcna2 | Potassium voltage-gated channel, Shaker-related subfamily, member 2 | 0.33 | 0.000641 | Potassium ion transport; synaptic transmission |
| ENSDART00000067514 | rbpms2a | RNA binding protein with multiple splicing 2a | 0.36 | 0.000558 | Nucleotide binding |
| Unknown | Unknown | 0.44 | 0.000407 | ||
| ENSDART00000018351 | zgc:65851 | Paralog of internexin neuronal intermediate filament protein, alpha | 0.28 | 0.001732 | Neurofilament cytoskeleton organization |
| ENSDART00000134832 | rbpms2b | RNA binding protein with multiple splicing 2b | 0.42 | 0.000690 | Nucleic acid binding |
| ENSDART00000114765 | kcnd1 | Potassium voltage-gated channel, Shal-related subfamily, member 1 | 0.42 | 0.000917 | Potassium ion transport; synaptic transmission |
| ENSDART00000113796 | cacnb3b | Calcium channel, voltage-dependent, beta 3b | 0.42 | 0.001316 | Calcium ion transmembrane transport |
| ENSDART00000064012 | ca4a | Carbonic anhydrase IV a | 0.52 | 0.000690 | Carbonate dehydratase activity; zinc ion binding |
| ENSDART00000113081 | gpr158 | G-protein coupled receptor 158 | 0.51 | 0.000947 | G-protein coupled receptor activity |
| ENSDART00000124112 | pou4f2 | POU domain, class 4, transcription factor 2 | 0.30 | 0.006744 | DNA-dependent transcription |
| ENSDART00000031167 | tfap2d | Transcription factor AP-2 delta | 0.43 | 0.001926 | DNA-dependent transcription |
| ENSDART00000031091 | vsnl1a | Visinin-like 1a | 0.45 | 0.001678 | Calcium ion binding |
| ENSDART00000077838 | ryr3 | Ryanodine receptor 3 | 0.51 | 0.001445 | Calcium ion transmembrane transport |
| ENSDART00000098599 | si:ch211-151 h10.2 | Uncharacterised protein | 0.44 | 0.002573 | |
| ENSDART00000127084 | LOC100537452 | Uncharacterized protein | 0.45 | 0.002423 | |
| ENSDART00000084303 | kcnq3 | Potassium voltage-gated channel, KQT-like subfamily, member 3 | 0.45 | 0.002495 | Potassium ion transport; synaptic transmission |
| ENSDART00000055281 | kcnc3b | Potassium voltage-gated channel, Shaw-related subfamily, member 3b | 0.53 | 0.001560 | Potassium ion transport; synaptic transmission |
| ENSDART00000126365 | smoc1 | SPARC related modular calcium binding 1 | 0.41 | 0.004141 | Extracellular matrix; calcium ion binding |
| ENSDART00000090092 | ank1 | Ankyrin 1, erythrocytic | 0.43 | 0.003530 | Cytoskeletal adaptor activity |
| ENSDART00000023562 | syt2 | Synaptotagmin 2 | 0.50 | 0.002264 | Calcium ion binding; synaptic transmission |
| ENSDART00000105932 | si:dkeyp-110e4.11 | Uncharacterised protein | 0.43 | 0.004012 |
Ranked by combined –log10 p-value and ≥1.5 absolute fold change
aGO terms determined by ZFIN and Uniprot databases
To determine the biological and functional implications of these expression changes by analysing over- versus under-expressed genes separately, we performed GO analysis and functional enrichment on our differentially expressed genes using WebGestalt and IPA, respectively. Figures 2 and 3 show the trimmed enriched GO terms for over- and under-expressed genes. Additional file 2: Table S1 and Additional file 3: Table S2 contain the untrimmed enriched GO terms, including IDs and p-values, with the full gene lists available in the Supporting data.
Fig. 2.
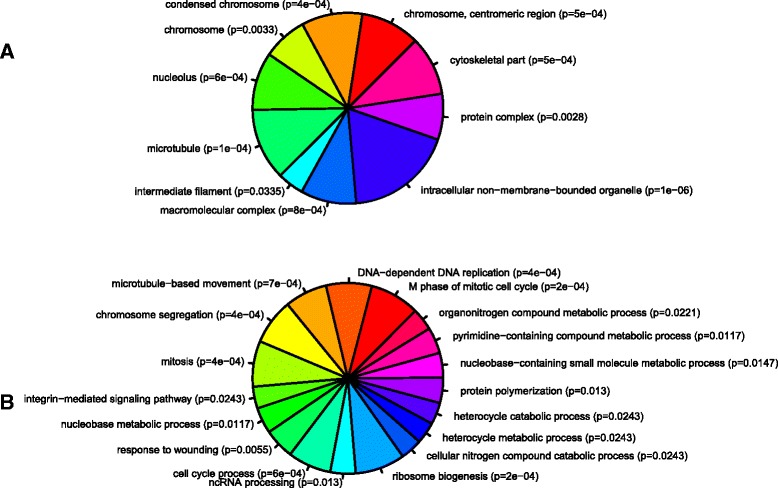
Gene ontology of over-expressed genes. GO analysis of enriched (a) cellular components and (b) biological processes of the over-expressed gene set. Each part represents -log2 of the p-value associated with the GO category from WebGestalt. WebGestalt results were filtered through GOTrim and presented only GO terms with ≥5 genes. Molecular function not shown as no terms passed this criteria.
Fig. 3.
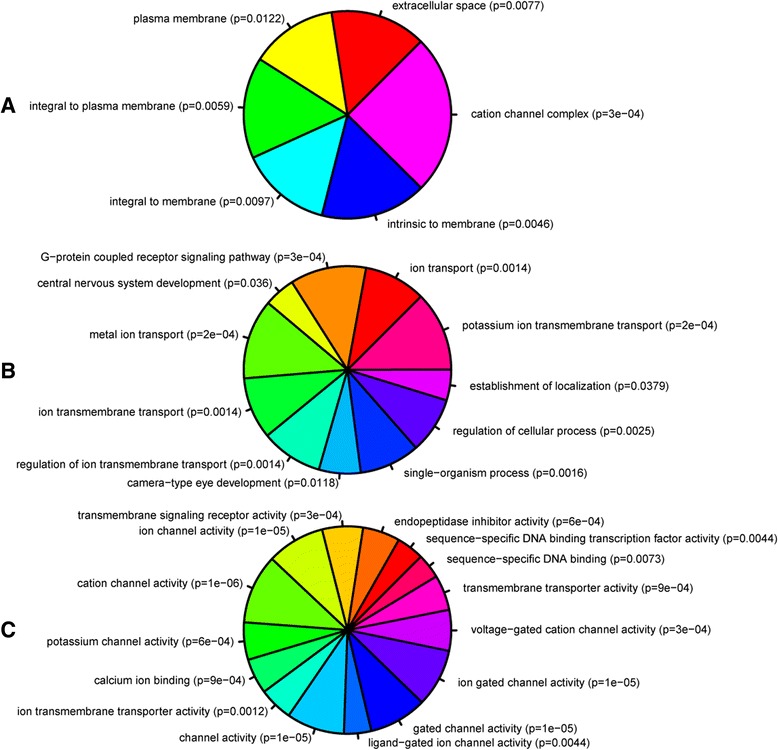
Gene ontology of under-expressed genes. GO analysis of enriched (a) cellular component, (b) biological processes, and (c) molecular functions of the under-expressed gene set. Each part represents -log2 of the p-value associated with the GO category from WebGestalt. WebGestalt results were filtered through GOTrim and presented only GO terms with ≥5 genes.
Enriched processes associated with over-expressed genes (Fig. 2) included ribosomal complex biogenesis, microtubule-based processes, and cell cycle activities. Other relevant processes enriched in our dataset included response to wounding (which included commonly upregulated genes, i.e. sox11b, mmp9, prph, gap43), and intracellular mediation of axon guidance signals (e.g. crmp2, crmp3, crmp5a). Our data also highlight for the first time that genes involved in non-coding RNA processing were enriched following optic nerve crush, implying that non-coding RNAs might orchestrate the regeneration process.
GO terms over-represented in the under-expressed genes (Fig. 3) revealed processes associated with eye development, DNA binding activity, G-protein coupled receptor signaling, and ion transport. In particular, transcription factors involved in eye development showed decreased expression after injury (e.g. vax1, irx4a, pou4f2, isl2b, tbr1b, tfap2d). The genes involved in the G-protein signaling pathways appeared to be associated with neuropeptide signaling (e.g. tacr2, drd2a, avpr2l, pdyn, p2rx1). Furthermore, there was a significant reduction in expression of ion transport genes, focusing on potassium and calcium channel activity.
Pathway analysis of the 804 probe identifiers uploaded into IPA resulted in only a subset being used, as 481 probes were annotated but only 431 mapped to a biological function. The over-expressed genes were associated with 71 significantly enriched canonical pathways (Fig. 4a), with the top ten pathways including cell cycle regulation, 14-3-3 and apoptosis signalling, consistent with GO term results. There were fewer canonical pathways enriched in the under-expressed gene set (7 pathways), but these pathways were associated with inflammatory and immune responses, as well as calcium signaling (Fig. 4b). The majority of inflammation-associated genes were found in the acute phase response pathway (comprising serping1, serpina1, a2m and hpx). Genes specific to calcium signaling included chrna6, chrna7, ryr1, camkk1.
Fig. 4.

Enriched IPA canonical pathways of genes that were differentially expressed after injury. (a) Over-expressed and (b) under-expressed genes are shown
Further functional analysis revealed the relationship between genes, by placing associated genes into networks related to specific biological processes. Over-expressed genes fell into networks associated with dermatological diseases and conditions, developmental and hereditary disorders Additional file 4: Figure S2. In contrast, under-expressed genes were associated with networks involved in neurological disease, development and function of the nervous system, and organ morphology Additional file 5: Figure S3.
Changes in miRNA expression and integration with mRNA profile identify candidate miRNA-mRNA target pairs involved in nerve regeneration
In order to examine the regulation of gene expression by miRNAs after optic nerve injury, we assessed changes in miRNA expression using the same RNA pool used for mRNA profiling. Of the 217 zebrafish specific miRNAs on the platform, we found 3 to be significantly altered 3 days after nerve crush (Table 3; all miRNA data provided in the Supporting data). We focused our subsequent investigation on two of the miRNAs over-expressed after nerve injury, miR-29b and miR-223, as their increased expression (16 and 55 % increase, respectively) is pertinent when considering the role of miRNAs is to negatively regulate their target genes [18], and unlike miR-21 [19], they have not been extensively studied in the brain.
Table 3.
Top miRNAs differentially expressed after optic nerve
| miRBase ID | miRNA name | Fold change | Adjusted P-value |
|---|---|---|---|
| MIMAT0001290 | dre-miR-223 | 1.551 | 0.0165 |
| MIMAT0001787 | dre-miR-21 | 1.319 | 0.0361 |
| MIMAT0001801 | dre-miR-29b-2 | 1.159 | 0.0481 |
To identify genes that are regulated by these miRNAs, we computationally identified target mRNAs that contain a putative miRNA binding site within their 3’UTR using TargetScan Fish [20], one of the few predictive databases to identify miRNA targets for species other than humans and rodents. To select gene candidates that were most likely to be regulated by each miRNA, we used stringent parameters whereby at least one miRNA binding site had a context + score of ≤0.30. This approach resulted in 427 and 505 unique putative Ensembl target genes for miR-29b and miR-223, respectively. We then performed GO analysis on our list of predicted target genes to determine if these miRNAs where associated with any particular biological function. These results suggest a propensity for miR-29b to target genes associated with DNA modification and extracellular matrix (ECM) activities (Fig. 5, Additional file 6: Table S3), in contrast to miR-223 whose putative targets appeared to fall into nucleoside catabolic processes (particularly purine metabolism) and GTPase regulators (Fig. 6, Additional file 7: Table S4). Interestingly, glycoprotein metabolism processes (GO: 0009100) were also highlighted, with an emphasis on heparan sulfate proteoglycan (HSPG) subtypes (p = 0.00390). We then integrated the predicted gene list with our own differentially expressed gene set and this revealed 11 predicted target genes for miR-29b and 13 for miR-223. We refined this list by focusing on target genes whose expression was inversely correlated with the miRNA expression (i.e. potentially downregulated), and using a Targetscan context + score of < −0.30 for at least one binding site, we identified five predicted gene targets for miR-29b and four for miR-223 (Table 4).
Fig. 5.
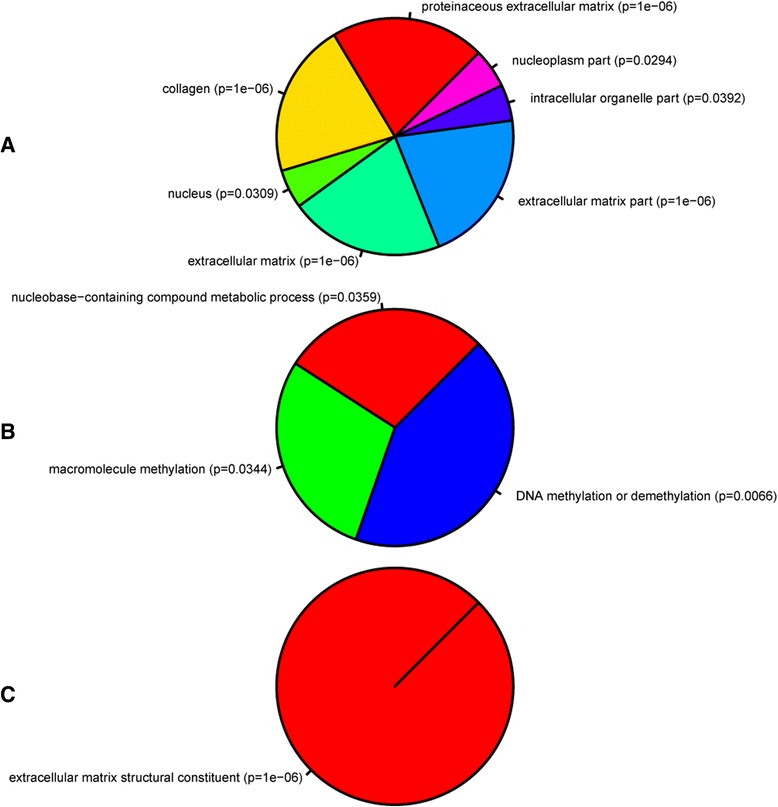
Gene ontology of miR-29b predicted targets from Targetscan Fish. Representation of GO terms associated with (a) cellular component, (b) biological process, and (c) molecular function. Predicted gene targets contained at least one miRNA binding site with a context score of ≤ −0.30. Each part represents -log2 of the p-value of biological process and cellular component from the set of significant biological processes and cellular components. The p-values were retrieved from gene ontology analysis in WebGestalt. A list of genes in each GO category is in Additional file 6: Table S3
Fig. 6.
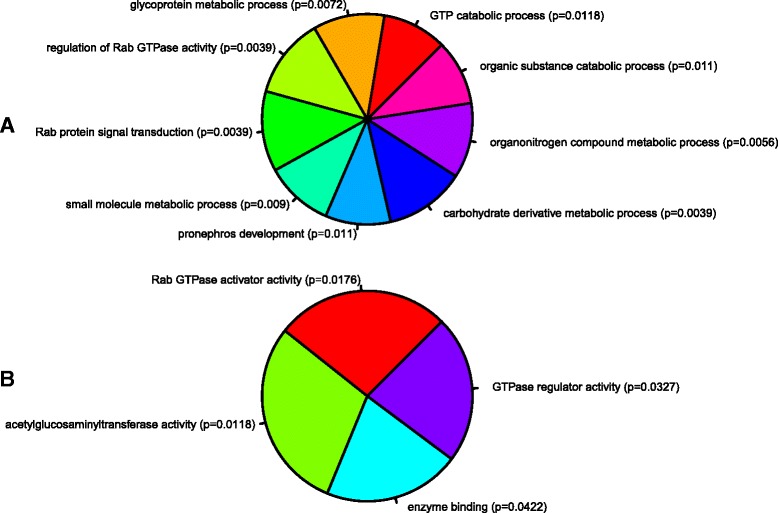
Gene ontology of miR-223 predicted targets from Targetscan Fish. Representation of GO terms associated with (a) biological process, and (b) molecular function. Predicted gene targets contained at least one miRNA binding site with a context score of ≤ −0.30. Each part represents -log2 of the p-value of biological process and cellular component from the set of significant biological processes and cellular components. The p-values were retrieved from gene ontology analysis in WebGestalt. A list of genes in each GO category is in Additional file 7: Table S4
Table 4.
Putative miR-29b and miR-223 targets downregulated after optic nerve crush
| Ensembl ID | Gene | mRNA changea | miRNA | Context + scoreb | No. binding sites |
|---|---|---|---|---|---|
| ENSDART00000087565 | eva1a | 0.61 | miR-29b | −0.46 | 1 (8mer) |
| eva-1 homolog a | (miR-223) c | (−0.09) | 1 (7mer-1A) | ||
| ENSDART00000050945 | layna | 0.45 | miR-29b | −0.50 | 1 (8mer) |
| Layilin a | (miR-223) c | (−0.16; −0.17) | 2 (7mer-1A) | ||
| ENSDART00000064163 | nefmb | 0.28 | miR-29b | −0.41 | 1 (8mer) |
| Neurofilament, medium polypeptide, b | |||||
| ENSDART00000018351 | ina (zgc:65851) | 0.50 | miR-29b | −0.41; −0.05 | 2 (8mer; 7mer-1A) |
| Internexin neuronal intermediate filament alpha | (miR-223) c | (−0.02; >0.03; >0.01) | 3 (8mer; 7mer-m8; 7mer-1A) | ||
| ENSDART00000021556 | si:ch211-51a6.2 | 0.57 | miR-29b | −0.33 | 1 (7mer-m8) |
| Homolog of prss12 | miR-223 | −0.32; −0.07 | 2 (7mer-m8; 7mer-1A) | ||
| ENSDART00000126365 | smoc1 | 0.41 | miR-223 | −0.39; > − 0.02 | 2 (8mer; 7mer-m8) |
| SPARC related modular calcium binding 1 | |||||
| ENSDART00000124670 | lrrn3 | 0.65 | miR-223 | −0.32 | 1 (8mer) |
| Leucine rich repeat neuronal protein 3-like | |||||
| ENSDART00000081039 | d sb:cb252 | 52.9 | miR-223 | −0.31 | 1 (7mer-m8) |
| Homolog of es1-like |
aFold change represents microarray expression. All genes had an adjusted p-value ≤0.05
bSee Methods for summary of Targetscan Fish context + score
cPredicted binding sites of miR-223 in genes with a context scores less stringent then our cut-off (i.e., ≤ − 0.30)
dMicroarray results revealed sb:cb252 was the most upregulated gene on microarray, yet it was also one of the top predicted miR-223 target genes by Targetscan Fish; we chose to included it in subsequent experiments to determine if it was a valid miR-223 target
Validation of putative target genes of miR-223 and miR-29b
To validate our computationally predicted miRNA-gene targets, we first confirmed changes in their expression using RT-qPCR (Fig. 7), which supported our microarray expression data for all but one miRNA target, sb:cb252. This gene was one of the top predicted targets for miR-223 identified in Targetscan (context + score = −0.31), but our array results showed strong over-expression (53-fold). However, RT-qPCR validation revealed a decrease in expression, supporting the idea that it is negatively regulated by miR-223 as predicted by Targetscan Fish; thus the gene was included in all subsequent analyses. To functionally verify the putative miRNA-mRNA interaction, we cloned part of the 3’UTR sequence that contained the miRNA binding site of each gene into a pmirGLO vector, which contains both luciferase and renilla reporter genes (Fig. 8). A mutated version of the miRNA seed site was also created. HEK293 cells were transfected with either a wild type (WT) or mutated construct (MT), along with a miRNA mimic (miR-29/miR-223) or negative control (miR-NC). Luciferase results revealed significant inhibition of luciferase activity for miR-29b with all of its predicted gene targets (i.e., eva1a, layna, nefmb, ina and si:ch211-51a6.2). However, miR-223 showed statistically significant binding only to smoc1 and sb:cb252 (Fig. 9).
Fig. 7.
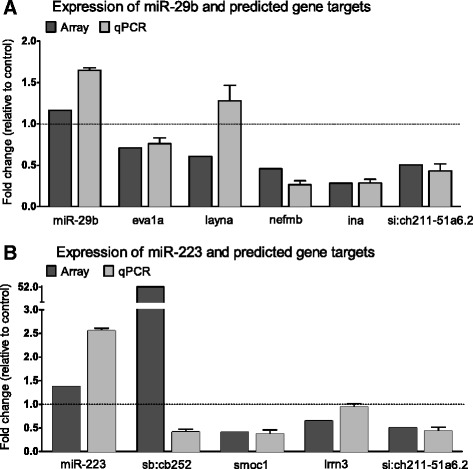
Validation of miR-29b and miR-223 putative gene targets from integration analysis. Microarray and corresponding RT-qPCR expression of genes predicted to be targeted by miR-29b (a) or miR-223 (b). The latter is presented as expression fold change (2-ΔΔCt) relative to PPIA (mean ± SD; n = 4 groups of pooled retinal RNA containing 4 animals)
Fig. 8.

Predicted miRNA binding sites within 3’UTR of predicted target genes. Sequence of miR-29b (a) and miR-223 (b) binding sites within 3’UTR of predicted target genes (nt, nucleotide position in 3’ UTR). Seed region is bolded. Mutations predicted to disrupt miRNA-mRNA binding were made in the seed region, mutated nucleotides underlined
Fig. 9.
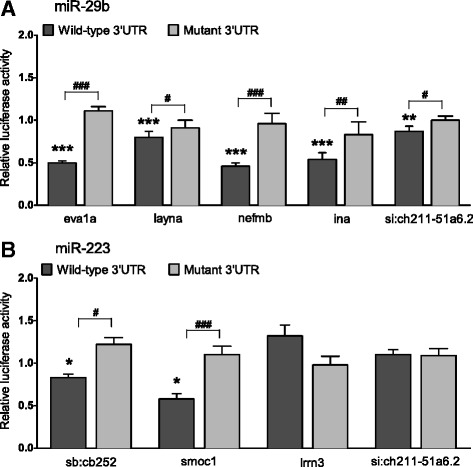
Validation of miRNA binding to 3’ UTR of putative target genes. Luciferase reporter assay of HEK293 cells cotransfected with pmirGLO plasmid containing the WT or MT 3’UTR miRNA seed sequence from each gene, and either miR-29b, miR-223 or miR-NC (scrambled control). Samples were analysed 48 h after transfection and data normalised to the pmirGLO only transfection. Columns represent the luciferase activity of either WT or MT constructs with miR-29b (a) or miR-223 (b), relative to transfection with the same construct and miR-NC. Data represents the mean ± SEM, n = 3 independent experiments containing 4 replicates each. Student’s t test comparing WT or MT construct with miRNA to miR-NC indicated as *p < 0.05, **p < 0.01, ***p < 0.001. Student’s t test comparing WT with miRNA to corresponding MT construct indicated as #p < 0.05, ## p < 0.01, ###p < 0.001
Discussion
Here we used microarrays to examine and integrate expression of miRNA and mRNA in zebrafish retina after optic nerve crush to identify potential regulatory mechanisms involved in central nerve regeneration. Our integrated approach to studying gene regulation highlighted two miRNAs that target genes in key biological processes associated with cell survival/apoptosis, ECM-cytoskeleton signaling, and HSPG binding. Our study has provided unique information about the cellular context in which these genetic regulatory changes occur at a critical time point in the regeneration pathway.
Zebrafish are a good model in which to delineate genes associated with regeneration and contrast with mammalian studies. However, it appears there are a lot more genetic similarities between the species than first thought, with overlap between zebrafish and mammals observed in both up- and downregulated genes [13, 21–23], and even in the expression of genes associated with inhibition of regeneration (e.g., socs3 and sfpq; [24, 25]). This further supports the idea that multiple pathway analysis is required, as differences are likely to be subtle and occur at multiple points across several processes [3, 26]. It is also likely that a successful response results from the synergistic activity of several cell types, given the interaction between RGCs and other retinal cell types that are reported to assist in a positive injury response. Our approach of profiling the expression of the whole retina takes into consideration the changes that occur to in the cells adjacent to the RGCs, such as Muller glia and amacrine cells. Interestingly, the GO processes altered in our dataset overlap with several mentioned in previous zebrafish studies that have utilised other ocular tissues (isolated RGCs and whole eye; [13, 21, 27]). However there were some processes that appeared unique to this study, in particular upregulation of non-coding RNAs, which supports our original hypothesis that gene regulatory processes are key to promoting successful regeneration in zebrafish.
The miRNAs: mir-29b and miR-223
The role of microRNAs in lower vertebrates with a known regenerative ability is gaining a lot of attention, with several recent studies identifying miRNAs associated with spinal cord repair (e.g., miR-125b in axolotl, miR-133b in zebrafish; [28, 29]) and appendage regeneration (e.g. miR-196 in axolotl tail, miR-203 in zebrafish fin; [30, 31]). Less is known about miRNA-mediated regeneration within the eye, with most focus on the role of miRNAs in Muller glia cells [32, 33]. In this study, we show for the first time that miR-29b and miR-223, are significantly over-expressed after optic nerve crush. In zebrafish, miR-29b is part of the miR-29 family that comprises three intergenic members that map to chromosome 4 (miR-29a and miR-29b-2 located within 10Kb of each other; miR-29b-2 referred to as miR-29b herein), and chromosome 6 (miR-29b-1; [34]). miR-29b has been associated with ECM remodeling, and shown to have both pro- and anti-apoptotic properties depending on the CNS injury/disease model [35–38]. The other miRNA highlighted in this study, miR-223, maps to chromosome 5 in zebrafish, and has been implicated in a wide range of pathophysiologies, including cancer, muscular dystrophy and atherosclerosis [39]. Within the CNS, it has been implicated in inflammatory processes including autoimmune disorders and mammalian CNS injury [40–42]. Our gene ontology analysis confirmed a role for miR-223 in the zebrafish immune response following injury but additionally highlighted enhanced cell survival and altered cytoskeleton/ECM response. The small number of significantly altered miRNAs found in this study may result from looking at one specific time point. On the other hand, the fact that a single miRNA is able to regulate a large number of genes might negate the need for changes in many miRNAs in order to bring about widespread biological change [10]. Further, it is likely that these two miRNAs interact with each other in a synergistic manner, as evidenced by the overlapping targeting of the same genes when less stringent cut-offs were applied during our integration analysis.
miRNA target genes: cell survival from altered mitochondrial function
After optic nerve injury, the viability of RGCs is paramount for axonal regeneration and restoration of function. Mammalian studies have shown a high degree of cell death in the retina 2–3 days after injury, in contrast to the limited death observed in zebrafish [43, 44]. This restrained cell death may be associated with several downregulated genes in our dataset that we functionally validated for the first time as targets genes of miR-29b and miR-223, including eva1a, layna, si:ch211-51a6.2 (a homolog of prss12), smoc1 and es1-like homolog, sb:cb252.
Eva1a is a strongly conserved transmembrane protein localized to the lysosome and endoplasmic reticulum membrane where it is thought to function as a regulator of programmed cell death processes as well as necrosis [45–47]. Another target that may play a role in mitochondrial function (and thus potentially cell survival) is the miR-223 target, sb:cb252. This uncharacterized gene shares significant sequence homology with its homolog, zebrafish es1 and human C21orf33 [48, 49]. The exact role of both sb:cb252 and es1 remains unknown, however they are postulated to be involved in mitochondrial function and es1 has been observed in zebrafish retina after a retinal lesion [50]. Downregulation of these targets may protect RGCs against cell death, consistent with the >90 % survival observed in zebrafish RGCs after crush [51].
ECM-cell signaling and indirect cytoskeleton modification
Interaction between the cell membrane and actin cytoskeleton is essential for many cellular processes, including cell shape, adhesion, migration and signal transduction [52]. The role of ECM-cell signaling in regeneration is particularly pertinent as a means of restructuring the cellular architecture in response to injury. The role of miRNAs, including the miR-29 family, in modifying ECM-cell signaling has recently been highlighted [53]. Interestingly, three of the miRNA targets we validated are associated with this process, layna, si:ch211-51a6.2 and smoc1.
Layilin (layna) is a transmembrane receptor that mediates hyaluronan signaling by binding to the ECM cytoskeletal linker proteins, talin, radixin and merlin [54, 55], thus destabilising the cytoskeleton [56, 57]. Downregulation of layna in zebrafish RGCs after crush may therefore reduce hyaluronan signaling and stabilise the cytoskeleton, promoting the early stages of axonal outgrowth. Interestingly, there were no significant changes in the expression of other hyaluronan receptors within our dataset (e.g. CD44, MyD88, TLR-4).
Another biological process highlighted in our study is heparan sulfate proteoglycan (HSPG) signalling, which is known to influence CNS regeneration [58]. miR-29b and miR-223 target genes share common functions surrounding the HSPG, agrin. The miR-29b target, si:ch211-51a6.2, is not characterised in zebrafish but is a homologue of the serine protease, neurotrypsin (prss12). Neurotrypsin cleaves agrin into two fragments which are involved in neural plasticity, axonal outgrowth and synaptogenesis (shorter C-terminal peptide; [59–61]), as well as cytoskeleton remodelling (longer N-terminal peptide: [62, 63]). Furthermore, the miR-223 target, secreted modular calcium binding 1 (smoc1), is a member of the SPARC (secreted protein acidic and rich in cysteine) family and binds to HSPGs (including agrin) to modulate cell adhesion [64, 65]. Although it is not clear how downregulation of their expression would impact on the regenerative process, key roles for these proteins in mediating the regenerative response in fish are supported by our finding that the agrin interactions canonical pathway is enriched with upregulated genes in our dataset. Interestingly, smoc1 has also been implicated in Smad1-dependent bone morphogenetic protein (BMP) signaling which regulates axonal regrowth in injured dorsal root ganglion neurons [66–68]. Thus the downregulation of smoc1 may also be part of BMP activation of pro-regenerative transcription program or manipulation of cytoskeletal dynamics at the growth cone [69].
Direct cytoskeleton modification
In addition to indirectly reorganizing the cytoskeleton by modulating membrane signaling mechanisms, our data highlight ways in which miR-29b may directly target members of the intermediate filaments of the cytoskeleton, by regulating expression of internexin (ina) and neurofilament-medium homolog b (nefmb) [70]. The latter is one of two homologs found in zebrafish (nefma and nefmb), however nefmb is likely to represent the zebrafish version of nefh, as it shares a significant homology to human NFH. Although some neurofilament proteins are upregulated during successful regeneration in the mammalian peripheral nervous system, and in lower vertebrates (e.g. prph, nefm), the overexpression of other intermediate filaments has been associated with several neurodegenerative and CNS diseases [71–73], with specifically nefh and ina potentially being biomarkers for the likes of Amyotrophic Lateral Sclerosis and neuropsychiatric Systemic Lupus Erythematosus [74, 75]. Both have been associated with the slowing of neurofilament axonal transport, resulting in neuronal aggregates which damage the endoplasmic reticulum and mitochondria in transit, ultimately leading to cell death [75–77]. Thus their downregulation during the initial stages of axonal regeneration in our study may promote regeneration by facilitating transport of neurofilament components to required cellular areas. It is interesting to note that other microtubule and actin components of the cytoskeleton, including b-actin, a- and b-tubulin, nefm and prph, were upregulated after injury in our dataset. This suggests that the specific downregulation of ina and nefmb may play a significant role in zebrafish injury response.
Zebrafish as a comparative model for miRNA role in CNS regeneration
Examining the role of miRNAs in regenerative permissive species, such as zebrafish, is an important step in delineating their role in higher order vertebrate injuries. Whilst the miRNA sequence is strongly conserved between species, this does not necessarily guarantee conservation of their function [78]. In many cases the target genes are not conserved, with the turnover of miRNA binding sites within the 3’UTR of genes thought to be a significant driver of evolutionary processes [79]. However, the two miRs examined in this study are predicted to target human and rat orthologs of the genes we validated here (Table 5), suggesting that the retention of miRNA binding sites across species may provide an opportunity to manipulate gene regulation in mammals based on our findings.
Table 5.
Predicted miR-29b and miR-223 binding of human and rat orthologs to the validated zebrafish genes
| Zfish refseq | Zfish ensembl | Zebrafish gene | Human ortholog (context score)a | Rat ortholog (context score)a |
|---|---|---|---|---|
| NM_001076587 | ENSDART00000087565 | eva1a | miR-29b (−0.21) | miR-223 (−0.32) |
| XM_688304 | ENSDART00000050945 | layna | miR-223 (−0.41) | miR-29b (−0.14) |
| miR-223 (−0.05) | ||||
| NM_001123280 | ENSDART00000064163 | nefmb | No direct ortholog | No direct ortholog |
| NEFH: miR-223 (−0.25) | ||||
| NM_199534 | ENSDART00000018351 | ina | miR-29b (−0.17) | miR-29b (−0.15) |
| XM_685557 | ENSDART00000021556 | si:ch211-51a6.2 | No direct ortholog | - |
| PRSS12: miR-223 (−0.08) | ||||
| NM_001201393 | ENSDART00000126365 | smoc1 | miR-223 (−0.11) | miR-223 (−0.11) |
| XM_003201552 | ENSDART00000124670 | lrrn3 | - | - |
| XM_003199845 | ENSDART00000081039 | sb:cb252 | No ortholog | No ortholog |
aContext score calculated by Targetscan for human and rat predictions (Grimson et al. [91])
Conclusion
Identifying the molecular and cellular factors associated with the successful regenerative response in zebrafish may aid in identifying therapeutic targets in the damaged mammalian CNS [6]. Our results provide a basis from which to investigate the cellular processes required for central nervous system regeneration, and further studies will examine more extensively the complete repertoire of mRNA targets of miR-29b and miR-223 (e.g., using high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) [80]). The synergistic activities of multiple miRNAs, and the redundancy in their regulation of specific target genes and signalling pathways, provides an opportunity to reengage 3’UTR targets in order to instigate a fish-like regenerative response in the mammalian CNS [4].
Methods
Animals and surgery
Wild type adult zebrafish (9–12 months old, we did not distinguish between males and females), Danio rerio, were maintained at 27 °C on a 12 h light–dark cycle. Optic nerve crush was performed as previously described [44]. Briefly, fish were anesthetized by immersion in 0.1 % Tricaine methanesulfonate (MS222; Sigma Aldrich) in Holtfreters solution pH 7.2 [81]. The right eye was deflected forward and connective tissue cut to expose the optic nerve, which was crushed with fine forceps (Dumont no5) 1 mm from the back of the eye; this procedure severs all RGC axons but leaves the nerve sheath intact as a conduit for regeneration. Three days after crush injury, fish were euthanised by overdose in MS222 and the eyes were removed. Retinae were dissected out and stored in RNAlater (Ambion) according to manufacturer’s instructions. The right retinae from intact, unoperated fish were used as controls. All procedures conformed to the NHMRC guidelines for the use of animals and were approved by the Animal Ethics Committee of the University of Western Australia.
RNA extraction
In order to generate enough sample material to be used for microarray and miRNA arrays, retinae from 4 fish were pooled. Four biological replicates were performed for each array type (i.e., 4 microarrays for crush and control tissue, each containing pooled retinal RNA from 4 fish). Total RNA from pooled tissue was extracted using Trizol reagent (Life Technologies) according to manufacturer’s instructions, and subsequently column-purified with miRNeasy kits (Qiagen). The concentration of RNA was determined using a NanoDrop spectrophotometer (Thermo Fischer) and RNA integrity was assessed using a Bioanalyzer 2100 (Agilent).
Microarray gene expression profiling and analysis
Microarray processing of Agilent Zebrafish 4x44K V3 GE array (one-colour labeling, hybridization and scanning) was performed by the Ramaciotti Centre for Gene Function Analysis (University of New South Wales, Australia), according to the manufacturer’s (Agilent) instructions. These arrays contain 43,603 probesets, representing 19,405 unique Unigene gene targets. Pre-processing of the array data included background correction using a normal-exponential convolution model [82] and quantile normalisation [83]. The limma Bioconductor package [84] was used for differential expression analysis. Probesets with an absolute fold change of ≥1.5 (log2 fold change 0.585) adjusted p-value ≤0.05 were considered as differentially expressed [85]. The microarray data are available from the Gene Expression Omnibus GSE70261, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70261.
Gene ontology and pathway analysis
To examine the biological and functional implications of differentially expressed genes, we stratified the differentially expressed genes into those that were over-expressed or over-expressed and performed gene set enrichment analysis in both, utilising the WebGestalt suite for gene ontology terms [86], and Ingenuity Pathway Analysis software (IPA), for canonical pathways and gene network analysis (http://www.ingenuity.com). For the purposes of quickly identifying key non-redundant GO terms, we utilised the GO Trimming [87] method (soft trim, 100 %) and only plotted terms with 5 or more genes, as evidenced in Figs. 2, 3, 5 and 6. We provide the full GO terms lists in the Supporting data.
microRNA expression profiling and analysis
Total RNA from the same sample used in microarray profiling was profiled by Exiqon Services (http://www.exiqon.com). Total RNA (~425 ng of each sample) was labeled using the miRCURY LNA microRNA Hi-Power Labeling kit with Hy3 (Exiqon) according to the manufacturer’s instructions. The Hy3-labeld samples were hybridized to the miRCURY LNA microRNA Array v.11—Other species (Exiqon), which contains probes targeting 217 mature miRNAs from Danio rerio miRNAs miRBase v18. The hybridization was performed according to the manufacturer’s instructions on a Tecan HS480 hybridization station (Tecan, Austria) and microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc., USA). Image analysis was carried out using the ImaGene 9.0 software (BioDiscovery, Inc., USA). The data were pre-processed using the robust multi-array average expression measure [88]. One control array was removed after performing QC using the ArrayQualityMetrics bioconductor package [89]. As with the mRNA analysis, we used limma for differential expression analysis, and considered miRNA targets with an adjusted p-value ≤0.05 of interest.
Platform integration
To identify putative miRNA-mRNA target pairs of interest, we integrated the mRNA and miRNA platform data, and filtered with other key datasets. Briefly, we utilised Targetscan Fish (version 6.2; [20] as the source of target prediction information, and cross-referenced the top miRNAs (by unadjusted miRNA p-value of ≤0.05) with mRNA array targets (by fold change >1.5 and adjusted p-value ≤0.05). We extracted conserved and non-conserved pairs from Targetscan Fish where the mRNA-miRNA pairs were inversely correlated (to reflect the typical miRNA-mRNA relationship), and context scores were ≤ −0.3, a generally accepted filter for prioritising higher likelihood binding sites from computational predictions. Targetscan predictions in vertebrates are ranked on the predicted efficacy of targeting as calculated using the context + scores of the binding sites, which takes into account the type of seed pairing site, miRNA-target complementarity outside the seed region., local AU content, site position within 3’UTR, target site abundance, and seed pairing stability [20, 90, 91]. We integrated Uniprot [92] and GO terms, and filtered putative target pairs by those with the most complete annotations and supporting information. We further examined our two top putative miRNAs of importance by selecting the set of genes targeted genome-wide by each miRNA, with high quality (single site) context scores ≤ −0.3, and performed GO biofunction enrichment (using Webgestalt) to identify if there were common functions targeted by each miRNA.
RT-qPCR
Reverse transcription of mRNA used 250 ng of total RNA and was performed according to manufacturer’s instructions using GoScript reverse transcription kit with random hexamer primers (Promega). Quantitative PCR was performed in triplicate on a Rotor Gene 6000 (Qiagen) using GoTaq SYBR Green Mastermix (Promega) according to manufacturer’s instructions. Primer sequences used to confirm gene expression are provided in Additional file 8: Table S5. Data were normalized to the housekeeping gene ppia and relative expression was calculated using the 2-ΔΔCt method [93].
Reverse transcription of miRNA used 10 ng of total RNA and was performed according to manufacturer’s instructions using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems) and TaqMan MicroRNA Assay primers for mature miR-29b, miR-223 and U6 snRNA (Assay ID 000413, 000526, 001973, respectively; Applied Biosystems). Quantitative PCR was performed using TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems) according to the manufacturer’s instructions. Levels of miR-29 and miR-223 were normalized to U6 snRNA and relative fold change between control and crush tissue was determined using 2-ΔΔCt method.
Luciferase reporter plasmids
Oligonucleotides containing either the putative binding site of miR-29b or miR-223 from the target gene 3’ UTRs (predicted by Targetscan Fish; Additional file 9: Table S6) were annealed and ligated using NheI and SalI restriction enzymes sites downstream of the luciferase reporter gene in pmirGLO Duel Luciferase miRNA target Expression Vector (Promega). An internal NotI site was included in the oligonucleotide sequence for clone confirmation, and sequence and orientation was verified by DNA sequencing. Mutated binding site constructs contained 4 base substitutions within the seed site predicted to disrupt miRNA binding. Where multiple bindings sites in the 3’UTR of a miRNA was predicted, the binding site with the lowest Targetscan Fish context score was used.
Transfection and luciferase assays
The HEK293 cell line was cultured at 37C in 5 % CO2 with Dulbecco’s modified Eagle’s medium supplemented with 10 % foetal bovine serum and 1 % penicillin/streptomycin. Synthetic miRNA molecules (mirVana miRNA mimics) corresponding to miR-223 (Product ID: MC10903), miR-29b (Product ID: MC10432) and a negative control miRNA (miR-NC: Product ID 4464076) were obtained from Ambion (Life Technologies). HEK293 cells were seeded at 3 × 104cells/well in 96-well white plate (Greiner) and were transfected 24 h later with miRNA mimics diluted in Optimem and Lipofectamine 2000 (Life Technologies). Cells were cotransfected with 40 ng of each construct and 25nM miRNA mimic. Firefly and Renilla luciferase activities were assayed 48 h after transfection using a Dual Glo Luciferase Assay System (Promega) with a luminometer (Enspire, PerkinElmer).
Statistical analysis
For all statistical analyses not described elsewhere, we utilised Prism software (GraphPad Software). A two-tailed Student t-test was used when two groups were compared.
Availability of supporting data
Microarray datasets are available from the Gene Expression Omnibus GSE70261. Additional supporting data are available from http://dx.doi.org/10.6070/H4GH9FZ9.
Acknowledgements
We are grateful to Carole Bartlett for assistance with the optic nerve crush procedure and to Rick Roberts for animal husbandry. JR is a NHMRC Senior Research Fellow. KWC is supported by the McCusker Charitable Foundation Bioinformatics Centre. Supported by the Neurotrauma Research Program WA funded through the Road Trauma Trust Association, but does not reflect views or recommendations of the Road Safety Council.
Abbreviations
- BMP
Bone morphogenetic protein
- CNS
Central nervous system
- DNA
Deoxyribonucleic acid
- ECM
Extracellular matrix
- GO
Gene ontology
- GTPase
Guanosine triphosphate hydrolase
- HITS-CLIP
High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation
- HSPG
Heparan sulfate proteoglycan
- IPA
Ingenuity pathway analysis
- miR-NC
Negative control miRNA construct
- mRNA
Messenger ribonucleic acid
- miRNA
Micro ribonucleic acid
- MT
Mutated construct
- NHMRC
National Health and Medical Research Council (Australia)
- ON
Optic nerve
- RGC
Retinal ganglion cell
- RNA
Ribonucleic acid
- RT-qPCR
Reverse transcription—quantitative polymerase chain reaction
- SPARC
Secreted protein acidic and rich in cysteine
- UTR
Untranslated region
- WT
Wild type
Additional files
Volcano plot of differential gene expression in zebrafish retina after optic nerve crush. Each probe on the array is represented by a single dot, with red dots signifying the 804 differentially expressed transcripts. P- values are presented as –log10 values, expression differences presented as log2 fold changes. We set cut off limits at p < 0.05 and absolute fold change ≥1.5. By these parameters, 459 transcripts were over-expressed and 198 were under-expressed after optic nerve injury. (TIFF 64 kb)
Unfiltered enriched GO terms associated with over-expressed gene set. Table contains each GO category and p-value. (XLS 29 kb)
Unfiltered enriched GO terms associated with under-expressed gene set. Table contains each GO category and p-value. (XLS 33 kb)
Top gene network of over-expressed genes. Top scoring IPA gene network diagram of over-expressed gene set. The enriched networks are associated with functions involved in Dermatological Diseases and Conditions, Developmental Disorder, and Hereditary Disorders, with a network score of 49. (TIFF 561 kb)
Top gene network of under-expressed genes. IPA gene network diagram of under-expressed gene set. The enriched networks are associated with functions involved in Neurological Disease, Nervous System Development and Function, and Organ Morphology, with a network score of 45. (TIFF 730 kb)
Unfiltered enriched GO terms associated with miR-29b predicted target genes sourced from Targetscan Fish database. Table contains each GO category and p-value. (XLS 21 kb)
Unfiltered enriched GO terms associated with miR-223 predicted target genes sourced from Targetscan Fish database. Table contains each GO category and p-value. (XLSX 9 kb)
qPCR primers for validating putative miRNA target genes. (DOCX 12 kb)
Oligonucleotide sequences used in luciferase constructs. (DOCX 13 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
PIF, KWC and JR designed the study and drafted the manuscript. PIF and JR performed the surgeries. PIF performed RNA extraction, RT-qPCR, plasmid construction and luciferase assays. KWC and DA performed statistical analysis. KWC performed bioinformatics analysis on mRNA and miRNA array data. PIF, KWC, ARH and JR analysed and interpreted results. KMG provided specific input into miRNA experimental design and results analysis. All authors read and approved the final manuscript.
Contributor Information
Paula I. Fuller-Carter, Email: paula.fuller@uwa.edu.au
Kim W. Carter, Email: Kim.Carter@telethonkids.org.au
Denise Anderson, Email: Denise.Anderson@telethonkids.org.au.
Alan R. Harvey, Email: alan.harvey@uwa.edu.au
Keith M. Giles, Email: keith.giles@nyumc.org
Jennifer Rodger, Email: jennifer.rodger@uwa.edu.au.
References
- 1.Kiernan JA. Hypothesis concerned with axonal regeneration in the mammalian nervous system. Biol Rev. 1979;54:155–97. doi: 10.1111/j.1469-185X.1979.tb00871.x. [DOI] [PubMed] [Google Scholar]
- 2.Beazley LD, Dunlop SA. Degeneration and regeneration in the nervous system. Singapore: Harwood Academic Press; 2000. Evolutionary hierarchy of optic nerve regeneration: implications for cell survival, axon outgrowth and map formation; pp. 119–52. [Google Scholar]
- 3.Becker T, Becker CG. Axonal regeneration in zebrafish. Curr Opin Neurobiol. 2014;27C:186–91. doi: 10.1016/j.conb.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Kusik BW, Hammond DR, Udvadia AJ. Transcriptional regulatory regions of gap43 needed in developing and regenerating retinal ganglion cells. Dev Dyn. 2010;239(2):482–95. doi: 10.1002/dvdy.22190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benowitz LI, Goldberg DE, Irwin N. Inosine stimulates axon growth in vitro and in the adult CNS. Prog Brain Res. 2002;137:389–99. doi: 10.1016/S0079-6123(02)37030-4. [DOI] [PubMed] [Google Scholar]
- 6.Williams RR, Venkatesh I, Pearse DD, Udvadia AJ, Bunge MB. MASH1/Ascl1a leads to GAP43 expression and axon regeneration in the adult CNS. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0118918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Madathil SK, Nelson PT, Saatman KE, Wilfred BR. MicroRNAs in CNS injury: potential roles and therapeutic implications. Bioessays. 2011;33:21–6. doi: 10.1002/bies.201000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–20. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12(12):846–60. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 12.Fischer D, Petkova V, Thanos S, Benowitz LI. Switching mature retinal ganglion cells to a robust growth state in vivo: gene expression and synergy with RhoA inactivation. J Neurosci. 2004;24:8726–40. doi: 10.1523/JNEUROSCI.2774-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veldman MB, Bemben MA, Thompson RC, Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 14.Harvey AR, Hu Y, Leaver SG, Mellough CB, Park K, Verhaagen J, et al. Gene therapy and transplantation in CNS repair: the visual system. Prog Retin Eye Res. 2006;25(5):449–89. doi: 10.1016/j.preteyeres.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Dunlop SA, Tee LB, Stirling RV, Taylor AL, Runham PB, Barber AB, et al. Failure to restore vision after optic nerve regeneration in reptiles: interspecies variation in response to axotomy. J Comp Neurol. 2004;478(3):292–305. doi: 10.1002/cne.20299. [DOI] [PubMed] [Google Scholar]
- 16.Bernhardt RR. Cellular and molecular bases of axonal regeneration in the fish central nervous system. Exp Neurol. 1999;157:223–40. doi: 10.1006/exnr.1999.7059. [DOI] [PubMed] [Google Scholar]
- 17.Kato S, Matsukawa T, Koriyama Y, Sugitani K, Ogai K. A molecular mechanism of optic nerve regeneration in fish: the retinoid signaling pathway. Prog Retin Eye Res. 2013;37:13–30. doi: 10.1016/j.preteyeres.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Farh KK-H, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science (New York, NY) 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 19.Ge XT, Lei P, Wang HC, Zhang AL, Han ZL, Chen X, et al. miR-21 improves the neurological outcome after traumatic brain injury in rats. Sci Rep. 2014;4:6718. doi: 10.1038/srep06718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, et al. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–66. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCurley AT, Callard GV. Time course analysis of gene expression patterns in zebrafish eye during optic nerve regeneration. J Exp Neurosci. 2010;2010:17–33. [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma TP, McDowell CM, Liu Y, Wagner AH, Thole D, Faga BP, et al. Optic nerve crush induces spatial and temporal gene expression patterns in retina and optic nerve of BALB/cJ mice. Mol Neurodegeneration. 2014;9:14. doi: 10.1186/1750-1326-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agudo M, Pérez-Marín MC, Lönngren U, Sobrado P, Conesa A, Cánovas I, et al. Time course profiling of the retinal transcriptome after optic nerve transection and optic nerve crush. Mol Vis. 2008;14:1050–63. [PMC free article] [PubMed] [Google Scholar]
- 24.Hellstrom M, Muhling J, Ehlert EM, Verhaagen J, Pollett MA, Hu Y, et al. Negative impact of rAAV2 mediated expression of SOCS3 on the regeneration of adult retinal ganglion cell axons. Mol Cell Neurosci. 2011;46(2):507–15. doi: 10.1016/j.mcn.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Elsaeidi F, Bemben M, Zhao X-F, Goldman D. Jak/Stat signaling stimulates zebrafish optic nerve regeneration and overcomes the inhibitory actions of Socs3 and Sfpq. J Neurosci. 2014;34:2632–44. doi: 10.1523/JNEUROSCI.3898-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Kesteren RE, Mason MR, Macgillavry HD, Smit AB, Verhaagen J. A gene network perspective on axonal regeneration. Front Mol Neurosci. 2011;4:46. doi: 10.3389/fnmol.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker CG, Becker T. Growth and pathfinding of regenerating axons in the optic projection of adult fish. J Neurosci Res. 2007;85:2793–9. doi: 10.1002/jnr.21121. [DOI] [PubMed] [Google Scholar]
- 28.Diaz Quiroz JF, Tsai E, Coyle M, Sehm T, Echeverri K. Precise control of miR-125b levels is required to create a regeneration-permissive environment after spinal cord injury: a cross-species comparison between salamander and rat. Dis Model Mech. 2014;7:601–11. doi: 10.1242/dmm.014837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Y-m, Gibbs KM, Davila J, Campbell N, Sung S, Todorova TI, et al. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur J Neurosci. 2011;33:1587–97. doi: 10.1111/j.1460-9568.2011.07643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sehm T, Sachse C, Frenzel C, Echeverri K. miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Dev Biol. 2009;334:468–80. doi: 10.1016/j.ydbio.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci U S A. 2008;105:18384–9. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajaram K, Harding RL, Bailey T, Patton JG, Hyde DR. Dynamic miRNA expression patterns during retinal regeneration in zebrafish: reduced dicer or miRNA expression suppresses proliferation of Müller glia-derived neuronal progenitor cells. Dev Dyn. 2014;243(12):1591–605. doi: 10.1002/dvdy.24188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Müller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–7. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thatcher EJ, Bond J, Paydar I, Patton JG. Genomic organization of zebrafish microRNAs. BMC Genomics. 2008;9:253. doi: 10.1186/1471-2164-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kole AJ, Swahari V, Hammond SM, Deshmukh M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011;25:125–30. doi: 10.1101/gad.1975411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S-y, Lee JH, Ha M, Nam J-w, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–9. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 37.Silva VAO, Polesskaya A, Sousa TA, Corrêa VMA, André ND, Reis RI, et al. Expression and cellular localization of microRNA-29b and RAX, an activator of the RNA-dependent protein kinase (PKR), in the retina of streptozotocin-induced diabetic rats. Mol Vis. 2011;17:2228–40. [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Jin Y. Genetic dissection of axon regeneration. Curr Opin Neurobiol. 2011;21:189–96. doi: 10.1016/j.conb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taïbi F, Metzinger-Le Meuth V, Massy Z, Metzinger L. miR-223: an inflammatory oncomiR enters the cardiovascular field. Biochim Biophys Acta. 2014;1842:1001–9. doi: 10.1016/j.bbadis.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 40.Bergman P, James T, Kular L, Ruhrmann S, Kramarova T, Kvist A, et al. Next-generation sequencing identifies microRNAs that associate with pathogenic autoimmune neuroinflammation in rats. J Immunol. 2013;190:4066–75. doi: 10.4049/jimmunol.1200728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yunta M, Nieto-Díaz M, Esteban FJ, Caballero-López M, Navarro-Ruíz R, Reigada D, et al. MicroRNA dysregulation in the spinal cord following traumatic injury. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Z, Zhong L, Xian R, Yuan B. MicroRNA-223 regulates inflammation and brain injury via feedback to NLRP3 inflammasome after intracerebral hemorrhage. Mol Immunol. 2015;65(2):267–76. doi: 10.1016/j.molimm.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 43.Berkelaar M, Clarke DB, Wang YC, Bray GM, Aguayo AJ. Axotomy results in delayed death and apoptosis of retinal ganglion cells in adult rats. J Neurosci. 1994;14(7):4368–74. doi: 10.1523/JNEUROSCI.14-07-04368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodger J, King CE, Lukehurst S, Chen PB, Dunlop SA, Beazley LD, et al. Changing Pax6 expression correlates with axon outgrowth and restoration of topography during optic nerve regeneration. Neuroscience. 2006;142(4):1043–54. doi: 10.1016/j.neuroscience.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 45.He P, Peng Z, Luo Y, Wang L, Yu P, Deng W, et al. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy. 2009;5(1):52–60. doi: 10.4161/auto.5.1.7247. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Khatibi NH, Hu Q, Yan J, Chen C, Han J, et al. Transmembrane protein 166 regulates autophagic and apoptotic activities following focal cerebral ischemic injury in rats. Exp Neurol. 2012;234:181–90. doi: 10.1016/j.expneurol.2011.12.038. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Yu C, Lu Y, He P, Guo J, Zhang C, et al. TMEM166, a novel transmembrane protein, regulates cell autophagy and apoptosis. Apoptosis. 2007;12(8):1489–502. doi: 10.1007/s10495-007-0073-9. [DOI] [PubMed] [Google Scholar]
- 48.Chang H, Gilbert W. A novel zebrafish gene expressed specifically in the photoreceptor cells of the retina. Biochem Biophys Res Commun. 1997;237(1):84–9. doi: 10.1006/bbrc.1997.7081. [DOI] [PubMed] [Google Scholar]
- 49.Lafreniere RG, Rochefort DL, Kibar Z, Fon EA, Han F, Cochius J, et al. Isolation and characterization of GT335, a novel human gene conserved in Escherichia coli and mapping to 21q22.3. Genomics. 1996;38(3):264–72. doi: 10.1006/geno.1996.0627. [DOI] [PubMed] [Google Scholar]
- 50.Cameron DA, Gentile KL, Middleton FA, Yurco P. Gene expression profiles of intact and regenerating zebrafish retina. Mol Vis. 2005;11:775–91. [PubMed] [Google Scholar]
- 51.Zou S, Tian C, Ge S, Hu B. Neurogenesis of retinal ganglion cells is not essential to visual functional recovery after optic nerve injury in adult zebrafish. PLoS One. 2013;8(2):e57280. doi: 10.1371/journal.pone.0057280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011;124:999–1006. doi: 10.1242/jcs.081513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edeleva EV, Shcherbata HR. Stress-induced ECM alteration modulates cellular microRNAs that feedback to readjust the extracellular environment and cell behavior. Front Genet. 2013;4:305. doi: 10.3389/fgene.2013.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bono P, Cordero E, Johnson K, Borowsky M, Ramesh V, Jacks T, et al. Layilin, a cell surface hyaluronan receptor, interacts with merlin and radixin. Exp Cell Res. 2005;308:177–87. doi: 10.1016/j.yexcr.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 55.Bono P, Rubin K, Higgins JM, Hynes RO. Layilin, a novel integral membrane protein, is a hyaluronan receptor. Mol Biol Cell. 2001;12:891–900. doi: 10.1091/mbc.12.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borowsky ML, Hynes RO. Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J Cell Biol. 1998;143:429–42. doi: 10.1083/jcb.143.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wegener KL, Basran J, Bagshaw CR, Campbell ID, Roberts GCK, Critchley DR, et al. Structural basis for the interaction between the cytoplasmic domain of the hyaluronate receptor layilin and the talin F3 subdomain. J Mol Biol. 2008;382:112–26. doi: 10.1016/j.jmb.2008.06.087. [DOI] [PubMed] [Google Scholar]
- 58.Lipton SA, Wagner JA, Madison RD, D’Amore PA. Acidic fibroblast growth factor enhances regeneration of processes by postnatal mammalian retinal ganglion cells in culture. Proc Natl Acad Sci U S A. 1988;85(7):2388–92. doi: 10.1073/pnas.85.7.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gschwend TP, Krueger SR, Kozlov SV, Wolfer DP, Sonderegger P. Neurotrypsin, a novel multidomain serine protease expressed in the nervous system. Mol Cell Neurosci. 1997;9:207–19. doi: 10.1006/mcne.1997.0616. [DOI] [PubMed] [Google Scholar]
- 60.Hilgenberg LGW, Su H, Gu H, O’Dowd DK, Smith MA. Alpha3Na+/K + −ATPase is a neuronal receptor for agrin. Cell. 2006;125:359–69. doi: 10.1016/j.cell.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 61.Matsumoto-Miyai K, Sokolowska E, Zurlinden A, Gee CE, Lüscher D, Hettwer S, et al. Coincident pre- and postsynaptic activation induces dendritic filopodia via neurotrypsin-dependent agrin cleavage. Cell. 2009;136:1161–71. doi: 10.1016/j.cell.2009.02.034. [DOI] [PubMed] [Google Scholar]
- 62.Reif R, Sales S, Hettwer S, Dreier B, Gisler C, Wölfel J, et al. Specific cleavage of agrin by neurotrypsin, a synaptic protease linked to mental retardation. FASEB J. 2007;21:3468–78. doi: 10.1096/fj.07-8800com. [DOI] [PubMed] [Google Scholar]
- 63.Wlodarczyk J, Mukhina I, Kaczmarek L, Dityatev A. Extracellular matrix molecules, their receptors, and secreted proteases in synaptic plasticity. Dev Neurobiol. 2011;71:1040–53. doi: 10.1002/dneu.20958. [DOI] [PubMed] [Google Scholar]
- 64.Klemenčič M, Novinec M, Maier S, Hartmann U, Lenarčič B. The heparin-binding activity of secreted modular calcium-binding protein 1 (SMOC-1) modulates its cell adhesion properties. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vannahme C, Smyth N, Miosge N, Gösling S, Frie C, Paulsson M, et al. Characterization of SMOC-1, a novel modular calcium-binding protein in basement membranes. J Biol Chem. 2002;277:37977–86. doi: 10.1074/jbc.M203830200. [DOI] [PubMed] [Google Scholar]
- 66.Parikh P, Hao Y, Hosseinkhani M, Patil SB, Huntley GW, Tessier-Lavigne M, et al. Regeneration of axons in injured spinal cord by activation of bone morphogenetic protein/Smad1 signaling pathway in adult neurons. Proc Natl Acad Sci U S A. 2011;108:E99–107. doi: 10.1073/pnas.1100426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas JT, Canelos P, Luyten FP, Moos M. Xenopus SMOC-1 Inhibits bone morphogenetic protein signaling downstream of receptor binding and is essential for postgastrulation development in Xenopus. J Biol Chem. 2009;284:18994–9005. doi: 10.1074/jbc.M807759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vuilleumier R, Springhorn A, Patterson L, Koidl S, Hammerschmidt M, Affolter M, et al. Control of Dpp morphogen signalling by a secreted feedback regulator. Nat Cell Biol. 2010;12:611–7. doi: 10.1038/ncb2064. [DOI] [PubMed] [Google Scholar]
- 69.Zhong J, Zou H. BMP signaling in axon regeneration. Curr Opin Neurobiol. 2014;27C:127–34. doi: 10.1016/j.conb.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young KG, Kothary R. Cytoskeleton of the nervous system. 2011;3:379–410. doi:10.1007/978-1-4419-6787-9.
- 71.Gervasi C, Thyagarajan A, Szaro BG. Increased expression of multiple neurofilament mRNAs during regeneration of vertebrate central nervous system axons. J Comp Neurol. 2003;461:262–75. doi: 10.1002/cne.10695. [DOI] [PubMed] [Google Scholar]
- 72.Jacobs AJ, Swain GP, Snedeker JA, Pijak DS, Gladstone LJ, Selzer ME. Recovery of neurofilament expression selectively in regenerating reticulospinal neurons. J Neurosci. 1997;17:5206–20. doi: 10.1523/JNEUROSCI.17-13-05206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cairns NJ, Zhukareva V, Uryu K, Zhang B, Bigio E, Mackenzie IR, et al. alpha-internexin is present in the pathological inclusions of neuronal intermediate filament inclusion disease. Am J Pathol. 2004;164(6):2153–61. doi: 10.1016/S0002-9440(10)63773-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ganesalingam J, An J, Bowser R, Andersen PM, Shaw CE. pNfH is a promising biomarker for ALS. Amyotroph Lateral Scler Frontotemporal Degeneration. 2013;14:146–9. doi: 10.3109/21678421.2012.729596. [DOI] [PubMed] [Google Scholar]
- 75.Lu XY, Chen XX, Huang LD, Zhu CQ, Gu YY, Ye S. Anti-alpha-internexin autoantibody from neuropsychiatric lupus induce cognitive damage via inhibiting axonal elongation and promote neuron apoptosis. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ackerley S, Thornhill P, Grierson AJ, Brownlees J, Anderton BH, Leigh PN, et al. Neurofilament heavy chain side arm phosphorylation regulates axonal transport of neurofilaments. J Cell Biol. 2003;161:489–95. doi: 10.1083/jcb.200303138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garcia ML, Barry DM. Cytoskeleton of the nervous system. In: Nixon RA, Yuan A, editors. New York, NY: Springer New York; 2011. p. 279–95.
- 78.Niwa R, Slack FJ. The evolution of animal microRNA function. Curr Opin Genet Dev. 2007;17:145–50. doi: 10.1016/j.gde.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 79.Xu J, Zhang R, Shen Y, Liu G, Lu X, Wu CI. The evolution of evolvability in microRNA target sites in vertebrates. Genome Res. 2013;23(11):1810–6. doi: 10.1101/gr.148916.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39(16):6845–53. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodger J, Goto H, Cui Q, Chen PB, Harvey AR. cAMP regulates axon outgrowth and guidance during optic nerve regeneration in goldfish. Mol Cell Neurosci. 2005;30(3):452–64. doi: 10.1016/j.mcn.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 82.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics (Oxford, England) 2007;23:2700–7. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 83.Yang YH, Thorne NP. Normalization for two-color cDNA microarray data. In: Goldstein D, editor. Science and statistics: a festschrift for terry speed. 2003. pp. 403–18. [Google Scholar]
- 84.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 85.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–84. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 86.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jantzen SG, Sutherland BJ, Minkley DR, Koop BF. GO trimming: systematically reducing redundancy in large Gene Ontology datasets. BMC Res Notes. 2011;4:267. doi: 10.1186/1756-0500-4-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 89.Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics--a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25(3):415–6. doi: 10.1093/bioinformatics/btn647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–46. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grimson A, Farh KK-h, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Uniprot. http://www.uniprot.org. 2014.
- 93.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]


